Abstract
Methane emission from paddy fields may be reduced by the addition of electron acceptors to stimulate microbial populations competitive to methanogens. We have studied the effects of ferrihydrite and gypsum (CaSO4 · 2H2O) amendment on methanogenesis and population dynamics of methanogens after flooding of Italian rice field soil slurries. Changes in methanogen community structure were followed by archaeal small subunit (SSU) ribosomal DNA (rDNA)- and rRNA-based terminal restriction fragment length polymorphism analysis and by quantitative SSU rRNA hybridization probing. Under ferrihydrite amendment, acetate was consumed efficiently (<60 μM) and a rapid but incomplete inhibition of methanogenesis occurred after 3 days. In contrast to unamended controls, the dynamics of Methanosarcina populations were largely suppressed as indicated by rDNA and rRNA analysis. However, the low acetate availability was still sufficient for activation of Methanosaeta spp., as indicated by a strong increase of SSU rRNA but not of relative rDNA frequencies. Unexpectedly, rRNA amounts of the novel rice cluster I (RC-I) methanogens increased significantly, while methanogenesis was low, which may be indicative of transient energy conservation coupled to Fe(III) reduction by these methanogens. Under gypsum addition, hydrogen was rapidly consumed to low levels (∼0.4 Pa), indicating the presence of a competitive population of hydrogenotrophic sulfate-reducing bacteria (SRB). This was paralleled by a suppressed activity of the hydrogenotrophic RC-I methanogens as indicated by the lowest SSU rRNA quantities detected in all experiments. Full inhibition of methanogenesis only became apparent when acetate was depleted to nonpermissive thresholds (<5 μM) after 10 days. Apparently, a competitive, acetotrophic population of SRB was not present initially, and hence, acetotrophic methanosarcinal populations were less suppressed than under ferrihydrite amendment. In conclusion, although methane production was inhibited effectively under both mitigation regimens, different methanogenic populations were either suppressed or stimulated, which demonstrates that functionally similar disturbances of an ecosystem may result in distinct responses of the populations involved.
Over 50% of the global annual methane emissions are of anthropogenic origin and the cultivation of irrigated rice may account for up to 20% of this efflux (20). Methane is an important greenhouse gas (10) with an atmospheric mixing ratio of 1.745 ppm at present that has increased by 7 ppb year−1 on average over the last 10 years (20). In the context of planetary warming, the reduction of global methane efflux is of great importance, and several mitigation strategies to reduce rice paddy emissions have been proposed (for recent reviews, see references 24 and 60). The degradation of organic matter in anoxic rice field soil is accomplished by hydrolytic, fermenting, and anaerobically respiring microorganisms, including methanogens (7). Directly after flooding of rice field soils, common electron donors (i.e., acetate and H2) for anaerobic respiration are present in excess due to the vigorous fermentation of organic matter, and methanogenesis occurs in parallel to sulfate and iron reduction (1, 40, 47). Once electron acceptors other than CO2 have been reduced in the bulk volume of the soil, methanogenesis is the dominating terminal respiratory process (7). When electron donors for respiratory processes become limiting, methanogenesis can be suppressed by the supplementation of alternative electron acceptors such as Fe(III) or sulfate (1). This mitigation strategy is based on the thermodynamic theory which predicts that the energetically more favorable electron acceptor will be utilized first under substrate limiting conditions (61). Microorganisms which can reduce the energetically more favorable electron acceptor [e.g., nitrate, Fe(III), sulfate] will outcompete those using the less favorable electron acceptor (e.g., CO2) (28). The thermodynamically superior functional group is believed to have a higher affinity (e.g., lower Km) (23, 46), a higher Vmax (58), and/or a lower threshold for common substrates (1, 8, 27, 49).
The mitigative potential of electron acceptor supplementation in the form of amorphous Fe(III) oxides (i.e., ferrihydrite [1, 5, 21]), sulfate-containing fertilizers (i.e., ammonium sulfate [9, 25, 50]), or other sulfate-containing amendments (i.e., gypsum or phosphogypsum [12, 26]) has been demonstrated in field and laboratory studies and reductions in CH4 emission rates of >70% have been reported (13, 21). The outcompetition of methanogens for common substrates like H2 or acetate by ferric iron-reducing bacteria (FRB) or sulfate-reducing bacteria (SRB) is well understood on a thermodynamic and process-based level. Conversely, little is known about the effects of alternative electron acceptor supplementation on the community structure of methanogens. How rapidly can inhibitory effects be observed, and are they reflected in the structure and dynamics of the methanogenic population or in rRNA expression levels? This knowledge is important for the better understanding of mitigation mechanisms and hence also for possible implementations for rice agriculture.
The aim of this study was to monitor the effects of different mitigation strategies on the methanogenic community in rice field soil using tools of molecular microbial ecology. Slurries of rice field soil were amended with ferrihydrite and gypsum and, together with an unamended control series, were incubated anoxically for 30 days. Small subunit (SSU) ribosomal DNA (rDNA) and rRNA dynamics of major methanogenic lineages, including the novel methanogens of rice cluster I (RC-I) (17, 31), were followed by terminal restriction fragment length polymorphism (T-RFLP) profiling. Furthermore, we determined SSU rRNA expression levels by PCR-independent quantitative rRNA hybridization. Our results revealed pronounced differences in methanogenic community structure and activity which were well correlated to biogeochemical measurements and dependent on soil treatment.
MATERIALS AND METHODS
Soil slurry experiments and chemical analyses.
Rice field soil from a field of the Italian Rice Research Institute near Vercelli (Italy) was sampled in 1997 and stored as described previously (32). Soil slurry experiments were set up by adding 10 ml of sterile, anoxic, double-distilled water to 10 g of dry sieved rice field soil in 60-ml serum vials. Supplementary electron acceptors were amended by adding 1.5% (wt/wt; 140 μmol g−1) ferrihydrite [Fe(OH)3], prepared as described by Jäckel and Schnell (21), or 0.15% (8.7 μmol g−1) gypsum (CaSO4 · 2H2O; Fluka, Buchs, Switzerland) to the dry soil. Triplicate vials for each time point (0 to 30 days) were sealed with butyl rubber septa, flushed with N2, and incubated statically at 25°C. At the end of incubation, the headspace of each vial was analyzed for gases and slurry aliquots were taken and stored at −20°C for chemical analysis as previously described (32) and at −80°C for nucleic acid extraction.
Extraction of nucleic acids.
DNA from soil samples was extracted as described previously (18) and purified using polyvinyl-polypyrrolidone spin columns (19). Total RNA was extracted using the protocol of Lüdemann et al. (30), with minor modifications. Six-hundred-microliter slurry aliquots were placed into 2-ml FastRNA tubes with lysing matrix B (Qbiogene, Heidelberg, Germany) containing 700 μl of precooled TPM buffer (50 mM Tris-HCl [pH 7.5], 1.7% [wt/vol] polyvinylpyrrolidone, 10 mM MgCl2) (15). The mixture was shaken for 40 s at maximum speed in a FastPrep 120 bead beater (Qbiogene) and cell debris and soil particles were sedimented by centrifugation (4 min; 20,000 × g; 4°C). The supernatant was saved, while the pellet was reextracted with 700 μl of a phenol-based lysis buffer (50 mM Tris-HCl [pH 7.5], 10 mM EDTA, 1% [wt/vol] sodium dodecyl sulfate [SDS], 6% [vol/vol] water-saturated phenol) by a second round of bead beating. After centrifugation, the combined supernatants were extracted with 500 μl each of water-saturated phenol, phenol-chloroform-isoamyl alcohol (25:24:1 [vol/vol/vol]), and finally chloroform-isoamyl alcohol (24:1 [vol/vol]) using 2-ml Phase Lock Gel Heavy cups (Eppendorf, Hamburg, Germany) for the last two steps to facilitate the clean separation of aqueous and organic phases and thereby maximize RNA yield. Total nucleic acids were precipitated from the aqueous phase with 3 volumes of ethanol and 0.1 volume of 3 M sodium acetate by freezing (−80°C overnight) and centrifugation (1 h; 20,000 × g; 4°C). The precipitate was washed once with 250 μl of cooled 70% ethanol, dried, and resuspended in 50 μl of EB buffer (10 mM Tris-HCl [pH 8.5]) (Qiagen, Hilden, Germany). RNA extracts were further purified by gel filtration with Sephadex G-75 spin columns (36) to remove residual humic acids. Extracted nucleic acids were checked by standard gel electrophoresis on a 1% agarose gel by using DNA SmartLadder (Eurogentec, Seraing, Belgium) or Escherichia coli 16S and 23S rRNA (Roche Diagnostics, Mannheim, Germany) as a reference and by ethidium bromide staining. Furthermore, nucleic acid extracts were quantified by standard UV photometry (BioPhotometer; Eppendorf).
PCR and RT-PCR amplification.
PCR amplification of archaeal SSU rRNA genes for T-RFLP was done with slurry DNA extracts as described before (32) using the primer combination of Ar109f (5′-ACK GCT CAG TAA CAC GT-3′) (16) and 5′ 6-carboxyfluorescein-labeled Ar912rt (5′-GTG CTC CCC CGC CAA TTC CTT TA-3′) (MWG Biotech, Ebersberg, Germany). This modification of the original Ar915r primer (54) carries a 3-bp elongated 3′ end and was designed to minimize nonspecific primer binding during reverse transcription (RT), especially as the selectivity of the original oligonucleotide has recently been shown to be questionable (53). The elongation of the primer has little effect on the absolute numbers of archaeal target sequences with zero to four mismatches in our ARB database (data not shown) but drastically reduces the numbers of nontarget bacterial matches with three and four mismatches (from n = 186 to 8 and 4,213 to 195, respectively). Consequently, it allows a more efficient discrimination between archaeal target and bacterial nontarget templates. This is especially important during RT, since RNA-DNA duplexes are more stable than DNA-DNA structures (48) and a low-stringency temperature of 48°C had to be used. The specificity of archaeal rRNA-targeted T-RFLP fingerprints was verified by comparison of rRNA- and rDNA-derived profiles generated with both the original Ar915r and the modified Ar912rt oligonucleotides. While only defined terminal restriction fragments (T-RFs) were detected for rDNA profiles with both primers (profiles were practically identical), the new primer eliminated putative nontarget amplicons (e.g., undefined T-RFs) below the detection threshold for rRNA profiles (data not shown).
For RT, coextracted DNA was removed from the slurry RNA extracts by DNase digestion of 1:10-diluted extracts. We added 5 μl of soil RNA to 5 μl of 10× DNase digestion buffer (200 mM Tris-HCl [pH 7.5], 100 mM MgCl2, 20 mM CaCl2), 10 U of RNase-free DNase I (Roche Diagnostics), and diethyl pyrocarbonate-treated H2O to a total volume of 50 μl. Incubation was performed for 1 h at 37°C, and heating to 80°C for 10 min stopped the reaction. Complete removal of DNA from the RNA extracts was verified by PCR (see above). RT-PCR was performed using the Access one-tube RT-PCR system (Promega, Hilden, Germany). The reaction mixture contained, in a total volume of 50 μl, 1× avian myeloblastosis virus-Tfl reaction buffer, 1 mM MgSO4, 200 μM deoxynucleoside triphosphates, 0.5 μM (each) primer (see above), 5 U of avian myeloblastosis virus reverse transcriptase, and 5 U of Tfl DNA polymerase. One microliter of DNase-treated, 1:10-diluted soil RNA extract was added as template. RT was carried out for 45 min at 48°C, followed immediately by 28 amplification cycles (30 s, 94°C; 45 s, 52°C; 90 s, 68°C) and a terminal extension step (5 min, 68°C). After amplification, PCR aliquots (5 μl) were visualized by standard agarose gel electrophoresis. Amplicons were purified with the MinElute PCR purification kit (Qiagen), re-eluted in 25 μl of EB buffer, and quantified by UV photometry.
T-RFLP analysis.
Since rDNA-targeted T-RFLP analysis has been shown earlier to be highly reproducible with replicate DNA extractions (33, 37), DNA was extracted only once from each time point and analyzed by triplicate T-RFLP profiling. T-RFLP analyses of rRNA were from triplicate RNA extractions for each time point. Restriction digests (TaqI; Promega) and T-RFLP analysis were performed as previously described (31). The digest (1.25 μl) was mixed with 0.95 μl of formamide loading dye (Amersham Pharmacia Biotech, Freiburg, Germany) and 0.3 μl of GeneScan-1000 (ROX) size standard (Applied Biosystems, Weiterstadt, Germany), denatured for 3 min at 95°C, and placed immediately on ice. T-RFLP analysis was performed on an ABI Prism 377 DNA sequencer (Applied Biosystems) in GeneScan mode. The fragment mixture (1.9 μl) was loaded into the wells of a 36-well shark tooth comb on a 36-cm 5% (wt/vol) polyacrylamide gel containing 6 M urea and 1× TBE buffer (89 mM Tris-borate, 2 mM EDTA).
Statistical analysis of T-RFLP data.
For statistics, T-RFLP data were evaluated as recently described (33, 44) using SYSTAT 10 software (SPSS Inc., Chicago, Ill.). Principal component analysis (PCA) was performed on T-RFLP profiles (14) with samples (soil treatment, time point, and targeted nucleic acids) as rows and the relative fluorescence intensities of all T-RFs as columns. A covariance data matrix was extracted with pairwise deletion and varimax factor rotation. Data reduction provided a two-factorial ordering of the variance of T-RFLP profiles, which was plotted on a map.
Quantitative rRNA hybridization.
Nonradioactive slot blot quantification of environmental RNA extracts was performed using the digoxigenin (DIG) system (Roche Diagnostics) as recommended by the manufacturer and described by Manz et al. (34), with minor modifications. In order to analyze all 18 different rRNA extracts (three treatments, six time points) together with an appropriate standard dilution series on one membrane for each probe, the RNAs originally extracted in triplicate from each sample were blotted in duplicate using a Hoefer PR 648 slot blot manifold (48 slots; Amersham Pharmacia Biotech). All hybridization, washing, and detection steps were done in 15-cm hybridization tubes (Ochs, Göttingen, Germany) in an OV4 hybridization oven (Whatman Biometra, Göttingen, Germany). After prehybridization, 50 pmol of DIG-labeled probe (MWG Biotech) was hybridized to the membrane-bound rRNA overnight at 40°C in 3 ml of hybridization buffer (5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 5× Denhardt's solution, 1% blocking reagent [Roche], 0.1% N-lauroylsarcosine, 0.1% SDS). Washing steps were carried out for 30 min in 1× SSC-1% SDS (pH 7) twice at 40°C and once at the probe-specific stringent washing temperatures (45, 54, 57). DIG-labeled probes were then detected with Fab fragments of anti-DIG antibodies conjugated to alkaline phosphatase (Roche Diagnostics) and the ECF fluorescence substrate (Amersham Pharmacia Biotech) on a Storm 860 gel and blot imaging system (Molecular Dynamics, Sunnyvale, Calif.) (55). Blot images were quantitatively evaluated using ImageQuant 5.0 (Molecular Dynamics).
Oligonucleotide probes and reference RNAs.
The following oligonucleotide probes were used in this study: S-*-Univ-1392-a-A-15 (38), S-D-Arch-0915-a-A-20 (54), S-F-Mbac-0310-a-A-22, S-F-Msae-0825-a-A-23, S-G-Msar-0821-a-A-24 (45), and S-*-RCI-0645-a-A-23 (57). E. coli 16S and 23S rRNAs (Roche) were used as universal standards. Methanobacterium bryantii DSM 863T, Methanosaeta concilii DSM 3671T, and Methanosarcina barkeri DSM 800T were obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ; Braunschweig, Germany) and grown in standard media as recommended by the DSMZ. RNA was extracted from harvested cell pellets following the above protocol for use as archaeal and family-specific standards. For the quantification of RC-I Archaea, 16S rRNA of a nearly full-length RC-I clone (AS04-16; accession number AJ308972) was in vitro transcribed (42) as previously described (57).
RESULTS
Biogeochemistry of reduction processes.
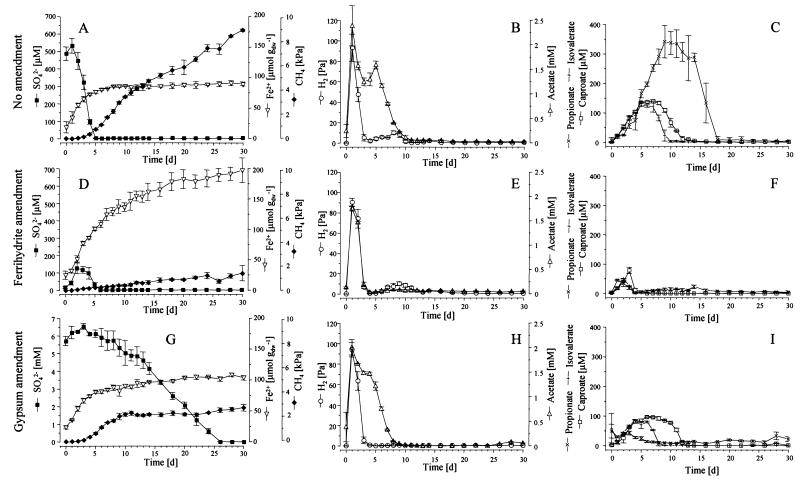
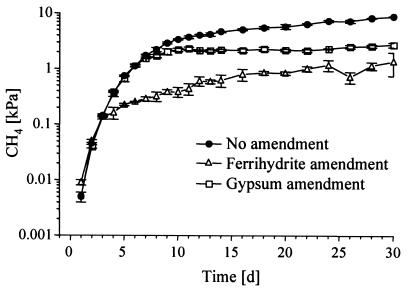
Reduction processes and transient accumulation of hydrogen, acetate, propionate, isovalerate, and caproate in all three parallel experiments (unamended and ferrihydrite and gypsum amended) were followed over 30 days (Fig. 1). The duration and rates of reduction processes are summarized in Table 1. Fe(III) reduction in the unamended soil was terminated within 8 days (Fig. 1A). The addition of ferrihydrite (140 μmol g−1) elongated the initial phase of rapid Fe(III) reduction (Fig. 1D), and steady reduction rates were 4.8 μmol day−1 g−1 on average (Table 1). Pore water sulfate concentrations in the unamended soil were depleted within 5 days (Fig. 1A). The addition of ferrihydrite reduced maximum sulfate concentrations to ∼120 μM (Fig. 1D), while amendment of gypsum drastically elevated sulfate concentrations to up to 6.5 mM (Fig. 1G). High sulfate reduction rates were observed only after day 8 and were 0.5 μmol day−1 g−1 on average (Table 1). After day 26, the amended sulfate was depleted. Methane formation started directly after flooding (47) in all three experiments and increased to similar levels of ∼140 Pa on day 3 (Fig. 2). In the unamended soil, methane mixing ratios continued to increase and production rates were a constant 0.5 μmol day−1 g−1 on average after day 10. Under ferrihydrite amendment, methane production was strongly suppressed after day 3 (Fig. 1D and 2), but mixing ratios still increased with low constant production rates of 80 nmol day−1 g−1 thereafter. In the gypsum-amended soil, an inhibition of methane production became apparent only after day 6 but was absolute between days 11 and 22 (Fig. 1G and 2; Table 1). Only after the end of sulfate reduction (day 26) did methane mixing ratios start to increase again in low amounts. Total methane emissions during the experiment were reduced by 85% under ferrihydrite amendment and by 69% under gypsum amendment.
FIG. 1.
Time course of reduction processes in unamended (A, B, C), ferrihydrite-amended (D, E, F), and gypsum-amended (G, H, I) rice field soil slurries. (A, D, G) Concentrations of SO42− and Fe2+ and CH4 partial pressure. (B, E, H) H2 partial pressure and concentration of acetate. (C, F, I) Concentrations of propionate, isovalerate, and caproate (means ± standard deviations [SD]; n = 3).
TABLE 1.
Rates of CH4 production, Fe3+ reduction, and SO42− reduction in unamended and ferrihydrite- and gypsum-amended rice field soil slurries within 30 days of anoxic incubation; inhibition time points of different processes; and contribution of different respiratory processes to total reducing equivalents formed
| Soil treatment | Process (day inhibition started) | Product formation [μmol day−1 g−1 (days)]
|
Reductant formed (μmol g−1)a | Total reducing equivalents formed [μmol g−1 (%)]b | |||
|---|---|---|---|---|---|---|---|
| Maximum | Initial | Steady-state | Inhibited | ||||
| No amendment | CH4 production | 1.25 (9) | 0.83 (4-10) | 0.5 (11-30) | 15.8 | 126.2 (56.8) | |
| Fe3+ reduction | 20.1 (2) | 11.6 (1-5) | 89.5 | 89.5 (40.3) | |||
| SO42− reduction | 0.33 (4) | 0.19 (1-5) | 0.8 | 6.4 (2.9) | |||
| Ferrihydrite amendment | CH4 production (3) | 0.38 (28) | 0.15 (3) | 0.08 (3-30) | 2.5 | 19.6 (9.0) | |
| Fe3+ reduction | 26.0 (3) | 14.2 (1-7) | 4.8 (7-20) | 197.1 | 197.1 (90.3) | ||
| SO42− reduction | 0.10 (4) | 0.05 (2-6) | 0.2 | 1.6 (0.7) | |||
| Gypsum amendment | CH4 production (7) | 0.84 (6) | 0.53 (4-10) | <0.01 (11-22) | 4.9 | 39.0 (17.9) | |
| Fe3+ reduction | 18.6 (2) | 11.4 (1-5) | 103.5 | 103.5 (47.4) | |||
| SO42− reduction | 0.69 (14) | 0.27 (4-12) | 0.5 (13-26) | 9.5 | 76.0 (34.8) | ||
Amount of either CH4 or Fe2+ formed or SO42− reduced.
The amount of product formed by each process was normalized to reducing equivalents of [H] as follows: from CH4 production (8 [H]), dissimilatory Fe3+ reduction (1 [H]), or SO42− reduction (8 [H]) to S2−. SO42− reduction was measured by following the disappearance of SO42−. The contribution of NO3− reduction was neglected (<20 μM present initially).
FIG. 2.
Time course of CH4 partial pressures (logarithmic scale) in unamended and electron acceptor-amended rice field soil slurries (means ± SD; n = 3).
Hydrogen, produced by vigorous fermentation processes directly after flooding, accumulated up to ∼92 Pa on day 1 in all parallel experiments (Fig. 1). It was depleted to ∼1 Pa on day 4 in all three experiments, but remained below 1 Pa (0.4 Pa on average after day 5) only under gypsum amendment (Fig. 1H). In the other two series, H2 partial pressures increased again to a second transient maximum of ∼10 Pa around day 9 and decreased to low levels of ∼0.5 Pa towards the end of incubation (Fig. 1B and E). Maximum acetate concentrations (∼2 mM) also accumulated on day 1 after flooding (Fig. 1). Acetate consumption was most efficient in the ferrihydrite series, and concentrations rapidly decreased to a constant ∼60 μM from day 4 onward (Fig. 1E). In the other two series, depletion was much slower and reached concentrations below 100 μM only after 8 to 10 days. But while acetate slowly decreased from 60 to 25 μM after day 11 in the unamended slurries (Fig. 1B), it was depleted below the detection limit (∼5 μM) between days 12 and 26 in the gypsum series (Fig. 1H). Other volatile fatty acids, like propionate, isovalerate, and caproate, transiently accumulated to substantial concentrations in the unamended experiment (Fig. 1C). Under ferrihydrite supplementation, depletion of all three compounds was accomplished on day 4 (Fig. 1F), while in the gypsum series, only the consumption of propionate was similarly efficient (Fig. 1I).
T-RFLP fingerprinting of archaeal SSU rDNA and rRNA dynamics.
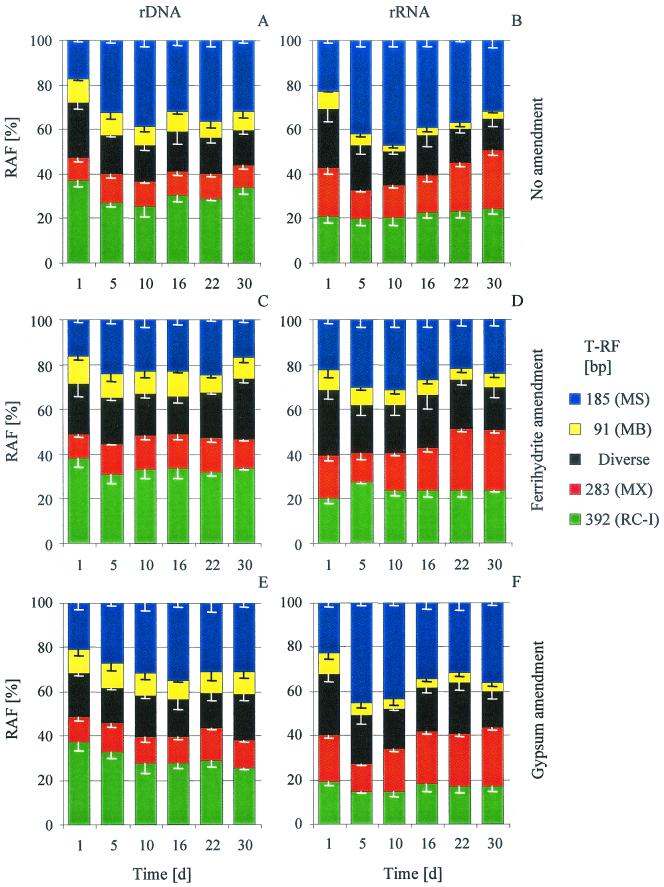
Nucleic acids of all three flooding experiments were extracted on days 1, 5, 10, 16, 22, and 30 after flooding and analyzed by T-RFLP. Relative population dynamics within the archaeal communities of the three flooding experiments as revealed by combined rDNA- and rRNA-targeted T-RFLP analysis are shown in Fig. 3. Structure and compositional shifts of the natural archaeal community (Fig. 3A) were similar to those found recently (32, 43). Most prominent were the T-RFs characteristic for RC-I (392 bp) and Methanosarcina spp. (185 bp), both contributing ∼30% relative amplicon frequency (RAF) on average. While the methanosarcinal T-RF more than doubled from 17 to 39% between day 1 and day 10, the RC-I T-RF decreased from 37 to 25% concomitantly. Also abundant were T-RFs of 283 bp (Methanosaetaceae, ∼11%) and 91 bp (Methanobacteriaceae, ∼9%). The T-RFLP profiling of SSU rRNA revealed even more pronounced dynamics of Methanosarcina spp., since their T-RF increased to 47% RAF on day 10 (Fig. 3B). The methanosaetal T-RF was always more abundant in rRNA profiles and, in contrast to stable rDNA abundances, doubled its RAF from 13% on day 5 to 26% on day 30. The RC-I T-RF remained stable (∼21%) but was always less abundant in rRNA profiles. rRNA-based RAFs of the Methanobacteriaceae were only initially comparable to rDNA ratios but decreased to ∼3% from day 10 onward.
FIG. 3.
Archaeal population dynamics in unamended (A, B), ferrihydrite-amended (C, D), and gypsum-amended (E, F) rice field soil slurries as determined by SSU rDNA (A, C, E) and SSU rRNA (B, D, F) targeted T-RFLP analysis. Shown is the percentage of individual T-RFs relative to the total integrated fluorescence (mean − SD; n = 3 T-RFLP replicates for rDNA and 3 extraction replicates for rRNA). Only T-RFs representing major methanogenic lineages are shown, and minor archaeal T-RFs described previously (6, 32, 44) have been combined as “Diverse.” MB, Methanobacteriaceae; MS, Methanosarcinaceae; MX, Methanosaetaceae; RC-I, rice cluster I; RAF, relative amplicon frequency.
Under ferrihydrite amendment, SSU rDNA-based population dynamics of Archaea as observed in the unamended rice field soil were largely suppressed (Fig. 3C). Only methanosarcinal RAFs increased temporarily from 16 to 24% between days 1 and 5. All other major T-RFs were relatively stable during incubation. On the rRNA level, similarly reduced methanosarcinal dynamics were observed. Surprisingly, the rRNA-based RAF of Methanosaeta spp. showed an even more pronounced increase towards the end of the experiment (Fig. 3D) than in unamended soil and reached ∼27% already after 22 days. Differences were also detected for the RC-I SSU rRNA RAF, which increased from 19 to 27% within the first 5 days after flooding and remained constant (∼23%) thereafter. The addition of gypsum, on the other hand, resulted in archaeal rDNA dynamics that were quite similar to those of unamended soil. Again, a strong but less marked increase of Methanosarcina species amplicon ratios was observed with time, while all other T-RFs remained relatively stable (Fig. 3E). Differences to the unamended soil became evident only by rRNA profiling (Fig. 3F). Methanosarcinal frequencies reached a maximum after a strong initial increase already on day 5, decreased thereafter, but increased again slightly on day 30. The frequencies of RC-I rRNA, however, were only ∼17% on average and were always reduced compared to both unamended and ferrihydrite-amended rice field soil slurries.
Statistical analysis.
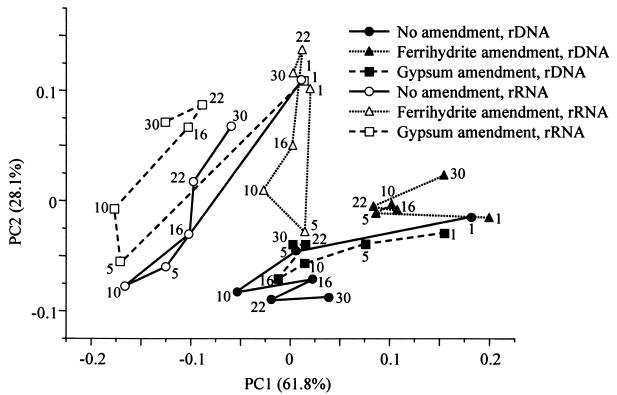
PCA is a preferred method for reduction of complex data sets and has been frequently used to facilitate the comparison of microbial communities with data derived from amplified gene restriction patterns (14, 41). PCA of T-RFLP profiles was done to visualize patterns of time-dependent community shifts within the three differently treated soils by generalizing variations in archaeal SSU rDNA and rRNA pools. To enhance visual clarity, PCA scores of averaged triplicate T-RFLP results were plotted. The total variance of T-RFLP fingerprints was reduced to two hypothetical principal components, which explained 62 and 28% of the variance, respectively. A factorial map (Fig. 4) shows the position, in ordination space, of archaeal SSU rDNA and rRNA fingerprints of the three soils and consecutive time points. PCA allowed for a clear separation of rDNA- and rRNA-derived profiles in ordination space. Furthermore, data points from day 1 after flooding were grouped for both rDNA and rRNA and thus indicate similar communities with almost identical T-RFLP patterns. Sequential rDNA dynamics within the natural and gypsum-amended slurries were also comparable, e.g., shifts in principal component ordination ran in parallel and end points were again closely grouped. However, PCA clearly showed that archaeal rDNA dynamics under ferrihydrite amendment were distinct and strongly reduced. For rRNA samples, successive shifts of principal component scores were more pronounced than for rDNA fingerprints (i.e., rRNA pools were more dynamic). Here, the unamended and gypsum-amended time series again displayed similar dynamics but were better discriminated than for rDNA profiles. Both were again clearly differentiated from the archaeal rRNA fingerprints of the ferrihydrite slurries.
FIG. 4.
Principal component ordination of the overall variance between averaged triplicate T-RFLP results generated with archaeal SSU rDNA and rRNA amplicons of natural and ferrihydrite- and gypsum-amended rice field soils. The abundance of all original T-RFs detected was included in this analysis. Successive time points (days) for each treatment and nucleic acid are connected by lines and indicated by numbers to illustrate time-dependent population shifts.
Quantitative SSU rRNA dynamics.
We performed quantitative hybridization probing of environmental rRNA extracts for the PCR-independent quantification of methanogen-specific rRNAs. In addition to universal and Archaea-specific probes, we used probes targeting the rRNA of Methanosarcina spp., members of the Methanosaetaceae and Methanobacteriaceae, and also the novel methanogens of RC-I (17, 31). Total amounts of extractable rRNA in the soil as detected by the universal probe decreased from ∼6.6 μg g−1 to ∼3.7 μg g−1 within the 30 days of incubation in all three flooding experiments (Fig. 5A). Total amounts of Archaea-specific rRNA (Fig. 5B) remained relatively constant (∼1.45 μg g−1) in unamended rice field soil over time, while they decreased in both ferrihydrite- and gypsum-amended slurries to ∼1 μg g−1 (day 30). Relative to the total extractable rRNA (universal probe), the percentage of archaeal rRNA was ∼22% on day 1 in all slurries and 36, 30, and 27% on day 30 in the unamended and ferrihydrite- and gypsum-amended slurries, respectively.
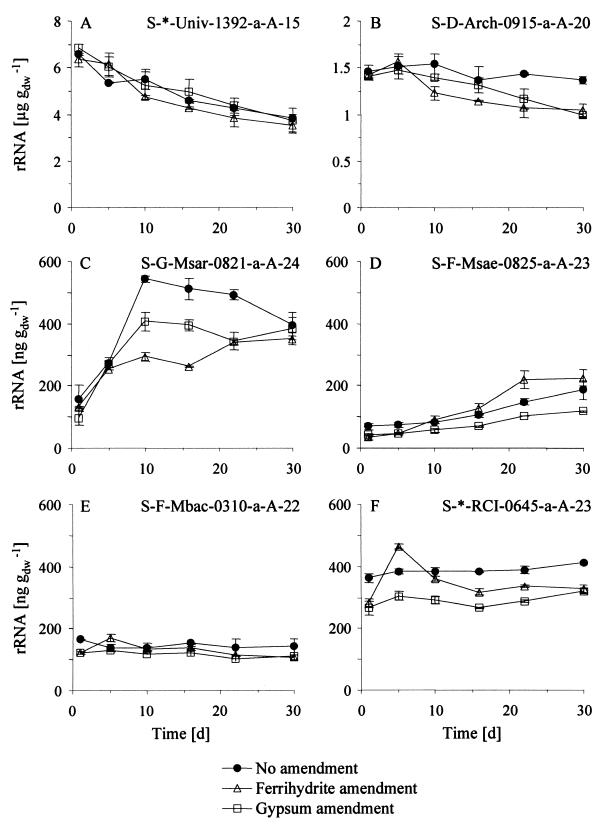
FIG. 5.
Methanogenic population dynamics in unamended and electron acceptor-amended rice field soil slurries as determined by quantitative slot blot hybridization of environmental rRNA extracts (means ± SD; n = 2 extractions per time point). (A) Total rRNA; (B) archaeal rRNA; (C) Methanosarcina spp.; (D) Methanosaetaceae; (E) Methanobacteriaceae; (F) rice cluster I. Abbreviations of probes are from reference 3.
Among the different methanogen-specific probes, the one targeting the rRNA of Methanosarcina spp. revealed the most significant quantitative dynamics (Fig. 5C). Between days 1 and 5, amounts and increases were similar in all three series. Obvious differences between the three treatments became evident on day 10 after flooding, when methanosarcinal rRNA quantities increased further to above 540 ng g−1 in the unamended slurries and to 405 ng g−1 after gypsum addition but remained below 300 ng g−1 in the ferrihydrite series. On day 30 after flooding, methanosarcinal rRNA quantities again approached similar amounts, of 350 to 400 ng g−1, in all three experiments. The rRNA dynamics of members of the Methanosaetaceae were different and less pronounced, as quantities increased steadily with incubation time in all three flooding series (Fig. 5D). Final amounts were lowest (115 ng g−1) under gypsum amendment and, surprisingly, highest under ferrihydrite addition (223 ng g−1). rRNA quantities of the Methanobacteriaceae, on the other hand, remained very stable with time in all slurries (∼126 ng g−1 on average), with the exceptions of slightly elevated levels (168 ng g−1) on day 5 under ferrihydrite amendment (Fig. 5E). Finally, we followed the rRNA dynamics of the yet uncultured methanogens of RC-I (31), using an oligonucleotide probe designed to specifically target this novel lineage (57). rRNA amounts of RC-I in the unamended soil were very stable, although they increased slowly from 361 to 409 ng g−1 during the experiment (Fig. 5F). Dynamics were similar in the gypsum-amended slurries; however, absolute levels were constantly ∼100 ng g−1 below those in natural soil. Surprisingly, under ferrihydrite addition, RC-I rRNA levels increased drastically from 280 to 462 ng g−1 between day 1 and day 5 after flooding and decreased again to ∼330 ng g−1 towards the end of the experiment. The strongest transient rRNA dynamics of the novel RC-I methanogens were thus detected under Fe(III) supplementation.
DISCUSSION
Methanogenic Archaea are important for the balance of fermentation processes in rice field soil since they consume the key intermediates H2 and acetate (7). Whereas the biogeochemistry of these processes has been studied extensively, we are only beginning to understand relationships between structure and function of methanogenic archaeal communities in the functionally highly dynamic rice field soil ecosystem. Recently, we found that the archaeal community structure in freshly flooded Italian rice field soil slurries was rather stable (32). Only Methanosarcina spp. were significantly stimulated by the availability of excess acetate and H2 present during the first 13 days after flooding as detected by T-RFLP analyses of SSU rDNA. In the present study, we induced major functional shifts on rice field soil microbial populations by amending either ferrihydrite or gypsum as electron acceptors to stimulate FRB or SRB as competitors of the methanogenic population for common electron donors (i.e., acetate and H2). These treatments had significant effects on both methane production and the dynamics of methanogenic populations.
The population dynamics of methanogens were followed by PCR-T-RFLP analysis of archaeal SSU rDNA and rRNA as well as by quantitative rRNA hybridization probing. All major trends detected by rRNA-based T-RFLP analysis were also found by the PCR-independent quantitative hybridization results. By analyzing rDNA and rRNA in parallel, we were able to compare population dynamics (SSU rDNA frequency) and activity shifts of community members as indicated by changes in rRNA content. We found that rRNA expression patterns may reveal population activity (4, 35) much more sensitively than is possible by following the relative SSU rDNA frequency. For example, the increase in relative rRNA abundance of Methanosarcina spp. (Fig. 3 and 5) in unamended soil was much more pronounced than the increase detectable for SSU rDNA (Fig. 3), which suggests both activation and growth of this population. The caveat of measuring the relative gene abundance may be that a strong increase of one population (i.e., Methanosarcina spp.) could mask a small absolute increase of the gene copy number in another population (32). In the case of Methanosaeta spp., we were unable to detect an increase of rDNA ratio by PCR-T-RFLP (Fig. 3), but both RT-PCR-T-RFLP (Fig. 3) and hybridization probing (Fig. 5) revealed a doubling of rRNA amounts. The most parsimonious explanation appears to be that Methanosaeta spp. were strongly activated but did not grow significantly during the first 30 days after flooding. In fact, these findings may represent an important principle for the population dynamics of methanogens in rice field soil: only the fast growing and metabolically versatile Methanosarcina spp. are capable of growing under the transiently high substrate concentrations within the first days after flooding (32). In laboratory culture, exponentially growing bacteria exhibit a tight coupling between growth rate and ribosome synthesis, whereas DNA synthesis rates are regulated more gradually (35). In natural environments, under conditions of low nutrient availability or starvation, it is largely unknown how the synthesis of ribosomes (and rRNA) is actually linked to growth (i.e., cell division, DNA replication) of especially slow growing and uncultivated microorganisms. We hypothesize that Methanosaeta spp. in rice field soil initially increase their rRNA content only, while their relative SSU rDNA frequency, and thus most likely their population size, remains rather constant. Similarly, the activation of Methanosaeta spp. in a bioreactor was initially detected only by a significant increase in rRNA abundance (11).
Addition of 1.5% ferrihydrite extended the phase of iron reduction to over 30 days compared to ∼8 days in unamended soil. This was accompanied by a rapid but incomplete inhibition of methanogenesis after day 3 and by significant effects on the population dynamics of methanogens, indicating that a potent population of FRB successfully competed with methanogens and was indigenously present in the soil. However, the mechanism of inhibition of methanogenesis by ferrihydrite addition in rice field soil is not totally clear. Previous studies have shown that ferric iron reducers can outcompete methanogens for H2 in rice field soil (1) by lowering the H2 partial pressure to thresholds (<0.4 Pa) that are thermodynamically not permissive for methanogenesis (29). In principle, competition for acetate should be similar to competition for H2 (5), but the experimental data available are contradictory with respect to the underlying mechanism of inhibition. FRB were shown to either maintain a lower threshold for acetate in Texas rice field soil than methanogens (52) or have a similar threshold as found in Italian rice field soil (2).
In our experiments, neither the H2 partial pressures nor the acetate thresholds were lower in ferrihydrite-amended slurries than in unamended soil, albeit methanogenesis was strongly (but not completely) inhibited (Table 1). The second transient accumulation of H2 (∼10 Pa on day 9), when iron was reduced at a high rate, indicates that H2 was a less competitive substrate than fatty acids for FRB. In contrast, acetate and other short- chain fatty acids were consumed more rapidly in ferrihydrite-amended slurries than in controls, but threshold concentrations for acetate were similar and did not decrease below 60 μM. This concentration is still permissive for acetotrophic methanogenesis by Methanosaeta spp. (22, 62). The lack of full inhibition of methanogenesis may be attributable to a decreasing bioavailability of Fe(III) as indicated by the progressive lowering of the iron reduction rate from day 7 onward (Fig. 3D). Only ∼200 μmol of Fe3+ g−1 of an available ∼240 μmol g−1 (∼100 μmol of indigenous Fe3+ g−1 plus 140 μmol of added Fe3+ g−1) was reduced after 30 days. It cannot be ruled out that the synthetic ferrihydrite utilized for amendment was less bioavailable for FRB due to a higher degree of crystallinity than the indigenous ferrihydrite (51). The low acetate concentrations observed corroborate both the increase of methanosaetal rRNA (Fig. 3D and 5D), which was strongest under ferrihydrite amendment, and the suppressed dynamics of Methanosarcina spp. (Fig. 3D), as indicated by rDNA and rRNA analysis. Most likely, the acetotrophic Methanosaeta spp. contributed substantially to the low amounts of methane produced in spite of ferrihydrite-mediated inhibition, whereas Methanosarcina spp. were apparently outcompeted for both hydrogen and acetate by FRB after day 4. The minor initial increase of Methanosarcina species ratios was likely to be related to excess substrate availability until day 4 (Fig. 1).
Unexpectedly, RC-I methanogens were significantly stimulated temporarily (up to day 5) only under ferrihydrite amendment as indicated by rRNA analysis (Fig. 3D and 5F), as were members of the Methanobacteriaceae, albeit to a lesser extent (Fig. 5E). Enrichment cultures of the novel RC-I methanogens have been shown to grow on H2-CO2 (31), and the members of the Methanobacteriaceae are also hydrogenotrophic (62). The transient stimulation of these hydrogenotrophic methanogens was paralleled by an increase in the total archaeal rRNA content between days 1 and 5, which was strongest under ferrihydrite amendment. In the same samples, no surplus increase of rRNA of the acetotrophic Methanosarcina and Methanosaeta spp. was observed. Furthermore, in the ferrihydrite-amended slurries, initial sulfate concentrations were much lower than in the control, probably due to sulfate anion adsorption to ferric iron minerals (39). Hence, the activity of SRB, major competitors for H2 (8), was also reduced. Thus, under ferrihydrite amendment, it is likely that a much larger fraction of the initially produced H2 was available for hydrogenotrophic methanogens than in unamended soil. However, the strong transient increase, especially of RC-I rRNA, most likely cannot be explained considering the energy available from the small amount of methane actually produced within the first 5 days under ferrihydrite amendment. Rather, we speculate that under these conditions, energy conservation of hydrogenotrophic methanogens might have been coupled to Fe(III) reduction, a process which has been shown to occur in hyperthermophilic methanogens such as Methanopyrus kandleri and Methanococcus thermolithotrophicus (56). If this was true, based on the molecular fingerprinting of methanogenic populations, our results might indicate a previously unknown effect of ferric iron fertilization: besides being inhibited due to substrate competition for methanogenesis, methanogens might even be stimulated to reduce Fe(III) instead of CO2.
The addition of 0.15% gypsum extended the phase of sulfate reduction to over 26 days after flooding. However, the immediate metabolic potential of the intrinsic SRB population in Italian rice field soil was not sufficient to inhibit methanogens by substrate competition, as indicated by the delayed inhibition of methanogenesis from day 6 on only. SRB required more than twice as much time than FRB to induce similar inhibitory effects on the methanogens. In contrast to this longer lag phase, SRB were capable of completely inhibiting methanogenesis after day 10, most likely after a sufficient population size was established, until sulfate was finally depleted. Interestingly, the activation of SRB and the gradual inhibition of methanogens were paralleled by two phases of substrate depletion. H2 was depleted to mixing ratios as low as 0.3 Pa already after day 4 under gypsum amendment (Fig. 1H) and a second transient accumulation of up to 10 Pa as in the ferrihydrite-amended and unamended slurries was not observed. Thus, a competitive population of H2-utilizing SRB was indigenously present in Italian rice field soil and, upon addition of sulfate, capable of outcompeting methanogens for H2 directly, which has been observed earlier (1, 2). Conversely, acetate-utilizing SRB were capable of completely outcompeting acetotrophic methanogens only after ∼11 days of incubation, when acetate became depleted below detection (<5 μM). This was also the phase of constant sulfate reduction rates and complete inhibition of methanogenesis (Fig. 1G). Most likely, acetate-utilizing SRB were initially present at lower numbers than H2 utilizers or as spores, i.e., Desulfotomaculum spp. (59), which became active only after germination.
T-RFLP, PCA, and quantitative rRNA hybridization showed that methanogenic population dynamics under gypsum amendment were not as different from those of the unamended soil as from those under ferrihydrite addition. We relate this to the delayed inhibition of namely the acetotrophic methanogens, resulting in “intermediate” methanosarcinal rRNA dynamics and nearly unchanged rDNA dynamics. Interestingly, RC-I rRNA levels as revealed by T-RFLP and quantitative rRNA analysis were significantly lower under gypsum amendment, which is basically a reversion of the effect observed on day 5 under ferrihydrite addition and therefore underscores the potential of H2-utilizing SRB for outcompeting hydrogenotrophic methanogens.
In this study, we induced major functional shifts within the rice field soil microbial community by supplementing alternative electron acceptors in the form of ferrihydrite and gypsum and thereby promoting respiratory processes other than methanogenesis. The enhanced activity of FRB and SRB resulted in an almost complete inhibition of methanogenesis under conditions of limiting substrate and nonlimiting electron acceptor availability. Steady-state methane production rates in unamended slurries after day 10 (0.5 μmol day−1 g−1) corresponded to sulfate reduction rates in gypsum-amended slurries after day 8 (0.5 μmol day−1 g−1) and to iron reduction rates in ferrihydrite-amended slurries after day 7 (4.8 μmol day−1 g−1). Considering the electron uptake potential of eight electrons per CO2 and SO42− and one electron per Fe3+, only the amounts of sulfate reduced perfectly matched the quantity of methane, which was not produced under inhibition. The balance of ferrihydrite reduction showed that FRB oxidized electron donors other than acetate and H2, since more ferrihydrite was reduced than expected (Table 1); this may be another reason for the lower efficiency of inhibition of methanogenesis under ferrihydrite amendment. The total amount of gypsum added was only 1/10 that of the ferrihydrite amendment, but still the mitigative effects were comparable (69 and 85% methane reduction, respectively). More research is needed to fully evaluate the long-term effects of alternative electron acceptor supplementation and their mitigative potential for rice agriculture.
Acknowledgments
This study was supported by a grant of the Deutsche Forschungsgemeinschaft (DFG) within the SFB395 “Interactions, adaptations, and catalytic capabilities of soil microorganisms” and by the Max-Planck Society.
We thank Bianca Wagner for expert technical assistance and Ralf Conrad for continuing support.
REFERENCES
- 1.Achtnich, C., F. Bak, and R. Conrad. 1995. Competition for electron donors among nitrate reducers, ferric iron reducers, sulfate reducers, and methanogens in anoxic paddy soil. Biol. Fertil. Soils 19:65-72. [Google Scholar]
- 2.Achtnich, C., A. Schumann, T. Wind, and R. Conrad. 1995. Role of interspecies H2 transfer to sulfate and ferric iron-reducing bacteria in acetate consumption in anoxic paddy soil. FEMS Microbiol. Ecol. 16:61-69. [Google Scholar]
- 3.Alm, E. W., D. B. Oerther, N. Larsen, D. A. Stahl, and L. Raskin. 1996. The oligonucleotide probe database. Appl. Environ. Microbiol. 62:3557-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amann, R., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chidthaisong, A., and R. Conrad. 2000. Turnover of glucose and acetate coupled to reduction of nitrate, ferric iron and sulfate and to methanogenesis in anoxic rice field soil. FEMS Microbiol. Ecol. 31:73-86. [DOI] [PubMed] [Google Scholar]
- 6.Chin, K. J., T. Lukow, and R. Conrad. 1999. Effect of temperature on structure and function of the methanogenic archaeal community in an anoxic rice field soil. Appl. Environ. Microbiol. 65:2341-2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conrad, R. 1996. Soil microorganisms as controllers of atmospheric trace gases (H2, CO, CH4, OCS, N2O, and NO). Microbiol. Rev. 60:609-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conrad, R. 1999. Contribution of hydrogen to methane production and control of hydrogen concentrations in methanogenic soils and sediments. FEMS Microbiol. Ecol. 28:193-202. [Google Scholar]
- 9.Corton, T. M., J. B. Bajita, F. S. Grospe, R. R. Pamplona, C. A. Assis, R. Wassmann, R. S. Lantin, et al. 2000. Methane emission from irrigated and intensively managed rice fields in Central Luzon (Philippines). Nutr. Cycl. Agroecosys. 58:37-53. [Google Scholar]
- 10.Crutzen, P. J. 1991. Atmospheric chemistry—methane's sinks and sources. Nature 350:380-381. [Google Scholar]
- 11.Delbes, C., R. Moletta, and J. J. Godon. 2001. Bacterial and archaeal 16S rDNA and 16S rRNA dynamics during an acetate crisis in an anaerobic digestor ecosystem. FEMS Microbiol. Ecol. 35:19-26. [DOI] [PubMed] [Google Scholar]
- 12.Denier van der Gon, H. A. C., and H. U. Neue. 1994. Impact of gypsum application on the methane emission from a wetland rice field. Global Biogeochem. Cycles 8:127-134. [Google Scholar]
- 13.Denier van der Gon, H. A. C., P. M. van Bodegom, R. Wassmann, R. S. Lantin, and T. M. Metra-Corton. 2001. Sulfate-containing amendments to reduce methane emissions from rice fields: mechanisms, effectiveness and costs. Mitigat. Adapt. Strat. Global Change 6:71-89. [Google Scholar]
- 14.Dollhopf, S. L., S. A. Hashsham, and J. Tiedje. 2001. Interpreting 16S rDNA T-RFLP data: application of self-organizing maps and principal component analysis to describe community dynamics and convergence. Microb. Ecol. 42:495-505. [DOI] [PubMed] [Google Scholar]
- 15.Felske, A., B. Engelen, U. Nübel, and H. Backhaus. 1996. Direct ribosome isolation from soil to extract bacterial rRNA for community analysis. Appl. Environ. Microbiol. 62:4162-4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grosskopf, R., P. H. Janssen, and W. Liesack. 1998. Diversity and structure of the methanogenic community in anoxic rice paddy soil microcosms as examined by cultivation and direct 16S rRNA gene sequence retrieval. Appl. Environ. Microbiol. 64:960-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grosskopf, R., S. Stubner, and W. Liesack. 1998. Novel euryarchaeotal lineages detected on rice roots and in the anoxic bulk soil of flooded rice microcosms. Appl. Environ. Microbiol. 64:4983-4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henckel, T., M. Friedrich, and R. Conrad. 1999. Molecular analyses of the methane-oxidizing microbial community in rice field soil by targeting the genes of the 16S rRNA, particulate methane monooxygenase, and methanol dehydrogenase. Appl. Environ. Microbiol. 65:1980-1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henckel, T., U. Jäckel, S. Schnell, and R. Conrad. 2000. Molecular analyses of novel methanotrophic communities in forest soil that oxidize atmospheric methane. Appl. Environ. Microbiol. 66:1801-1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.IPCC. 2001. Climate change 2001: the scientific basis. In J. T. Houghton, Y. Ding, D. J. Griggs, M. Noguer, P. J. van der Linden, and D. Xiaosu (ed.), Contribution of Working Group I to the Third Assessment Report of the Intergovernmental Panel on Climate Change (IPCC). Cambridge University Press, Cambridge, England.
- 21.Jäckel, U., and S. Schnell. 2000. Suppression of methane emission from rice paddies by ferric iron fertilization. Soil Biol. Biochem. 32:1811-1814. [Google Scholar]
- 22.Jetten, M. S. M., A. J. M. Stams, and A. J. B. Zehnder. 1992. Methanogenesis from acetate—a comparison of the acetate metabolism in Methanothrix soehngenii and Methanosarcina spp. FEMS Microbiol. Rev. 88:181-197. [Google Scholar]
- 23.Kristjansson, J. K., P. Schönheit, and R. K. Thauer. 1982. Different Ks-values for hydrogen of methanogenic bacteria and sulfate reducing bacteria—an explanation for the apparent inhibition of methanogenesis by sulfate. Arch. Microbiol. 131:278-282. [Google Scholar]
- 24.Kumaraswamy, S., A. K. Rath, B. Ramakrishnan, and N. Sethunathan. 2000. Wetland rice soils as sources and sinks of methane: a review and prospects for research. Biol. Fertil. Soils 31:449-461. [Google Scholar]
- 25.Lindau, C. W., P. K. Bollich, R. D. DeLaune, A. R. Mosier, and K. F. Bronson. 1993. Methane mitigation in flooded Louisiana rice fields. Biol. Fertil. Soils 15:174-178. [Google Scholar]
- 26.Lindau, C. W., P. Wickersham, R. D. DeLaune, J. W. Collins, P. K. Bollick, L. M. Scott, and E. N. Lambremont. 1998. Methane and nitrous oxide evolution and N-15 and Ra-226 uptake as affected by application of gypsum and phosphogypsum to Louisiana rice. Agr. Ecosyst. Environ. 68:165-173. [Google Scholar]
- 27.Lovley, D. R. 1985. Minimum threshold for hydrogen metabolism in methanogenic bacteria. Appl. Environ. Microbiol. 49:1530-1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lovley, D. R. 1991. Dissimilatory Fe(III) and Mn(IV) reduction. Microbiol. Rev. 55:259-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lovley, D. R., and S. Goodwin. 1988. Hydrogen concentrations as an indicator of the predominant terminal electron-accepting reactions in aquatic sediments. Geochim. Cosmochim. Acta 52:2993-3004. [Google Scholar]
- 30.Lüdemann, H., I. Arth, and W. Liesack. 2000. Spatial changes in the bacterial community structure along a vertical oxygen gradient in flooded paddy soil cores. Appl. Environ. Microbiol. 66:754-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lueders, T., K. J. Chin, R. Conrad, and M. Friedrich. 2001. Molecular analyses of methyl-coenzyme M reductase α-subunit (mcrA) genes in rice field soil and enrichment cultures reveal the methanogenic phenotype of a novel archaeal lineage. Environ. Microbiol. 3:194-204. [DOI] [PubMed] [Google Scholar]
- 32.Lueders, T., and M. Friedrich. 2000. Archaeal population dynamics during sequential reduction processes in rice field soil. Appl. Environ. Microbiol. 66:2732-2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lukow, T., P. F. Dunfield, and W. Liesack. 2000. Use of the T-RFLP technique to assess spatial and temporal changes in the bacterial community structure within an agricultural soil planted with transgenic and non-transgenic potato plants. FEMS Microbiol. Ecol. 32:241-247. [DOI] [PubMed] [Google Scholar]
- 34.Manz, W., R. Amann, W. Ludwig, M. Wagner, and K. H. Schleifer. 1992. Phylogenetic oligonucleotide probes for the major subclasses of Proteobacteria—problems and solutions. Syst. Appl. Microbiol. 15:593-600. [Google Scholar]
- 35.Molin, S., and M. Givskov. 1999. Application of molecular tools for in situ monitoring of bacterial growth activity. Environ. Microbiol. 1:383-391. [DOI] [PubMed] [Google Scholar]
- 36.Moran, M. A., V. L. Torsvik, T. Torsvik, and R. E. Hodson. 1993. Direct extraction and purification of ribosomal RNA for ecological studies. Appl. Environ. Microbiol. 59:915-918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Osborn, A. M., E. R. B. Moore, and K. N. Timmis. 2000. An evaluation of terminal-restriction fragment length polymorphism (T-RFLP) analysis for the study of microbial community structure and dynamics. Environ. Microbiol. 2:39-50. [DOI] [PubMed] [Google Scholar]
- 38.Pace, N. R., D. A. Stahl, D. J. Lane, and G. J. Olsen. 1986. The analysis of natural microbial populations by ribosomal RNA sequences. Adv. Microb. Ecol. 9:1-55. [Google Scholar]
- 39.Parfitt, R. L., and R. S. C. Smart. 1978. Mechanism of sulfate adsorption on iron-oxides. Soil Sci. Soc. Am. J. 42:48-50. [Google Scholar]
- 40.Patrick, W. H., Jr., and C. N. Reddy. 1978. Chemical changes in rice soils, p. 361-379. Soils and rice. International Rice Research Institute, Los Banos, Philippines.
- 41.Poly, F., L. Ranjard, S. Nazaret, F. Gourbiere, and L. J. Monrozier. 2001. Comparison of nifH gene pools in soils and soil microenvironments with contrasting properties. Appl. Environ. Microbiol. 67:2255-2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Polz, M. F., and C. M. Cavanaugh. 1997. A simple method for quantification of uncultured microorganisms in the environment based on in vitro transcription of 16S rRNA. Appl. Environ. Microbiol. 63:1028-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ramakrishnan, B., T. Lueders, R. Conrad, and M. Friedrich. 2000. Effect of soil aggregate size on methanogenesis and archaeal community structure in anoxic rice field soil. FEMS Microbiol. Ecol. 32:261-270. [DOI] [PubMed] [Google Scholar]
- 44.Ramakrishnan, B., T. Lueders, P. F. Dunfield, R. Conrad, and M. W. Friedrich. 2001. Archaeal community structures in rice soils from different geographical regions before and after initiation of methane production. FEMS Microbiol. Ecol. 37:175-186. [Google Scholar]
- 45.Raskin, L., J. M. Stromley, B. E. Rittmann, and D. A. Stahl. 1994. Group-specific 16S ribosomal-RNA hybridization probes to describe natural communities of methanogens. Appl. Environ. Microbiol. 60:1232-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robinson, J. A., and J. M. Tiedje. 1984. Competition between sulfate-reducing and methanogenic bacteria for H2 under resting and growing conditions. Arch. Microbiol. 137:26-32. [Google Scholar]
- 47.Roy, R., H. D. Klüber, and R. Conrad. 1997. Early initiation of methane production in anoxic rice soil despite the presence of oxidants. FEMS Microbiol. Ecol. 24:311-320. [Google Scholar]
- 48.Saenger, W. 1984. Principles of nucleic acid structure. Springer, Berlin, Germany.
- 49.Schönheit, P., J. K. Kristjansson, and R. K. Thauer. 1982. Kinetic mechanisms for the ability of sulfate reducers to out-compete methanogens for acetate. Arch. Microbiol. 132:285-288. [Google Scholar]
- 50.Schütz, H., A. Holzapfel-Pschorn, R. Conrad, H. Rennenberg, and W. Seiler. 1989. A 3-year continuous record on the influence of daytime, season, and fertilizer treatment on methane emission rates from an Italian rice paddy. J. Geophys. Res. 94:16405-16416. [Google Scholar]
- 51.Schwertmann, U. 1988. Some properties of soil and synthetic iron oxides, p. 203-250. In J. W. Stucki, B. A. Goodman, and U. Schwertmann (ed.), Iron and soil clay minerals. Reidel Publishing Company, Dordrecht, The Netherlands.
- 52.Sigren, L. K., S. T. Lewis, F. M. Fisher, and R. L. Sass. 1997. Effects of field drainage on soil parameters related to methane production and emission from rice paddies. Global Biogeochem. Cycles 11:151-162. [Google Scholar]
- 53.Simon, H. M., J. A. Dodsworth, and R. M. Goodman. 2000. Crenarchaeota colonize terrestrial plant roots. Environ. Microbiol. 2:495-505. [DOI] [PubMed] [Google Scholar]
- 54.Stahl, D. A., and R. Amann. 1991. Development and application of nucleic acid probes, p. 205-248. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons Ltd., Chichester, England.
- 55.Stubner, S., and K. Meuser. 2000. Detection of Desulfotomaculum in an Italian rice paddy soil by 16S ribosomal nucleic acid analyses. FEMS Microbiol. Ecol. 34:73-80. [DOI] [PubMed] [Google Scholar]
- 56.Vargas, M., K. Kashefi, E. L. Blunt-Harris, and D. R. Lovley. 1998. Microbiological evidence for Fe(III) reduction on early Earth. Nature 395:65-67. [DOI] [PubMed] [Google Scholar]
- 57.Weber, S., T. Lueders, M. W. Friedrich, and R. Conrad. 2001. Methanogenic populations involved in the degradation of rice straw in anoxic paddy soil. FEMS Microbiol. Ecol. 38:11-20. [Google Scholar]
- 58.Widdel, F. 1988. Microbiology and ecology of sulfate- and sulfur-reducing bacteria, p. 469-585. In A. J. B. Zehnder (ed.), Biology of anaerobic microorganisms. Wiley Interscience, New York, N.Y.
- 59.Wind, T., S. Stubner, and R. Conrad. 1999. Sulfate-reducing bacteria in rice field soil and on rice roots. Syst. Appl. Microbiol. 22:269-279. [DOI] [PubMed] [Google Scholar]
- 60.Yagi, K., H. Tsuruta, and K. Minami. 1997. Possible options for mitigating methane emission from rice cultivation. Nutr. Cycl. Agroecosyst. 49:213-220. [Google Scholar]
- 61.Zehnder, A. J. B., and W. Stumm. 1988. Geochemistry and biochemistry of anaerobic habitats, p. 1-38. In A. J. B. Zehnder (ed.), Biology of anaerobic microorganisms. Wiley Interscience, New York, N.Y.
- 62.Zinder, S. H. 1993. Physiological ecology of methanogens, p. 128-206. In J. G. Ferry (ed.), Methanogenesis. Chapman & Hall, New York, N.Y.