Abstract
The effect of polypeptide denaturation of Bacillus thuringiensis Cry1A toxins or purified Manduca sexta 120-kDa aminopeptidase N on the specificities of their interactions was investigated. Ligand and dot blotting experiments were conducted with 125I-labeled Cry1Ac, Cry1Ac mutant 509QNR-AAA511 (QNR-AAA), or 120-kDa aminopeptidase N as the probe. Mutant QNR-AAA does not bind the N-acetylgalactosamine moiety on the 120-kDa aminopeptidase. Both 125I-Cry1Ac and 125I-QNR-AAA bound to 210- and 120-kDa proteins from M. sexta brush border membrane vesicles and purified 120-kDa aminopeptidase N on ligand blots. However, on dot blots 125I-QNR-AAA bound brush border vesicles but did not bind purified aminopeptidase except when aminopeptidase was denatured. In the reciprocal experiment, 125I-aminopeptidase bound Cry1Ac but did not bind QNR-AAA. 125I-aminopeptidase bound Cry1Ab to a limited extent but not the Cry1Ab domain I mutant Y153D or Cry1Ca. However, denatured 125I-aminopeptidase detected each Cry1A toxin and mutant but not Cry1Ca on dot blots. The same pattern of recognition occurred with native (nondenatured) 125I-aminopeptidase probe and denatured toxins as the targets. The broader pattern of toxin-binding protein interaction is probably due to peptide sequences being exposed upon denaturation. Putative Cry toxin-binding proteins identified by the ligand blot technique need to be investigated under native conditions early in the process of identifying binding proteins that may serve as functional toxin receptors.
Bacillus thuringiensis Cry1 proteins are specifically and highly toxic to lepidopteran insects (21). Cry1 proteins are expressed by the bacterium as 130-kDa protoxins and then occluded in large bipyramidal crystals. Feeding larvae ingest crystals that dissolve in the alkaline midgut, releasing soluble protoxin. The protoxin is processed by gut proteinases to active toxin that binds to receptors located in the brush border epithelium. The toxin inserts into the membrane and oligomerizes, forming ion channels. These events culminate in midgut cell lysis and insect mortality. The B. thuringiensis toxin mode of action has recently been reviewed (1).
Toxin binding to receptors in the insect midgut is an important determinant of insect susceptibility and a mechanism by which insects become resistant to B. thuringiensis Cry toxins. For example, the Cry1A-resistant YHD2 strain of Heliothis virescens has a transposon insertion in a cadherin-like protein, a known class of Cry1 receptors (6). The implication is that knockout of the receptor causes the observed lack of Cry protein binding (11) and insect resistance.
In addition to cadherin-like proteins, aminopeptidases and glycoconjugates have been identified as binding molecules that in some cases serve as receptors for B. thuringiensis Cry1 proteins in lepidopteran species (reviewed in reference 7). The ligand blotting technique is frequently the first step in identifying a toxin-binding molecule. In this technique brush border proteins are separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and blotted onto a membrane filter. Toxin-binding molecules are identified on the filter by using radiolabeled toxin or an indirect toxin detection technique. The ligand blotting technique detects Cry toxin binding to molecules under denaturing conditions.
Where investigated, Cry1 proteins often bind the same proteins under denaturing and native conditions. For example, Cry1Ab binds the 210-kDa cadherin-like protein under denaturing and nondenaturing conditions (23). Also, Cry1Ac binds to 120-kDa aminopeptidase N (APN) under both denaturing and nondenaturing conditions (20). However, Cry1Aa binds to the 175-kDa cadherin-like protein from Bombyx mori under native conditions but not under denaturing conditions. As technologies such as phage display are applied to the investigation of binding epitopes on proteins (8), the concept of where the epitope (i.e., peptide) is located on a native protein becomes especially relevant.
Cry1Ac recognizes an N-acetylgalactosamine (GalNAc) on APN via a sugar-binding pocket located in domain III of the toxin (3, 5). Cry1Ac mutant 509QNR-AAA511 (QNR-AAA) is a mutant with a disrupted sugar-binding pocket (10). QNR-AAA has reduced binding to brush border membrane vesicles (BBMV) but is only twofold less toxic than Cry1Ac to Manduca sexta (10). Surface plasmon resonance (SPR) binding analyses revealed that Cry1Ac mutant QNR-AAA did not bind APN under nondenaturing conditions. Ligand blot analyses with M. sexta BBMV and purified APN revealed that QNR-AAA did not bind APN but bound a 210-kDa protein, in agreement with SPR analyses (10). The results that prompted the present study came from a ligand blot experiment that we conducted in which Cry1Ac and QNR-AAA were probed against M. sexta BBMV and purified APN. Unexpectedly, both Cry1Ac and QNR-AAA bound the 210-kDa protein and the 120-kDa APN.
Since our ligand blotting results disagreed with results reported elsewhere (10), we designed experiments to investigate the apparent inconsistency. We investigated the possibility that denaturation of a target binding molecule altered binding specificity. The hypothesis that denatured (unfolded) proteins have binding epitopes not exposed on native proteins was tested. Native and denatured target proteins (BBMV and purified APN) were probed with Cry1Ac and QNR-AAA. For comparison, we included Cry1Ab, which also binds APN (17), and Cry1Ab mutant Y153D. Mutant Y153D is impaired in irreversible binding and membrane insertion and is not toxic to M. sexta (4, 9). When denaturation of target molecules caused altered binding specificity, we reversed the approach by denaturing the toxin probe (Cry1Ac or QNR-AAA). Denaturation of either toxin or target proteins resulted in altered specificity but not a complete loss of specificity.
MATERIALS AND METHODS
Bacterial strains and toxin purification.
B. thuringiensis strain HD-73 producing Cry1Ac was obtained from the Bacillus Genetic Stock Center (Columbus, Ohio). A B. thuringiensis strain producing Cry1Ca was obtained from Ecogen, Inc. (Langhorne, Pa.). Escherichia coli strain NRD-12 expressing a cry1Ab gene was kindly provided by Luke Masson (Biotechnology Research Institute, Montreal, Canada). E. coli strains producing Cry1Ac mutant toxin QNR-AAA and Cry1Ab mutant toxin Y153D were obtained from Donald Dean (Ohio State University, Columbus).
Toxins from B. thuringiensis were isolated and purified as described previously (4). Cry1Ab, QNR-AAA, and Y153D toxins were purified from E. coli as described by Lee et al. (13). Protein concentration was determined by the Bradford protein assay using bovine serum albumin as the standard (2).
Iodination of toxins and APN.
Cry1Ac, Cry1Ac mutant QNR-AAA, and APN were iodinated with Na125I by using IODO-Beads (Pierce Co.) according to the manufacturer's instructions. Either 1 μg of toxin or 5 μg of APN was labeled with 0.5 mCi of Na125I (Amersham Pharmacia). Based on the input protein, the specific activities of labeled Cry1Ac and QNR-AAA were 27.3 and 25.6 μCi/μg, respectively, while the specific activity of APN was 10.3 μCi/μg.
Gel electrophoresis.
The purity of toxin and APN preparations was analyzed by SDS-8% PAGE. Five micrograms of purified protein or 105 cpm of radiolabeled toxin or APN per lane was electrophoresed. Gels were stained with Coomassie brilliant blue R-250 or exposed to Kodak XAR-5 film with an intensifying screen at −80°C for autoradiography.
Preparation of BBMV.
Midguts were dissected from fifth-instar M. sexta larvae, washed with ice-cold MET buffer (250 mM mannitol, 17 mM Tris-HCl [pH 7.5], 5 mM EGTA), immediately frozen on dry ice, and stored at −80°C until needed. BBMV were prepared by the MgCl2 precipitation method (25). The final BBMV pellet was suspended in ice-cold 10 mM Tris-HCl (pH 7.5). Rat intestinal BBMV were prepared as described by Steiger and Murer (22).
BBMV proteins were solubilized by mixing BBMV with 1% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS) in Tris-buffered saline (pH 7.5) for 30 min at 4°C. The mixture was centrifuged at 15,000 × g for 10 min, and the supernatant was collected. Solubilized BBMV for dot blotting were prepared immediately before use.
Purification of APN from M. sexta BBMV.
APN was purified by anion-exchange chromatography followed by hydrophobic interaction chromatography to remove associated lipids as reported previously (19).
Ligand and reverse-ligand blotting.
BBMV (5 μg) or toxins (5 μg) were separated by SDS-PAGE and then electroblotted onto polyvinylidene difluoride Q (PVDF) membrane filters (Millipore). The filters were blocked with 3% bovine serum albumin (BSA) in TBST (25 mM Tris-HCl [pH 7.4], 3 mM KCl, 135 mM NaCl, 0.1% Tween 20) for 1 h. After incubation with 106 cpm of 125I-labeled toxin or APN in 10 ml of 0.1% BSA-TBST for 3 h (1 h for 125I-APN), the filters were washed in 0.1% BSA-TBST three times for 15 min and exposed to film overnight at −80°C.
Dot and reverse-dot blotting.
Various concentrations of protein diluted in 100 μl of Tris-buffered saline were dot blotted on PVDF filters under a vacuum for 10 min by using a Bio-Rad apparatus. The filters were incubated with 106 cpm of 125I-labeled probe (toxin or APN) in the manner described above for ligand blotting. Proteins were denatured by adding SDS to a concentration of 1% and boiling for 5 min. Samples were clarified by centrifugation at 10,000 × g for 10 min prior to dot blotting.
RESULTS
Autoradiography of 125I-labeled proteins and ligand blotting with 125I-labeled Cry1Ac and QNR-AAA toxins.
125I-labeled APN, Cry1Ac, and QNR-AAA appeared as single bands when preparations were separated by SDS-PAGE and visualized on autoradiograms (Fig. 1A).
FIG. 1.
Autoradiograpy of 125I-labeled proteins (A) and ligand blot analyses of SDS-PAGE-separated BBMV proteins and purified APN with 125I-Cry1Ac (B) or 125I-QNR-AAA (C). (A) Lanes 1 to 3 contained 100,000 cpm of radiolabeled APN, Cry1Ac, and QNR-AAA, respectively. (B and C) Five micrograms each of M. sexta (lane 1) and rat (lane 2) BBMV proteins and 1 μg of purified APN (lane 3) were transferred to PVDF membranes, incubated with 125I-labeled toxin (0.5 nM), and detected by autoradiography. M. sexta BBMV proteins were also incubated with 0.5 nM 125I-toxin in the presence of 100 mM GalNAc and autoradiographed (lane 4).
The differences between the ligand blot results shown in Fig. 1 and previously published results prompted us to undertake this investigation. In previous studies, Cry1Ac recognized the 120-kDa APN and the 210-kDa cadherin-like protein on ligand blots (7). Additionally, Cry1Ac mutant QNR-AAA (which is impaired in GalNAc recognition) bound the 210-kDa protein but not the 120-kDa APN (10). In this study, 125I-Cry1Ac bound proteins as expected, including the absence of Cry1Ac binding to rat BBMV and competition for Cry1Ac binding by GalNAc (Fig. 1B). However, the QNR-AAA binding results shown in Fig. 1C were not expected. 125I-QNR-AAA bound to the 210-kDa protein, the 120-kDa APN, a 110-kDa protein, and a 45-kDa protein. GalNAc did not compete with QNR-AAA binding and apparently increased binding to the 110-and 45-kDa proteins. Apparently, our ligand blotting conditions contributed to an apparent change in QNR-AAA binding specificity. Since SPR analyses showed that QNR-AAA did not bind M. sexta APN under nondenaturing conditions (10), we considered the possibility that denatured proteins on ligand blots have binding sites not exposed on the native protein.
Dot blotting with 125I-labeled toxins.
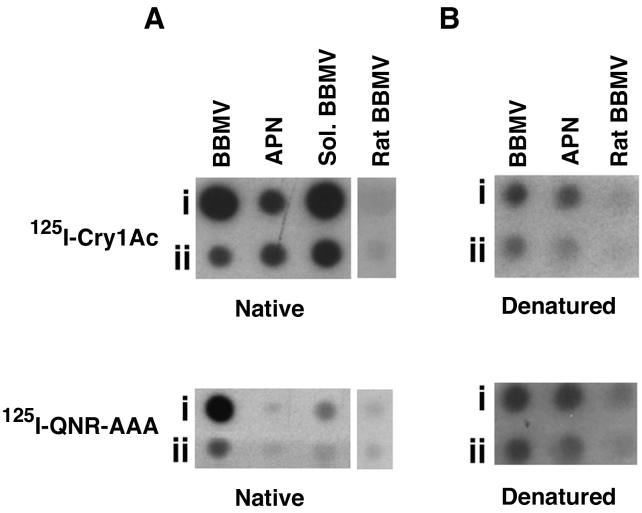
The effect of denaturation (SDS and heat treatment) on Cry1Ac and QNR-AAA binding specificity was tested by dot blot experiments. The results for Cry1Ac and QNR-AAA binding to native proteins are shown in Fig. 2A. M. sexta BBMV, CHAPS-solubilized BBMV, and purified APN were spotted onto a PVDF membrane and then probed with 125I-Cry1Ac or 125I-QNR-AAA. 125I-Cry1Ac bound BBMV, CHAPS-solubilized BBMV, and purified APN. 125I-QNR-AAA bound strongly to BBMV and less strongly to CHAPS-solubilized BBMV but did not bind purified APN.
FIG. 2.
Dot blot analyses of M. sexta BBMV, CHAPS-solubilized BBMV (Sol. BBMV), and purified APN with 125I-Cry1Ac or 125I-QNR-AAA. In most cases the amounts of proteins in rows i and ii were 5 and 1 μg, respectively; for purified APN the amounts of proteins were 2 and 1 μg in rows i and ii, respectively. Native (A) or denatured (B) proteins were spotted on PVDF membranes and damp dried under a vacuum. The membranes were incubated with 125I-Cry1Ac (0.5 nM) or 125I-QNR-AAA (0.5 nM) for 1 h and autoradiographed as described in Materials and Methods.
The effect of denaturation on QNR-AAA binding is shown in Fig. 2B. Denaturation did not alter the binding pattern of Cry1Ac, although the amounts of Cry1Ac binding to denatured BBMV and APN were reduced. However, 125I-QNR-AAA bound denatured APN in addition to BBMV and CHAPS-solubilized BBMV. 125I-labeled Cry1Ac or QNR-AAA did not bind denatured rat BBMV. Denaturation, in this case by SDS and heat treatment, altered QNR-AAA binding specificity.
Reverse-dot blotting with native and denatured 125I-APN.
Next, we probed a series of toxins on dot blots with either native or denatured receptor (APN) to determine the extent to which specificity for toxins was changed. The results of native APN binding are shown in Fig. 3A and B. 125I-APN bound strongly to Cry1Ac and weakly to Cry1Ab (note the faint signal) but not to QNR-AAA, the Cry1Ab domain I mutant Y153D, or Cry1Ca. As expected, GalNAc decreased 125I-APN binding to Cry1Ac. The dot blotting and reverse-dot blotting results were in agreement: APN has a specific pattern of binding specificity for wild-type Cry1 toxins, and native APN does not bind mutant QNR-AAA.
FIG. 3.
Reverse-dot blot analyses of purified toxins. Five micrograms (row i) and 1 μg (row ii) of purified toxins were dot blotted as described in the legend to Fig. 2. (A) Toxin on membrane exposed to 125I-APN. (B) Toxin on membrane exposed to 125I-APN plus 100 mM GalNAc. (C) Toxin on membrane exposed to denatured 125I-APN. 125I-APN was denatured as described in Materials and Methods. (D) Purified toxins were denatured as described in Materials and Methods before dot blotting onto the membrane and exposure to 125I-APN. All preparations were incubated for 1 h, washed, and autoradiographed.
Since BBMV proteins on ligand blots are denatured, we probed toxins on dot blots with denatured APN. 125I-APN was boiled in 1% SDS for 5 min before the dot blot was probed. Denatured 125I-APN bound strongly to all the toxins except Cry1Ca (Fig. 3C). While denaturation exposed binding sites on APN recognized by Cry1Ac and Cry1Ab toxins, binding sites were apparently not present on Cry1Ca. Since QNR-AAA bound denatured APN in reverse-dot blot experiments, the data agreed with the observation that denaturation altered QNR-AAA binding specificity (ligand and dot blot experiments).
Since denatured APN had an increased range of binding, we reversed the procedure by denaturing toxins before dot blotting and then tested for altered binding of APN. 125I-APN was probed against toxins that were denatured by boiling in 1% SDS before dot blotting. 125I-APN bound denatured Cry1Ac, Cry1Ab, and the Cry1A mutant toxins. 125I-APN did not bind denatured Cry1Ca (Fig. 3D). Denaturation of either APN or toxin decreased binding specificity. However, the absence of binding to denatured Cry1Ca is evidence that binding has some level or degree of specificity.
Reverse-ligand blotting with 125I-labeled APN.
Cry1 proteins on ligand blots were probed with 125I-APN. This experiment allowed us to test binding specificity under actual ligand blot conditions. 125I-APN bound Cry1Ac, Cry1Ac mutant QNR-AAA, Cry1Ab, and Cry1Ab mutant Y153D but not Cry1C (Fig. 4). There was no competition between GalNAc and labeled APN for binding to these toxins (data not shown). The reverse-ligand blot results are further evidence that denaturation of either toxin or brush border proteins induces protein interactions that do not occur under native conditions.
FIG. 4.
Reverse-ligand blot analyses of SDS-8%PAGE-separated purified toxins with 125I-APN (1 nM). Five micrograms of each purified toxin was transferred to a PVDF membrane filter, incubated with 125I-APN, washed, and autoradiographed.
DISCUSSION
We report that denaturation of either Cry1 toxin or brush border proteins alters toxin-binding specificity. Previous reports have suggested that alteration of toxin-binding specificity occurs under denaturing conditions. Nagamatsu et al. (18) observed that Cry1Aa bound the 175-kDa protocadherin of B. mori under native conditions but not under denaturing conditions. Vadlamudi et al. (23) demonstrated that Cry1Ab toxin could interact with both native and denatured 210-kDa cadherin-like protein. Lee and Dean (14) reported inconsistencies between BBMV binding study results and ligand blot results.
For identification of putative toxin receptors, native toxins and denatured BBMV proteins on ligand blots have been used (15, 23). In this study we attempted to determine if the state of the polypeptides (both toxin and receptor) affects their interactions. To address this issue, we performed combinations of binding experiments involving labeled receptor and toxins in their native and denatured forms. Table 1 summarizes the results of these experiments, including the state of the polypeptides in the experiments which we conducted and the resulting alterations in binding specificity. Table 1 shows whether toxin bound to APN and vice versa under different experimental conditions.
TABLE 1.
Summary of experiments conducted to investigate the interaction of B. thuringiensis Cry1A toxins with purified M. sexta 120-kDa APN
| Exptl method (radiolabeled protein[s]) | State of APN | State of toxin | Binding of toxin to APN | Binding of APN to Cry1Ac/binding of APN to QNR-AAA |
|---|---|---|---|---|
| Ligand blot (Cry1Ac/QNR-AAA) | Denatured | Native | Yes/Yesa | |
| Dot blot (Cry1Ac/QNR-AAA) | Native | Native | Yes/Noa | |
| Denatured | Native | Yes/Yesa | ||
| Reverse-dot blot (APN) | Native | Native | Yes/No | |
| Denatured | Yes/Yes | |||
| Reverse-dot blot (denatured APN) | Denatured | Native | Yes/Yes | |
| Reverse-ligand blot (APN) | Native | Denatured | Yes/Yes |
Results for Cry1Ac/results for QNR-AAA.
In the present study, Cry1Ac bound 210- and 120-kDa proteins in BBMV and purified APN on ligand blots, which is consistent with previously published results (Fig. 1) (7). Jenkins et al. (10) reported QNR-AAA binding only to the 210-kDa protein on ligand blots, whereas we detected QNR-AAA binding to the 210-, 120-, and 110-kDa proteins using the same technique. We also observed that QNR-AAA gave a stronger signal than Cry1Ac on ligand blots.
The Cry1Ac mutant QNR-AAA bound BBMV but not APN under native conditions (10). The lack of QNR-AAA binding to native APN on dot blots agreed with this finding. Since QNR-AAA is toxic to M. sexta (10), another protein must be a functional receptor in vivo. Evidence supports the hypothesis that the 210-kDa Bt-R1 protein is the remaining functional receptor (24). The possibility that another protein(s) (e.g., the 110-kDa protein) is the QNR-AAA binding protein(s) is currently being investigated in our lab. The reason for QNR-AAA binding to the 45-kDa protein and the reason for increased binding to the 110-kDa protein in the presence of GalNAc are unknown.
Since QNR-AAA bound to purified APN only under denaturing conditions (Table 1), it appears that QNR-AAA recognizes an epitope on APN that is exposed upon denaturation. From our ligand blot results obtained in the presence of GalNAc, it also appears that this second toxin recognition site on APN is GalNAc independent. Jenkins et al. (10) suggested that toxin interacts with APN initially via the GalNAc-dependent site on domain III, followed by domain II interactions. Although speculative, it appears that toxin can interact with APN under denaturing conditions directly via the GalNAc-independent domain II site.
Using labeled APN, we designed reverse-dot blot and reverse-ligand blot experiments to further test the hypothesis that denaturation affected binding specificity. Labeled APN bound strongly to Cry1Ac (Fig. 3A), and this binding was reduced in the presence of GalNAc (Fig. 3B). This result confirmed the ligand blotting results obtained with Cry1Ac and GalNAc and was evidence that radiolabeling of native APN did not alter the major binding properties of APN.
Labeled APN bound weakly to Cry1Ab but did not bind to QNR-AAA, Cry1Ab mutant Y153D, or Cry1Ca on dot blots (Fig. 3A). The absence of binding of labeled APN to QNR-AAA agreed with dot blot results which showed that labeled QNR-AAA did not bind purified APN. The limited APN binding to Cry1Ab was surprising as Cry1Ab bound to APN in an SPR analysis (16). However, Keeton et al. (12) did not detect Cry1Ab binding to APN on ligand blots. Y153D, the Cry1Ab domain I mutant, did not bind APN. As expected from the results of Luo et al. (15), Cry1Ca did not bind the 120-kDa APN.
When denatured toxins were probed with labeled APN on reverse-dot blots, APN bound to Cry1Ac, Cry1Ab, and the Cry1A mutants tested (Fig. 3D). These results were confirmed with reverse-ligand blots in which SDS-PAGE-separated Cry1A toxins were probed with labeled APN (Fig. 4). In both cases, APN did not bind to denatured Cry1Ca, indicating that the binding of APN to toxin was altered, yet retained specificity for Cry1A toxins. This is evidence that Cry1A toxins, but not Cry1Ca toxin, possess an APN-binding epitope(s) that becomes accessible upon denaturation. Denaturation apparently causes toxin to bind BBMV proteins that would not bind in the native toxin confirmation. Whether this phenomenon occurs naturally as toxin unfolds in vivo is unknown.
Our summary conclusion is that while the ligand blotting technique is a useful technique for identifying putative toxin-binding proteins in brush border membranes, the technique exposes a unique binding epitope(s) not present on the surface of the native BBMV proteins due to denaturation. Therefore, interpretation of ligand blot results for identification of putative binding proteins should be carried out with caution, and such results should be confirmed by other techniques.
REFERENCES
- 1.Aronson, A. I., and Y. Shai. 2001. Why Bacillus thuringiensis insecticidal toxins are so effective: unique features of their mode of action. FEMS Microbiol. Lett. 195:1-8. [DOI] [PubMed] [Google Scholar]
- 2.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 3.Burton, S. L., D. J. Ellar, J. Li, and D. J. Derbyshire. 1999. N-acetylgalactosamine on the putative insect receptor aminopeptidase N is recognized by a site on the domain III lectin-like fold of a Bacillus thuringiensis insecticidal toxin. J. Mol. Biol. 287:1011-1022. [DOI] [PubMed] [Google Scholar]
- 4.Daniel, A., D. H. Dean, and M. J. Adang. 2001. Analyses of the pore forming ability of Bacillus thuringiensis Cry1A mutant toxins using a light-scattering technique. Pestic. Biochem. Physiol. 70:7-18. [Google Scholar]
- 5.deMaagd, R. A., P. L. Bakker, L. Masson, M. J. Adang, S. Sangadala, W. Stiekema, and D. Bosch. 1999. Domain III of the Bacillus thuringiensis delta-endotoxin Cry1Ac is involved in binding to Manduca sexta brush border membranes and to its purified aminopeptidase N. Mol. Microbiol. 31:463-471. [DOI] [PubMed] [Google Scholar]
- 6.Gahan, L. J., F. Gould, and D. G. Heckel. 2001. Identification of a gene associated with Bt resistance in Heliothis virescens. Science 293:857-860. [DOI] [PubMed] [Google Scholar]
- 7.Garczynski, S. F., and M. J. Adang. 2000. Investigations of Bacillus thuringiensis Cry1 toxin receptor structure and function, p. 181-197. In J.-F. Charles, A. Delecluse, and C. Nielsen-Leroux (ed.), Entomopathogenic bacteria: from laboratory to field application. Kluwer Academic Press, Dordrecht, The Netherlands.
- 8.Gomez, I., D. I. Oltean, S. S. Gill, A. Bravo, and M. Soberon. 2001. Mapping the epitope in cadherin-like receptors involved in Bacillus thuringiensis Cry1A toxin interaction using phage display. J. Biol. Chem. 276:28906-28912. [DOI] [PubMed] [Google Scholar]
- 9.Hussain, S.-R. A., A. I. Aronson, and D. H. Dean. 1996. Substitution of residues on the proximal side of Cry1A Bacillus thuringiensis δ-endotoxins affects irreversible binding to Manduca sexta midgut membrane. Biochem. Biophys. Res. Commun. 226:8-14. [DOI] [PubMed] [Google Scholar]
- 10.Jenkins, J. L., M. K. Lee, S. Sangadala, M. J. Adang, and D. H. Dean. 1999. Binding of Bacillus thuringiensis Cry1Ac toxin to Manduca sexta aminopeptidase-N receptor is not directly related to toxicity. FEBS Lett. 462:373-376. [DOI] [PubMed] [Google Scholar]
- 11.Jurat-Fuentes, J. L., F. L. Gould, and M. J. Adang. 2000. High levels of resistance and cross-resistance to Bacillus thuringiensis Cry1 toxins in Heliothis virescens are due to reduced toxin binding and pore formation. Resist. Pest Manage. 11:23-24. [Google Scholar]
- 12.Keeton, T. P., B. R. Francis, W. S. A. Maaty, and L. A. Bulla, Jr. 1998. Effects of midgut-protein-preparative and ligand binding procedures on the toxin binding characteristics of BT-R1, a common high-affinity receptor in Manduca sexta for Cry1A Bacillus thuringiensis toxins. Appl. Environ. Microbiol. 64:2158-2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee, M., R. Milne, A. Ge, and D. Dean. 1992. Location of a Bombyx mori receptor binding region on a Bacillus thuringiensis delta-endotoxin. J. Biol. Chem. 267:3115-3121. [PubMed] [Google Scholar]
- 14.Lee, M. K., and D. H. Dean. 1996. Inconsistencies in determining Bacillus thuringiensis toxin binding sites relationship by comparing competition assays with ligand blotting. Biochem. Biophys. Res. Commun. 220:575-580. [DOI] [PubMed] [Google Scholar]
- 15.Luo, K., Y.-J. Lu, and M. J. Adang. 1996. A 106-kDa form of aminopeptidase is a receptor for Bacillus thuringiensis CryIC δ-endotoxin in the brush border membrane of Manduca sexta. Insect Biochem. Mol. Biol. 26:33-40. [Google Scholar]
- 16.Masson, L., Y.-J. Lu, A. Mazza, R. Brosseau, and M. J. Adang. 1995. The CryIA(c) receptor purified from Manduca sexta displays multiple specificities. J. Biol. Chem. 270:20309-20315. [DOI] [PubMed] [Google Scholar]
- 17.Masson, L., A. Mazza, R. Brousseau, and B. Tabashnik. 1995. Kinetics of Bacillus thuringiensis toxin binding with brush border membrane vesicles from susceptible and resistant larvae of Plutella xylostella. J. Biol. Chem. 270:11887-11896. [DOI] [PubMed] [Google Scholar]
- 18.Nagamatsu, Y., S. Toda, F. Yamaguchi, M. Ogo, M. Kogure, M. Nakamura, Y. Shibata, and T. Katsumoto. 1998. Identification of Bombyx mori midgut receptor for Bacillus thuringiensis insecticidal CryIA(a) toxin. Biosci. Biotechnol. Biochem. 62:718-726. [DOI] [PubMed] [Google Scholar]
- 19.Sangadala, S., P. Azadi, R. Carlson, and, M. J. Adang. 2000. Chemical analyses of Manduca sexta aminopeptidase N, co-purifying lipids and functional interaction with Bacillus thuringiensis Cry1Ac toxin. Insect Biochem. Mol. Biol. 32:97-107. [DOI] [PubMed] [Google Scholar]
- 20.Sangadala, S., F. Walters, L. H. English, and M. J. Adang. 1994. A mixture of Manduca sexta aminopeptidase and alkaline phosphatase enhances Bacillus thuringiensis insecticidal CryIA(c) toxin binding and 86Rb+-K+ leakage in vitro. J. Biol. Chem. 269:10088-10092. [PubMed] [Google Scholar]
- 21.Schnepf, E., N. Crickmore, J. Van Rie, D. Lereclus, J. Baum, J. Feitelson, D. R. Zeigler, and D. H. Dean. 1998. Bacillus thuringiensis and its pesticidal crystal. Microbiol. Mol. Biol. Rev. 62:775-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Steiger, B., and H. Murer. 1983. Heterogeneity of brush-border membrane vesicles from rat small intestine prepared by a precipitation method using Mg/EGTA. Eur. J. Biochem. 135:95-101. [DOI] [PubMed] [Google Scholar]
- 23.Vadlamudi, R. K., T. H. Ji, and L. A. Bulla, Jr. 1993. A specific binding protein from Manduca sexta for the insecticidal toxin of Bacillus thuringiensis subsp. berliner. J. Biol. Chem. 268:12334-12340. [PubMed] [Google Scholar]
- 24.Vadlamudi, R. K., E. Weber, I. Ji, T. H. Ji, and L. A. Bulla, Jr. 1995. Cloning and expression of a receptor for an insecticidal toxin of Bacillus thuringiensis. J. Biol. Chem. 270:5490-5494. [DOI] [PubMed] [Google Scholar]
- 25.Wolfersberger, M. G., P. Luthy, A. Maurer, P. Parenti, V. F. Sacchi, B. Giordana, and G. M. Hanozet. 1987. Preparation and partial characterization of amino acid transporting brush border membrane vesicles from the larval midgut of the cabbage butterfly (Pieris brassicae). Comp. Biochem. Physiol. A 86:301-308. [Google Scholar]