Abstract
Rhizobium etli type strain CFN42 contains six plasmids. We analyzed the distribution of genetic markers from some of these plasmids in bean-nodulating strains belonging to different species (Rhizobium etli, Rhizobium gallicum, Rhizobium giardinii, Rhizobium leguminosarum, and Sinorhizobium fredii). Our results indicate that independent of geographic origin, R. etli strains usually share not only the pSym plasmid but also other plasmids containing symbiosis-related genes, with a similar organization. In contrast, strains belonging to other bean-nodulating species seem to have acquired only the pSym plasmid from R. etli.
Rhizobia are able to establish a nitrogen-fixing symbiosis with the roots of leguminous plants. The genomes of these bacteria usually contain a chromosome and a variable number of plasmids. Most genes related to symbiotic functions are located on the symbiotic plasmid (pSym) (11).
Several species of rhizobia that nodulate common bean (Phaseolus vulgaris) have been described, and the origins of two of these species, Rhizobium etli and Rhizobium tropici, seem to be in Mesoamerica (15, 26). Rhizobium gallicum and Rhizobium giardinii have been found in some regions of Europe (1, 13, 27), whereas Rhizobium leguminosarum is widespread in European soils. A P. vulgaris-nodulating biovar has been described for all of these species except R. tropici, based on the presence and conservation of reiterations of the nitrogenase reductase gene (nifH). Although R. etli and R. tropici are abundant in soils in Mesoamerica, presumably the center of origin of P. vulgaris, these two species have also been found in Europe; in particular, R. etli has been isolated from different locations in Spain (13, 23) and from an Austrian soil (27).
The study of plasmid-encoded functions in Rhizobium has been focused mainly on the symbiotic plasmids. However, some work has been done on analysis of the cryptic extrachromosomal elements in R. leguminosarum bv. viciae (14), Rhizobium trifolii (3), and R. etli (5). The R. etli type strain (CFN42) contains six plasmids (p42a to p42f), whose sizes range from 150 to 600 kb. p42d (390 kb) corresponds to the pSym plasmid (21). Using this strain, some of us have shown that a plasmid distinct from the pSym, p42b, carries sequences required for lipopolysaccharide biosynthesis and for successful establishment of the symbiosis (5, 10); it has also been found that p42a is a self-transmissible plasmid, which is indispensable for mobilization of the pSym plasmid (4), and an unusual fixL homolog and a reiteration of the fixK and fixNOQP genes have been localized on another plasmid, p42f (12).
With the aim of gaining a comprehensive view of their genomic organization, we studied bean-nodulating Rhizobium strains belonging to different species and with diverse geographical origins to determine the presence and distribution of genetic regions found on the different plasmids of R. etli CFN42.
Plasmid pattern and localization of genetic markers
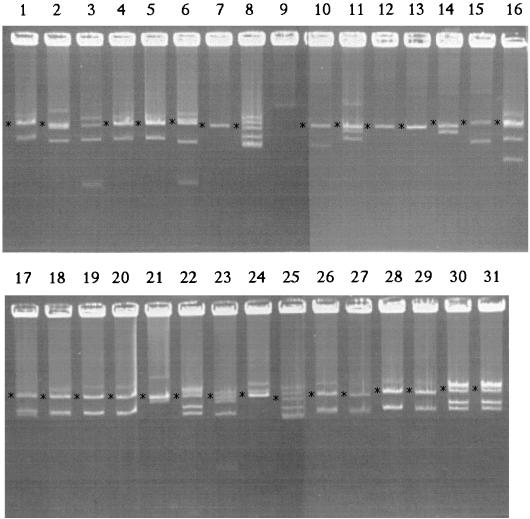
We initiated this study with a collection of 31 strains isolated from bean nodules. The rhizobial strains used in this work are listed in Table 1. Rhizobium strains were grown at 30°C in PY medium (17). Most of the strains came from different regions of Spain and were classified by using multilocus enzyme electrophoresis and ribosomal DNA (rDNA) analysis (13, 23). Bean-nodulating strains from other geographical areas were also included, as was a pea-nodulating R. leguminosarum bv. viciae strain (Table 1). We determined the plasmid patterns of the different strains (Fig. 1) in Eckhardt type gels (8, 14). As Fig. 1 shows, the number of plasmids per strain varied between one and six, with most strains containing four plasmids. The plasmid sizes were variable, and most were in the same range as the R. etli CFN42 plasmid sizes. Only five of the six plasmids present in CFN42 are apparent in the gel (Fig. 1, lanes 8 and 25) due to comigration of p42a and p42b. Some strains contained plasmids that were considerably smaller; these strains included FNJ11, 14C-1, GR62, and GR87. Other strains contained megaplasmids (>1,000 kb); these strains included GR03, GR42, GR18, GR93, and GR84 (Fig. 1), as well as Bra5 (data not shown). Interestingly, all of the strains containing megaplasmids except strain Bra5 belong to species other than R. etli.
TABLE 1.
Rhizobium strains
| Strain | Taxona | Geographical origin | Reference |
|---|---|---|---|
| CFN42 | R. etli | Mexico | 21 |
| CFNX5 | R. etli | Mexico | 24 |
| FNJ11 | R. etli | Mexico | 13 |
| GR10 | R. etli | Spain | 13 |
| GR14 | R. etli | Spain | 13 |
| GR75 | R. etli | Spain | 13 |
| GR56 | R. etli | Spain | 13 |
| GR87 | R. etli | Spain | 13 |
| 4C-2 | R. etli | Spain | 23 |
| 4PR-2 | R. etli | Spain | 23 |
| 6PR-1 | R. etli | Spain | 23 |
| 6C-1 | R. etli | Spain | 23 |
| 8C-3 | R. etli | Spain | 23 |
| 8NJ-2 | R. etli | Spain | 23 |
| 14C-1 | R. etli | Spain | 23 |
| 14PR-2 | R. etli | Spain | 23 |
| 16C-1 | R. etli | Spain | 23 |
| 16NJ-2 | R. etli | Spain | 23 |
| 17C-2 | R. etli | Spain | 23 |
| 17NJ-2 | R. etli | Spain | 23 |
| 21NJ-2 | R. etli | Spain | 23 |
| 21PR-1 | R. etli | Spain | 23 |
| CIAT894 | R. etli | Colombia | 19 |
| Bra5 | R. etli | Brazil | 19 |
| GR64 | S. fredii | Spain | 13 |
| GR03 | R. giardinii bv. phaseoli | Spain | 13 |
| GR93 | R. giardinii | Spain | 13 |
| GR42 | R. gallicum bv. phaseoli | Spain | 13 |
| GR18 | R. gallicum bv. phaseoli | Spain | 13 |
| GR60 | R. gallicum bv. phaseoli | Spain | 13 |
| GR45 | R. gallicum | Spain | 13 |
| GR84 | R. leguminosarum bv. phaseoli | Spain | 13 |
| VF39 | R. leguminosarum bv. viciae | Germany | 20 |
Based on 16S rDNA data, pSym characteristics, and host range.
FIG. 1.
Eckhardt type gel showing the plasmid patterns of Rhizobium strains isolated from bean nodules. Lane 1, strain GR10; lane 2, GR14; lane 3, GR62; lane 4, GR75; lane 5, GR56; lane 6, GR87; lane 7, GR03; lane 8, CFN42; lane 9, GR93; lane 10, GR64; lane 11, GR84; lane 12, GR42; lane 13, GR18; lane 14, GR60; lane 15, GR45; lane 16, FNJ11; lane 17, 4C-2; lane 18, 4PR-2; lane 19, 6PR-1; lane 20, 6C-1; lane 21, 8C-3; lane 22, 8NJ-2; lane 23, 14C-1; lane 24, 14PR-2; lane 25, CFN42; lane 26, 16C-1; lane 27, 16NJ-2; lane 28, 17C-2; lane 29, 17NJ-2; lane 30, 21NJ-2; and lane 31, 21PR-1. The positions of plasmids hybridizing with the nifH probe are indicated by asterisks.
We determined the localization of different genetic markers by hybridization of the blotted Eckhardt gels with [32P]CTP-labeled probes. All of the probes used corresponded to sequences localized on different plasmids from R. etli type strain CFN42; these probes included nifH on p42d (pSym), fixL and katG on p42f, lpsβ on p42b, and p42a. For p42a, two different probes were used: the whole plasmid (180 kb) and a cosmid clone containing only the transfer region (tra and trb genes, oriT, and oriV). Table 2 summarizes the results. The plasmids present in each strain were labeled according to size, starting with the smallest plasmid. Because the number of plasmids was different in different strains, plasmids carrying the same genetic marker may be designated with different letters. For reference, the positions of plasmids hybridizing with nifH are indicated in Fig. 1. All strains except GR93 (13) and GR62 contained a plasmid that hybridized with nifH. Strain GR62 also is not able to nodulate bean, suggesting that it has been naturally cured of pSym. Most of the R. etli strains (17 of 24 strains [71%]) had a plasmid containing the fixL marker, while none of the eight bean-nodulating strains belonging to other species contained this gene (Table 2). For many strains the plasmids carrying nifH and fixL migrated very close to each other (Table 2), so it was difficult to discern whether these genes were located on the same plasmid. In order to clarify if these genes were present on the same plasmid or on plasmids that migrated close together, we performed the following experiment. Strains GR14, GR10, GR75, 6C-1, 16C-1, and FNJ11 were labeled with Tn5mob by mating them with Escherichia coli S17/pSup5011 (28) and selecting Nalr Nmr transconjugants (nalidixic acid concentration, 20 μg ml−1; neomycin concentration, 60 μg ml−1). To identify the labeled plasmids, the Rhizobium derivatives were mated with strain GMI9023 (a plasmidless Agrobacterium tumefaciens strain [25]) by using pRK2013 (9) as a helper for conjugation. Rifr Nmr (rifampin concentration, 100 μg ml−1; neomycin concentration, 60 μg ml−1) transconjugants were selected on Luria-Bertani medium (16). Plasmids present in the transconjugants were visualized in Eckhardt type gels. The gels were blotted onto nylon membranes and hybridized with nifH and fixL probes. In all cases, individual transconjugants hybridized either to nifH or to fixL but never simultaneously to both genes, indicating that as in CFN42, these two genes were located on different plasmids.
TABLE 2.
Plasmid number and localization of nifH, fixL, lpsβ, p42a, and katG sequences in bean-nodulating Rhizobium strains
| Strain | Plasmid(s)a | Localization of:
|
|||||
|---|---|---|---|---|---|---|---|
| nifH | fixL | lpsβ | p42ab | trap42a | katG | ||
| CFN42c | a, b, c, d, e, fd | d | f | b | a (a) | a | f |
| FNJ11c,e | a, b, c, d, e | c | d | b | a (a, e) | a | |
| GR14e | a, b, c, d, e | b | c | a | |||
| 6C-1c | a, b, c, d, e | b | c | a | a | a | |
| CIAT894 | a, b, c, d, e | d | e | a | |||
| 16C-1c,e | a, b, c, d | b | c | a | a (a) | a | |
| Bra5 | a, b, c, d | b | chrf | ||||
| 14C-1c | a, b, c, d, e | d | b | (a) | e | ||
| 21NJ-2c | a, b, c, d | c | a | d | |||
| GR03c | a, b | a | |||||
| GR42c | a, b | a | a | ||||
| GR18c | a, b | a | a | a | |||
| GR84 | a, b, c, d | c | a | a | |||
| VF39 | a, b, c, d, e, f | d | c | c | |||
| GR10e | a, b, c, d | b | c | a | b or c | NTg | NT |
| GR75c,e | a, b, c, d | b | c | a | NT | NT | |
| GR56c,e | a, b, c, d | b | c | a | NT | NT | |
| GR87c | a, b, c, d, e | c | e | b (a, b) | NT | NT | |
| 4C-2c | a, b, c, dh | c | c | a | a (a) | NT | NT |
| 4PR-2c | a, b, c | b | b | a | a (a) | NT | NT |
| 6PR-1c | a, b, c, d | b | c | a | a | NT | NT |
| 8C-3c | a, b, c | b | b | chr | NT | NT | |
| 8NJ-2c | a, b, c, d, e | c | a | NT | NT | ||
| 14PR-2c | a, b, c | b | c | c | NT | NT | |
| 16NJ-2c | a, b, c | b | b | a | a | NT | NT |
| 17C-2c,e | a, b, c, d | b | c | a | a (a) | NT | NT |
| 17NJ-2c | a, b, c | b | b | a | a (a) | NT | NT |
| 21PR-1c | a, b, c, d | c | a | NT | NT | ||
| GR93c | a | NT | NT | ||||
| GR64 | a, b, c | b | NT | NT | |||
| GR60 | a, b, c | b | b | NT | NT | ||
| GR45c | a, b, c | b | a | NT | NT | ||
The same letter does not necessarily indicate that the plasmids are the same size in two strains.
Parentheses indicate that a plasmid is transmissible.
Strain analyzed to determine whether transmissible plasmids are present.
Underlining indicates that bands are very close together.
fixL and nifH were found to be localized in different plasmids (see text for details).
chr, chromosome.
NT, not tested.
Boldface type indicates that bands were very intense, possibly due to the presence of more than one plasmid.
Previous work (10) indicated that lpsβ is present in plasmids of R. etli and R. leguminosarum strains. We confirmed this for the strains analyzed in this study; R. etli and R. leguminosarum strains showed a positive signal, while strains belonging to other species were negative. Interestingly, two strains (8C-3 and Bra5), showed an lpsβ hybridization signal on the chromosome (Table 2). Plasmids hybridizing with p42a (whole plasmid) were found in approximately 60% of the strains (17 of 27 strains), representing all of the different species (Table 2). Except for strains GR84 and GR42, all of the strains that hybridized with the whole p42a plasmid also hybridized with a cosmid clone containing only the transfer region. Recently, it has been shown in our laboratory that single copies of the katG and oxyR genes of CFN42 are localized on plasmid p42f (C. Vargas, S. Encarnación, A. Dávalos, A. Reyes-Pérez, Y. Mora, A. Garcia-de los Santos, S. Brom, and J. Mora, unpublished data). However, only CFN42 and two other strains (14C-1 and 21NJ-2) showed a plasmid-localized hybridization signal with the katG probe (Table 2).
With few exceptions, we found a high level of conservation of plasmid-associated markers among R. etli strains; lpsβ, fixL, and nifH genes were always located on three distinct replicons, as in type strain CFN42. In a few cases, some genes were located in the chromosome instead of a plasmid (lpsβ genes in strain Bra5) or, alternatively, in a plasmid instead of the chromosome (katG and oxyR in CFN42 and two other strains), which may just reflect the known genomic plasticity of Rhizobium strains (24). While these plasmid-associated traits were very conserved among the R. etli strains, they were absent in strains belonging to other bean-nodulating species (R. giardinii, R. gallicum, Sinorhizobium fredii). In R. leguminosarum strains these genes were also organized differently; thus, a fixL gene was absent from R. leguminosarum bv. phaseoli strain GR84, whereas in R. leguminosarum bv. viciae strain VF39 fixL and lpsβ were located on the same plasmid.
Identification of conjugative plasmids
To determine the presence of transmissible plasmids, we randomly labeled the different strains with Tn5 by mating them with E. coli S17/pSup2021 (28) and selecting Nalr Nmr transconjugants. Tn5-labeled derivatives of each strain were used as donors in conjugations with A. tumefaciens strain GMI9023. The rationale for this experiment was that Tn5 integrated randomly into the chromosome or into the different plasmids present in a strain; its integration into a transmissible plasmid gave the plasmid a selective marker in conjugation experiments. Plasmids present in the selected transconjugants were visualized with Eckhardt gels. Nine of the 23 strains analyzed contained transmissible plasmids (Table 2). Usually, the conjugative plasmids were among the plasmids that hybridized with p42a. However, some plasmids that showed no similarity to p42a were identified as transmissible; these plasmids included plasmid a of 14C-1, plasmid e of FNJ11, and plasmid a of GR87 (Table 2). Two strains, FNJ11 and GR87, simultaneously contained two conjugative plasmids.
RFLP analysis of plasmid-localized genetic markers
In order to obtain further information concerning the genetic organization of bean-nodulating strains, we determined the restriction fragment length polymorphisms (RFLPs) of a number of plasmid-borne genes of strain CFN42. Genomic DNAs were digested with appropriate restriction enzymes, electrophoresed in 1% (wt/vol) agarose gels, blotted onto nylon membranes, and hybridized under stringent conditions using Rapid-hyb buffer. Probes were linearized by digesting them with appropriate restriction enzymes and were labeled with [32P]CTP using a Rediprime DNA labeling system (Amersham Pharmacia Biotech, Little Chalfont, Buckinghamshire, England). The pSym plasmid of CFN42 is characterized by the presence of three reiterated nifH sequences (21), one copy of the fixK and fixN genes (12) (referred to as fixKd and fixNd in Table 3), and replication-related repABC genes (22). In 70% (16 of 23) of the R. etli strains the hybridizing restriction fragments were identical for all markers, regardless of the geographic origins of the strains (Table 3). In some strains, including 6C-1 (Table 3) and 4C-2, 6PR-1, 17C-2, and 17NJ-2 (data not shown), the size of one of the three nifH reiterations differed slightly (5.2 kb instead of 4.7 kb). fixKd and fixNd were always located on a single restriction fragment that was the same size for all strains. Only three strains, CIAT894, 16C-1 (Table 3), and 16NJ-2 (data not shown), lacked the fixKd and fixNd copies. With only two exceptions (CFN42 and CIAT894), all of the R. etli strains carried the repABC genes on a similar (7.2-kb) restriction fragment (Table 3). Interestingly, the bean-nodulating strains belonging to R. giardinii (GR03), R. gallicum (GR42, GR18, GR60), and R. leguminosarum (GR84) were identical to R. etli strains for all of the pSym-associated markers. Furthermore, we determined that the pSym plasmids of strains GR56, 6PR-1, and 17C-2 (R. etli), GR03 (R. giardinii), GR18 (R. gallicum), and GR84 (R. leguminosarum) belong to the same incompatibility group as the pSym plasmid of CFN42. The experiment in which this was determined consisted of isolating transconjugants of the strains after mobilization of the pSym plasmid from strain CFNX5. The basis for this experiment was as follows. R. etli strains contain three reiterated nifH sequences, and strain CFNX5 is a derivative of CFN42 with a GDYN-1 element inserted into the nifHc reiteration (24), which generates a characteristic pattern of the nifH reiterations that is easily differentiated from the pattern of the wild-type strains. The transconjugants were analyzed for their nifH reiteration patterns by hybridization of EcoRI-digested DNA with a nifH probe. Consequently, analysis of the nifH reiteration patterns of the transconjugants allowed us to determine that for the six strains tested, the transconjugants lost the endogenous pSym plasmid upon introduction of CFNX5 pSym. Additionally, the transconjugants of GR56, 6PR-1, and 17C-2 conserved the hybridization signal for fixL, indicating that these strains also carry the fixL gene on a plasmid different from pSym.
TABLE 3.
RFLP analysis of Southern-blotted total DNA probed with plasmid or chromosomal genes from CFN42
| Strain | Hybridization patternsa
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| nifH | fixKd | fixNd | repABC | fixL | fixKf | fixNf | lpsβ1 | lpsβ2 | leuAb | phaCb | katGb | |
| CFN42 | AAA | A | A | A | A | A | A | A | A | I | I | Ic |
| FNJ11 | AAA | A | A | B | B | B | B | B | B | II | II | II |
| GR14d | AAA | A | A | B | B | B | B | C | C | II | II | II |
| 6C-1 | AAA | A | A | B | B | B | B | C | C | II | II | II |
| CIAT894 | AAA | NDe | ND | C | C | A | A | D | D | II | III | III |
| 16C-1 | ABA | ND | ND | B | B | B | B | C | C | II | II | II |
| Bra5 | AAA | A | A | B | ND | ND | ND | Ef | Ef | III | IV | IV |
| 14C-1 | AAA | A | A | B | ND | ND | ND | F | F | II | IV | Vc |
| 21NJ-2 | AAA | A | A | B | ND | ND | ND | A | A | I | V | VIc |
| GR03 | AAA | A | A | B | ND | ND | ND | ND | ND | IV | VI | ND |
| GR42 | AAA | A | A | B | ND | ND | ND | ND | ND | V | VII | ND |
| GR18 | AAA | A | A | B | ND | ND | ND | ND | ND | V | VII | ND |
| GR84 | AAA | A | A | B | ND | ND | ND | A | A | VI | VIII | VII |
| VF39 | C | ND | B | ND | D | Cg | Cg | A | A | VI | VIII | VIII |
Identical letters indicate that RFLPs were identical. Three nifH copies are present in bean-nodulating strains.
The probes were cosmid clones containing the genes.
Plasmid localization.
The RFLPs of plasmid markers in strain GR14 are similar to those found in 11 other Spanish strains isolated from various locations (see text for details).
ND, not detected.
Chromosomal localization.
Localized on the same plasmid as lpsβ genes.
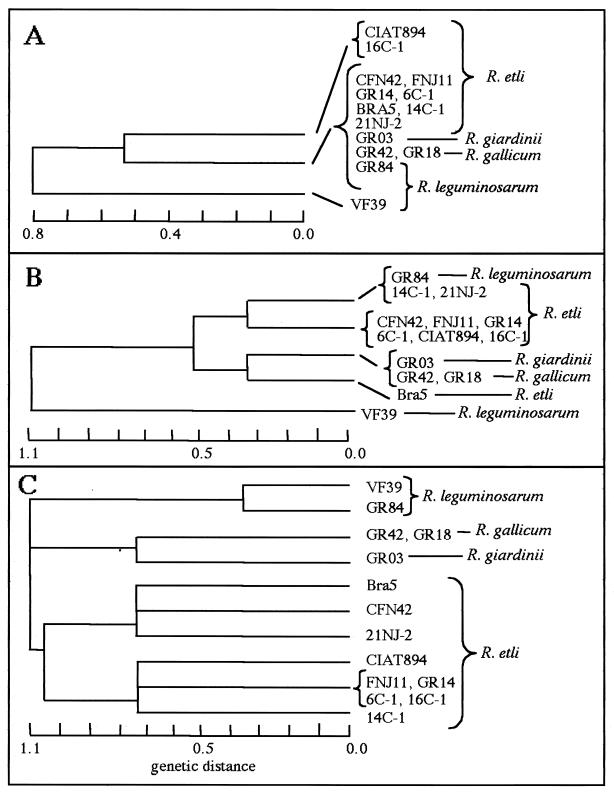
These data confirm and extend previous observations that P. vulgaris-nodulating strains belonging to several species (R. gallicum, R. giardinii, R. leguminosarum, and R. etli) share a symbiotic plasmid. The lack of variability found among pSym-associated markers is also shown in Fig. 2A, which shows the results of a cluster analysis of the pSym-associated RFLP data performed with the ETDIV and ETCLUS programs (13). All bean-nodulating strains belonging to either R. etli, R. giardinii, R. gallicum, or R. leguminosarum formed one group, which was separate from the pea-nodulating R. leguminosarum bv. viciae strain VF39.
FIG. 2.
Plasmid and chromosomal relatedness among different bean-nodulating strains. (A) Cluster analysis based on RFLPs of markers associated with the pSym plasmid of CFN42 (nifH, fixKd, fixNd, and repABC). (B) Cluster analysis based on RFLPs of markers associated with p42b (lpsβ1 and lpsβ2), p42d (nifH, fixKd, fixNd, and repABC), and p42f (fixL, fixKf, and fixNf). (C) Cluster analysis based on RFLP analysis of chromosomal markers (phaC, leuA, and katG). Species classification based on 16S rDNA analysis is indicated.
The distinctive traits of plasmid f of CFN42 are the presence of fixL and reiteration of the fixK and fixN genes (fixKf and fixNf in Table 3) (12). Most R. etli strains (70%) contained a plasmid equivalent to p42f with regard to organization of these genes. None of the bean-nodulating strains belonging to other species showed p42f-associated markers (Table 3).
p42b is distinguished by the presence of lpsβ genes. A p42b-like plasmid was confined to R. etli and R. leguminosarum strains. In all cases except strain 14C-1, lpsβ1 and lpsβ2 were localized in the same restriction fragment. R. leguminosarum bv. viciae strain VF39 also showed positive hybridization with the p42f-associated markers. However, a distinctive feature of this strain is that unlike R. etli strains, lpsβ, fixL, fixK, and fixN are localized on a single plasmid in VF39 (Table 3).
R. etli strains GR10, GR75, GR56, 4C-2, 4PR-2, 6PR-1, 6C-1, 8C-3, 14PR-2, 17C-2, and 17NJ-2 were similar to GR14 and 6C-1 in terms of pSym-, p42f-, and p42b-associated markers (data not shown).
In contrast to the results obtained when only pSym markers were used, the plasmid relatedness among the strains based on the presence of RFLP markers from p42b, p42d, and p42f shows the differences among species (Fig. 2B). As Fig. 2B shows, the majority of the R. etli strains are tightly clustered and are separated from R. giardinii and R. gallicum strains. However, some R. etli strains are in different groups; Bra5 is in one group, and another group is formed by 14C-1 and 21NJ-2. The latter group also includes R. leguminosarum bv. phaseoli strain GR84, while the pea-nodulating strain (VF39) appears in a separate group.
For all of the plasmid-borne genes analyzed, we also performed hybridization experiments with cosmid clones that contained the genes and adjacent genetic regions. The distribution of the hybridization patterns for the cosmid clones was similar to that of the individual genes (data not shown). These results further confirmed the identity of the pSym plasmids present in the different bean-nodulating species and the similarity between the p42b- and p42f-like plasmids shared by the R. etli strains.
RFLP analysis of chromosomal genetic regions
In order to compare genetic relationships among chromosomal and plasmid-localized genetic markers, we determined the RFLP patterns of EcoRI-restricted DNAs of the strains in hybridizations with cosmids containing chromosomal genetic regions of R. etli type strain CFN42. One of the cosmids used as a probe covered the genomic region containing the leuA leucine biosynthesis gene (J. M. Sanjuan-Pinilla, S. Brom, J. Olivares, and J. Sanjuan, unpublished data); another carried the phaC gene, involved in poly-β-hydroxybutyrate biosynthesis (6). We also included a cosmid containing the katG gene from CFN42; even though this cosmid contained DNA from plasmid p42f, we considered it chromosomal, because this was its location in most of the R. etli strains.
In each strain, between three and eight hybridizing fragments were detected for each of the cosmid probes. R. etli CIAT894, which does not contain a homolog of the CFN42 katG gene, showed only one hybridizing band when it was probed with the katG cosmid, whereas R. giardinii and R. gallicum strains showed no hybridization with this probe. The different RFLP patterns observed (Table 3) were used to perform a cluster analysis (Fig. 2C). The results revealed three main genetic groups: one group including the R. leguminosarum strains (either R. leguminosarum bv. phaseoli or R. leguminosarum bv. viciae), one group containing the strains belonging to R. gallicum and R. giardinii, and one group for the R. etli strains. A cluster analysis was also performed using only the RFLPs of the leuA and phaC cosmids, excluding katG. The results obtained were very similar to those shown in Fig. 2C (data not shown). Thus, this analysis clearly distinguished among different 16S rDNA species, regardless of the plasmid content. Despite the few chromosomal regions probed and the limited number of strains used in this analysis, the results indicate that there is wide chromosomal diversity among R. etli strains, in agreement with previous reports (7, 13). The high degree of conservation of both chromosomal and plasmid genetic markers among R. etli strains originally isolated from very distant geographical locations supports the suggestion that European representatives of R. etli are indeed descendants of American strains which separated only a few centuries ago, after the introduction of P. vulgaris into Europe (13).
Diverse data indicate that there is a close functional and structural relationship among R. etli and R. leguminosarum strains. In some cases, analysis of chromosomal 16S rDNA has indicated closer relationships between R. etli and R. leguminosarum strains than among different R. leguminosarum strains (7), although total DNA relatedness data clearly distinguish between these two species (29). In this work, all the R. etli chromosomal probes hybridized with total DNAs of R. leguminosarum strains, but the RFLP patterns were clearly distinct from the patterns for the R. etli strains. The lpsβ locus associated with p42b has been found in R. leguminosarum bv. viciae and R. leguminosarum bv. trifolii strains (10). Similarly, a gene encoding a unipolarly located cell surface-associated agglutinin, rapA1, was found to be present in R. leguminosarum bv. trifolii, R. leguminosarum bv. viciae, R. leguminosarum bv. phaseoli, and R. etli strains and to be absent in other rhizobial species (2). Also, the fixL gene present in p42f is homologous to a fixL gene from R. leguminosarum bv. viciae strain VF39 (12, 18), although we do not know if this gene is widely distributed among R. leguminosarum strains.
In this work, we found that the majority of R. etli strains, independent of geographical origin, share not only the presence but also the localization and organization of genetic regions associated with two other replicons present in type strain CFN42: p42b and p42f. These results suggest that at least some plasmid-encoded traits may also be valuable for discriminating between R. etli and strains belonging to other species that nodulate P. vulgaris. In agreement with this suggestion, it has been found that utilization of dulcitol, which Amarger and collaborators described as one of the most useful traits for differentiating the species that nodulate common bean (1), is associated with a plasmid (p42c) in R. etli CFN42 (4). Comparison of the distribution of pSym and the distribution of p42b- or p42f-associated traits suggests that pSym is more promiscuous than the other plasmids. Interestingly, we have observed that the pSym plasmid of CFN42 is transmissible, while we have not been able to detect transfer of p42b or p42f (4).
Acknowledgments
We are grateful to Laura Cervantes and Javier Rivera for excellent technical assistance, to Miguel Angel Cevallos for providing phaC and repABC probes, and to Jesús Caballero-Mellado for help with the cluster analysis.
This work was supported in part by grant IN202599 (DGAPA, UNAM) and by a CSIC-CONACYT cooperation grant.
REFERENCES
- 1.Amarger, N., V. Macheret, and G. Laguerre. 1997. Rhizobium gallicum sp. nov. and Rhizobium giardinii sp. nov., from Phaseolus vulgaris nodules. Int. J. Syst. Bacteriol. 47:996-1006. [DOI] [PubMed] [Google Scholar]
- 2.Ausmees, N., K. Jacobsson, and M. Lindberg. 2001. A unipolarly located, cell-surface-associated agglutinin, RapA, belongs to a family of Rhizobium-adhering proteins (Rap) in Rhizobium leguminosarum bv. trifolii. Microbiology 147:549-559. [DOI] [PubMed] [Google Scholar]
- 3.Baldani, J. I., R. W. Weaver, M. F. Hynes, and D. B. Eardly. 1992. Utilization of carbon substrates, electrophoretic enzyme patterns, and symbiotic performance of plasmid-cured clover rhizobia. Appl. Environ. Microbiol. 58:2308-2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brom, S., A. García de los Santos, L. Cervantes, R. Palacios, and D. Romero. 2000. In Rhizobium etli symbiotic plasmid transfer, nodulation competitivity and cellular growth require interaction among different replicons. Plasmid 44:34-43. [DOI] [PubMed] [Google Scholar]
- 5.Brom, S., A. García de los Santos, T. Stepkowsky, M. Flores, G. Dávila, D. Romero, and R. Palacios. 1992. Different plasmids of Rhizobium leguminosarum bv. phaseoli are required for optimal symbiotic performance. J. Bacteriol. 174:5183-5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cevallos, M. A., S. Encarnación, A. Leija, Y. Mora, and J. Mora. 1996. Genetic and physiological characterization of a Rhizobium etli mutant strain unable to synthesize poly-β-hydroxybutyrate. J. Bacteriol. 178:1646-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eardly, B. D., F. Wang, T. S. Whittam, and R. K. Selander. 1995. Species limits in Rhizobium populations that nodulate the common bean (Phaseolus vulgaris). Appl. Environ. Microbiol. 61:507-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eckhardt, T. 1978. A rapid method for the identification of plasmid deoxyribonucleic acid in bacteria. Plasmid 1:584-588. [DOI] [PubMed] [Google Scholar]
- 9.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.García-de los Santos, A., and S. Brom. 1997. Characterization of two plasmid-borne lpsβ loci of Rhizobium etli required for lipopolysaccharide synthesis and for optimal interaction with plants. Mol. Plant-Microbe Interact. 10:891-902. [DOI] [PubMed] [Google Scholar]
- 11.García-de los Santos, A., S. Brom, and D. Romero. 1996. Rhizobium plasmids in bacteria legume interactions. World J. Microbiol. Biotechnol. 12:119-125. [DOI] [PubMed] [Google Scholar]
- 12.Girard, L., S. Brom, A. Dávalos, O. López, M. Soberón, and D. Romero. 2000. Differential regulation of fixN-reiterated genes in Rhizobium etli by a novel fixL-fixK cascade. Mol. Plant-Microbe Interact. 13:1283-1292. [DOI] [PubMed] [Google Scholar]
- 13.Herrera-Cervera, J. A., J. Caballero-Mellado, G. Laguerre, H. V. Tichy, N. Requena, N. Amarger, E. Martínez-Romero, J. Olivares, and J. Sanjuan. 1999. At least five different rhizobial species nodulate Phaseolus vulgaris in a Spanish soil. FEMS Microbiol. Ecol. 30:87-97. [Google Scholar]
- 14.Hynes, M. F., and N. F. McGregor. 1990. Two plasmids other than the nodulation plasmid are necessary for formation of nitrogen-fixing nodules by Rhizobium leguminosarum. Mol. Microbiol. 4:567-574. [DOI] [PubMed] [Google Scholar]
- 15.Martínez-Romero, E., L. Segovia, F. M. Mercante, A. A. Franco, P. Graham, and M. A. Pardo. 1991. Rhizobium tropici, a novel species nodulating Phaseolus vulgaris L. beans and Leucaena sp. trees. Int. J. Syst. Bacteriol. 41:417-426. [DOI] [PubMed] [Google Scholar]
- 16.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 17.Noel, K. D., A. Sánchez, L. Fernández, J. Leemans, and M. A. Cevallos. 1984. Rhizobium phaseoli symbiotic mutants with transposon Tn5 insertions. J. Bacteriol. 158:148-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patchkowski, T., A. Schlüter, and U. B. Priefer. 1996. Rhizobium leguminosarum bv. viciae contains a second fnr/fixK gene and an unusual fixL homolog. Mol. Microbiol. 21:267-280. [DOI] [PubMed] [Google Scholar]
- 19.Piñero, D., E. Martínez, and R. K. Selander. 1988. Genetic diversity and relationships among isolates of Rhizobium leguminosarum bv. phaseoli. Appl. Environ. Microbiol. 54:2825-2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Priefer, U. 1989. Genes involved in lipopolysaccharide production and symbiosis are clustered on the chromosome of Rhizobium leguminosarum bv. viciae VF39. J. Bacteriol. 171:6161-6168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quinto, C., H. de la Vega, M. Flores, L. Fernández, T. Ballado, G. Soberón, and R. Palacios. 1982. Reiteration of nitrogen fixation gene sequences in Rhizobium phaseoli. Nature (London) 299:724-728.
- 22.Ramírez-Romero, M. A., P. Bustos, L. Girard, O. Rodríguez, M. A. Cevallos, and G. Dávila. 1997. Sequence, localization and characteristics of the replicator region of the symbiotic plasmid of Rhizobium etli. Microbiology 143:2825-2831. [DOI] [PubMed] [Google Scholar]
- 23.Rodríguez-Navarro, D. M., A. M. Buendía, M. Camacho, M. M. Lucas, and C. Santamaría. 2000. Characterization of Rhizobium spp. bean isolates from South-West Spain. Soil Biol. Biochem. 32:1601-1613. [Google Scholar]
- 24.Romero, D., S. Brom, J. Martínez-Salazar, M. L. Girard, R. Palacios, and G. Dávila. 1991. Amplification and deletion of a nod-nif region in the symbiotic plasmid of Rhizobium phaseoli. J. Bacteriol. 173:2435-2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosenberg, C., and T. Hughet. 1984. The pATC58 plasmid of Agrobacterium tumefaciens is not essential for tumor induction. Mol. Gen. Genet. 196:533-536. [Google Scholar]
- 26.Segovia, L., J. P. W. Young, and E. Martínez-Romero. 1993. Reclassification of American Rhizobium leguminosarum bv. phaseoli type I strains as Rhizobium etli sp. nov. Int. J. Syst. Bacteriol. 43:374-377. [DOI] [PubMed] [Google Scholar]
- 27.Sessitsch, A., A. D. L. Hardarson, and W. M. de Vos. 1997. Characterization of Rhizobium etli and other Rhizobium spp. that nodulate Phaseolus vulgaris L. in an Austrian soil. Mol. Ecol. 6:601-608. [Google Scholar]
- 28.Simon, R. 1984. High frequency mobilization of gram-negative bacterial replicons by the in vitro constructed Tn5-mob transposon. Mol. Gen. Genet. 196:413-420. [DOI] [PubMed] [Google Scholar]
- 29.Van Berkum, P., D. Beyene, and B. Eardly. 1996. Phylogenetic relationships among Rhizobium species nodulating the common bean (Phaseolus vulgaris L.). Int. J. Syst. Bacteriol. 46:240-244. [DOI] [PubMed] [Google Scholar]