Abstract
Streptococcus mutans, a major etiological agent of dental caries, causes demineralization of the tooth tissue due to the formation of acids from dietary carbohydrates. Dominant among the virulence determinants of this organism are aciduricity and acidogenicity, the abilities to grow at low pH and to produce acid, respectively. The mechanisms underlying the ability of S. mutans to survive and proliferate at low pH are currently under investigation. In this study we cultured S. mutans at pH 5.2 or 7.0 and extracted soluble cellular proteins. These were analyzed using high-resolution two-dimensional gel electrophoresis, and replicate maps of proteins expressed under each of the two conditions were generated. Proteins with modulated expression at low pH, as judged by a change in the relative integrated optical density, were excised and digested with trypsin by using an in-gel protocol. Tryptic digests were analyzed using matrix-assisted laser desorption ionization mass spectrometry to generate peptide mass fingerprints, and these were used to assign putative functions according to their homology with the translated sequences in the S. mutans genomic database. Thirty individual proteins exhibited altered expression as a result of culture of S. mutans at low pH. Up-regulated proteins (n = 18) included neutral endopeptidase, phosphoglucomutase, 60-kDa chaperonin, cell division proteins, enolase, lactate dehydrogenase, fructose bisphosphate aldolase, acetoin reductase, superoxide dismutase, and lactoylglutathione lyase. Proteins down-regulated at pH 5.2 (n = 12) included protein translation elongation factors G, Tu, and Ts, DnaK, small-subunit ribosomal protein S1P, large-subunit ribosomal protein L12P, and components of both phosphoenolpyruvate:protein phosphotransferase and multiple sugar binding transport systems. The identification of proteins differentially expressed following growth at low pH provides new information regarding the mechanisms of survival and has identified new target genes for mutagenesis studies to further assess their physiological significance.
Dental caries is one of the most prevalent oral diseases and constitutes a significant economic burden in terms of treatment costs, especially in the Western world, where consumption of a diet comprising relatively high levels of freely fermentable carbohydrates is significantly correlated with the disease development. Dental caries is multifactorial in nature, and caries incidence is related not only to host diet and salivary function but also to the establishment of a cariogenic microflora (54). Thus, bacterial fermentation of dietary carbohydrates, most notably sucrose, results in the production of acidic end products which initially demineralize enamel and later demineralize dentine, and if this process is left unchecked it results in the formation of a carious lesion. While recent studies have suggested that streptococci other than mutans streptococci may be significant in the development of some forms of this disease (2, 8, 43), mutans streptococci, especially Streptococcus mutans, are still considered major pathogens with respect to the etiology of caries (27).
S. mutans is considered a major pathogen by virtue of the fact that it is isolated in increased numbers from diseased sites in caries-prone individuals, is cariogenic in animal models, and possesses a range of physiological traits that make it particularly suited for the colonization of caries-prone tooth sites (7, 54). The virulence determinants of this organism have received much attention over several decades and have been investigated using a range of biochemical, physiological, and genetic techniques. The most extensively studied virulence determinants are the glucosyltransferases, which play a role in adhesion, contributing to the production of extracellular polysaccharide that localizes both the bacteria and their secreted products within the plaque matrix (27, 34, 35, 54). In addition, more recent studies have proposed that the surface-associated protein P1, an adhesin, contributes to the cariogenicity of S. mutans (15). Of at least equal importance in terms of the virulence of S. mutans are two other pathogenicity factors, namely, acidogenicity and aciduricity, the ability of bacteria to generate acidic end products and to survive at low pH, respectively. S. mutans is undoubtedly one of the most acidogenic of the species found within dental plaque, being capable of producing acid from fermentable carbohydrate at a higher rate and over a much greater pH range than most other streptococci (21). Tolerance of acidic environments and an adaptive response to exposure to low pH are also of major importance in the pathogenesis of dental caries. S. mutans employs a number of strategies to resist the effects of acid stress, and earlier studies have highlighted the role of the proton-translocating ATPase (H+-ATPase) as a key factor contributing to the aciduric response (18, 40). Thus, it is not only the production of acid, driving enamel demineralization, that makes this organism such a successful pathogen but also the aciduric characteristics of the species.
Transposon mutagenesis has identified individual S. mutans genes regulated by environmental stresses (17, 24), while antisense RNA methodologies have demonstrated that the sgp gene of S. mutans, which encodes a GTP-binding protein, is involved in the response to low pH (3, 4, 44). Differential-display reverse transcription-PCR (11) was used to investigate stress-responsive genes that were up-regulated following acid shock, resulting in the identification of, among other proteins, a putative branched-chain amino acid aminotransferase. Using a combination of radioisotopic labeling and one-dimensional polyacrylamide gel electrophoresis (PAGE), 36 proteins, perhaps including subunits of the H+-ATPase complex and the 60-kDa chaperonin, were found to be immediately up-regulated in S. mutans as a result of instantaneous exposure to low pH (28). More recently, isotope labeling in combination with two-dimensional (2-D) PAGE was used to monitor protein expression in acid- and nutrient-stressed S. mutans cells, and that study defined a group of 64 proteins, 25 of them acid specific, that were synthesized in greater amounts following acid stress and 49 proteins that were down-regulated. The identities of these stress-responsive proteins remain to be established (49).
While investigations of the effect of instantaneous acid shock on the synthesis of proteins may identify factors crucial to the survival of an organism, proteins expressed during growth in acidic conditions, affecting long-term survival and persistence, are most likely to contribute to the caries process. In this study, we adopted the view that the acid-tolerant response of S. mutans is not simply the ability of the organism to survive at low pH but also its ability to grow (54). Therefore, we have used 2-D PAGE and matrix-assisted laser desorption ionization-time-of-flight (MALDI-TOF) mass spectrometry to identify S. mutans proteins with altered expression during growth at pH 5.2 and 7.0.
MATERIALS AND METHODS
Bacterial strain and culture.
S. mutans strain UA159, sequenced at the University of Oklahoma (http://www.genome.ou.edu/smutans.html), was used throughout this study. The isolate was maintained by routine culture on Columbia agar (Oxoid Ltd., Basingstoke, Hampshire, United Kingdom) supplemented with 5% (vol/vol) defibrinated horse blood (TCS Microbiology, Buckingham, United Kingdom) and incubation in an anaerobic atmosphere comprising 10% CO2, 10% H2, and 80% N2 (MK3 Anaerobic Workstation; Don Whitley Scientific Ltd., Shipley, West Yorkshire, United Kingdom) at 37°C for 1 or 2 days. Inocula were prepared by suspending individual colonies in filter-sterilized 50 mM sodium phosphate buffer (pH 7.5) to give an A620 of approximately 1.5. This suspension was used as the inoculum for growth in nutrient-rich media. S. mutans was cultured in brain heart infusion broth (BHI) (Oxoid) prepared according to the manufacturer's instructions to give a pH of 7.0 or with the pH adjusted to 5.2 (2) by the addition of 0.2 M disodium hydrogen orthophosphate and 0.1 M citric acid (each at a final concentration of approximately 20 mM). Growth characteristics were determined by culture of S. mutans in 200-μl aliquots of medium in 96-well sterile microtiter trays with a 5% inoculum. Growth was monitored by measuring the A620 in a 96-well plate-reading spectrophotometer (iEMS Reader; Labsystems, Life Sciences International, Hampshire, United Kingdom) at 0.25-h intervals with incubation at 37°C and a 10-s period of shaking prior to each absorbance measurement. Cultures for protein extraction and subsequent analysis by 2-D PAGE were prepared by inoculation of BHI (pH 7.0 or 5.2) in 10-ml volumes and static, aerobic incubation at 37°C until mid-exponential phase was reached. Cells were pelleted by centrifugation (2,700 × g, 10 min), washed thrice in 50 mM sodium phosphate buffer (pH 7.5), and stored at −20°C until required for analysis. Growth experiments and protein extraction for each culture condition were performed on at least three occasions to enable a statistical analysis of the data. The pHs of supernatants at the time of harvest and of stationary-phase cultures were determined, and residual glucose was measured (kit no. 510-A; Sigma Chemical Company, Poole, Dorset, United Kingdom).
To assess adaptation to acid tolerance, 10-ml portions of mid-exponential-phase cultures from both growth conditions were taken in triplicate and pelleted by centrifugation as described above. Cell pellets were washed in BHI, resuspended in BHI (pH 7.0) or BHI adjusted to pH 4.0 with HCl, and incubated at 37°C for 1 h. Samples were taken before treatment and after 1 h and diluted serially with fastidious anaerobe broth (Lab M, Lancashire, United Kingdom). The cell suspensions were plated on Columbia agar supplemented with 5% (vol/vol) defribinated horse blood. The CFU were counted after a 2-day anaerobic incubation at 37°C, and results are expressed as the mean percentage (± standard deviation) of survivors. Pretreatment and posttreatment values were compared using Student's t test.
Analysis of soluble cellular proteins by 2-D PAGE.
Soluble cellular proteins of S. mutans were extracted from protoplasts by using a modification (55) of a method described previously (23). Briefly, cell pellets were washed in 0.9% NaCl; resuspended in 25 mM Tris-HCl (pH 7.5) containing 0.1 mg of lysozyme per ml, 0.26 mg of phenylmethylsulfonyl fluoride per ml, 50 μg of chloramphenicol per ml (to prevent bacterial protein synthesis), and 18% sucrose; and incubated at 37°C for 15 min. Protoplasts were pelleted by centrifugation and lysed by the addition of 50 mM Tris-HCl containing 0.3% sodium dodecyl sulfate (SDS)-0.3% dithiothreitol and treatment at 100°C for 5 min. Following vortexing to disperse the material, the sample was centrifuged (11,600 × g, 10 min) prior to the addition of magnesium chloride and endonuclease (Sigma) to the clear supernatant and incubation at 4°C for 10 min. Proteins were precipitated from solution by treatment with 4 volumes of ice-cold acetone and centrifugation (15,100 × g, 4°C, 10 min).
Samples were resuspended in a buffer containing 7 M deionized urea, 2 M thiourea, 2% Tergitol NP-40, 62 mM dithiothreitol, and 2% pH 3 to 10 carrier ampholytes (Bio-Rad Laboratories Ltd., Hemel Hempstead, Hertfordshire, United Kingdom) to give a protein concentration of approximately 1 μg/μl. Samples (285 μl) were applied to 17-cm immobilized pharmalyte gradient (IPG) strips (pH 3 to 6, 4 to 7, or 5 to 8 [linear]) (Bio-Rad), and the strips were rehydrated under active conditions according to the manufacturer's instructions in a Protean isoelectric focusing cell (Bio-Rad). Following rehydration, the strips were focused at 20°C with an initial voltage of 250 V for 15 min and ramping to 10,000 V over 5 h, after which this voltage was maintained until 60,000 V·h was attained. After the focusing stage, IPG strips were equilibrated sequentially in a buffer (Tris-HCl containing 6 M urea, 30% [vol/vol] glycerol, and 2% SDS) containing 1% dithiothreitol or 2.5% iodoacetamide for 15 min each. Strips were applied to SDS-12 to 14% gradient self-cast polyacrylamide gels (200 by 160 by 1.5 mm), and electrophoresis was carried out in a Protean II xi cell (Bio-Rad) at 35 mA per gel for ca. 5 h with 250 mM glycine-25 mM Tris-HCl-0.1% SDS as the running buffer. Low-molecular-weight markers (Sigma) were applied at the acidic end of the IPG strips. Gels were fixed and stained with colloidal Coomassie brilliant blue (38) (Sigma) or a modified silver stain (45).
Gels were scanned at 300 dots per in., and 2-D maps of cellular proteins were analyzed using 2D Advanced software (version 5.01; Phoretix, Newcastle upon Tyne, United Kingdom). Observed masses for resolved proteins were calculated by comparison of their mobilities with those of molecular weight markers, and pI values were calculated according to the linearity of the pH strips. Integrated optical densities (IOD) were obtained for each resolved protein following definition of spot boundaries, and these were expressed as a percentage of the total protein detected on each gel. Where proteins occurred as distinct isoforms, the sum of the individual IOD values was used to determine total expression of the given polypeptide. A total of at least three protein preparations, extracted from independent cultures, for each growth condition were analyzed by 2-D PAGE, and proteins subjected to further analysis were visible in at least two gels from any one growth condition. Proteins were considered to be differentially expressed if the mean percent IOD was up- or down-regulated 1.5-fold or greater under the two growth conditions, and these proteins were excised from gels and subjected to further analysis by peptide mass fingerprinting. Significant differences in protein expression levels were determined using Student's t test with a P value of <0.05. The most abundant proteins, in the mass and pI ranges investigated here, expressed when S. mutans was cultured at pH 5.2 or 7.0 had the greatest mean IOD when calculated as a percentage of the total IOD of all proteins.
Identification of resolved proteins by peptide mass fingerprinting.
Up- and down-regulated proteins were excised from gels stained with colloidal Coomassie brilliant blue and digested with trypsin by using an in-gel protocol (45, 55). Briefly, washed gel pieces were swollen in 50 mM ammonium bicarbonate containing sequencing-grade trypsin (Promega, Southampton, Hampshire, United Kingdom) at a concentration of 12.5 ng/μl, and enzymatic cleavage was carried out at 37°C overnight. Peptides were purified and desalted using ZipTips (C18 reverse-phase tips; Millipore Ltd., Watford, Hertfordshire, United Kingdom) according to the manufacturer's instructions and eluted in 1:1 acetonitrile-0.1% trifluoroacetic acid. Peptide solutions (0.5 μl) were applied to a stainless steel target plate and mixed in equal volume with a saturated solution of α-cyano-4-hydroxycinnamic acid in 70% acetonitrile-0.033% aqueous trifluoroacetic acid. Samples were allowed to air dry prior to acquisition of spectra on a Voyager Elite MALDI-TOF mass spectrometer (Applied Biosystems, Cheshire, United Kingdom) operating with delayed extraction in reflector mode. An average of 256 shots were acquired per spectrum, and all spectra were recorded using a laser (337-nm output, 3-ns pulse width) at an intensity of ca. 1,000. Peptide mass fingerprints were calibrated using a mixture of des-Arg1-bradykinin, angiotensin 1, and Glu1-fibrinopeptide with close external calibration, and Data Explorer software (Applied Biosystems) was used to label monoisotopic peaks.
Monoisotopic masses from peptide mass fingerprints were analyzed with MS-Fit (University of California Mass Spectrometry Facility; http:prospector.ucsf.edu/ucsfhtml3.4/msfit.htm), installed locally, with the scoring for missed cleavage sites set at 0. The mass accuracy was usually 200 ppm or less, and no upper or lower mass limits for the target proteins were applied. All searches were performed against the S. mutans partially completed annotated genomic database (http://igweb.integratedgenomics.com/IGwit/), which became available in October 2000 and presently comprises 1,795 open reading frames (ORFs). If no homologs were identified using the S. mutans database, the nonredundant National Center for Biotechnology Information database (http://www.ncbi.nlm.nih.gov/GenBank/) and the annotated genomic databases for Streptococcus pneumoniae and Streptococcus pyogenes were searched (http://igweb.integratedgenomics.com/IGwit). A putative function was assigned to a protein if it was returned with the highest-ranking molecular weight search score (a probability function based on the number of peptides matched and the mass accuracy) (39), with a minimum value of approximately 1,000 and where there was a clear delineation between this and the next highest ranking candidate. In addition, at least four peptides were required to match, and there was a sequence coverage of at least 10%. All proteins assigned functions in this manner were also required to have good agreement between the theoretical and observed masses and pI values.
RESULTS
Growth of S. mutans in media with initial pH values of 7.0 and 5.2.
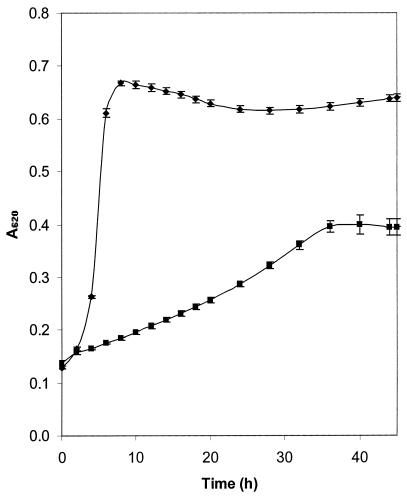
S. mutans UA159 was cultured at an initial pH of 7.0 or 5.2 in nutrient-rich medium (BHI) in culture volumes of 200 μl in order to determine the kinetics of bacterial growth. An arithmetic growth curve was obtained for low-pH cultures, and this is typical of partial inhibition by stress factors. No discernible lag phase was observed for growth under either condition, but doubling times were markedly different, being 1.0 h for pH 7.0 and 6.6 h for pH 5.2 (Fig. 1). The maximum A620 at the stationary phase also differed significantly (P < 0.05) for the two cultures, being 0.666 (±0.007) and 0.405 (±0.017) for pH 7.0 and 5.2, respectively, demonstrating that growth was slowed and reduced at the lower pH. Nonetheless, these data demonstrate that the isolate is aciduric and is capable of proliferation under these acidic conditions. When these experiments were extended to the investigation of bacterial growth in larger culture volumes, as used for the preparation of cellular material for analysis by 2-D PAGE, we demonstrated that at the mid-exponential phase of growth the pHs were 6.2 (±0.08) and 4.7 (±0.10) for cells grown in media with initial pH values of 7.0 and 5.2, respectively, and glucose remained (>5 mM) in the supernatants of both cultures, suggesting that the cells, as used for analysis by 2-D PAGE, were not nutrient limited. At stationary phase, all glucose had been utilized and the terminal pHs were 5.3 (±0.02) and 4.5 (±0.01) for cultures grown in media with initial pH values of 7.0 and 5.2, respectively. Cells from acid-grown cultures, taken at mid-exponential phase, demonstrated increased acid tolerance, with 65.3% (±4.1) of the cells surviving for 1 h at pH 4.0 compared to 36.6% (±13.8) of the cells from the cultures initially at pH 7.0 (P < 0.05). Growth was observed in the cultures suspended in the BHI at pH 7.0 from both growth conditions.
FIG. 1.
Growth of S. mutans at pH 7.0 and 5.2. S. mutans strain UA159 was cultured in 200-μl aliquots of BHI adjusted to pH 7.0 (♦) or pH 5.2 (▪). Growth was measured by monitoring the A620, and error bars indicate standard deviations from experiments carried out in triplicate.
Resolution of S. mutans cellular proteins by 2-D PAGE.
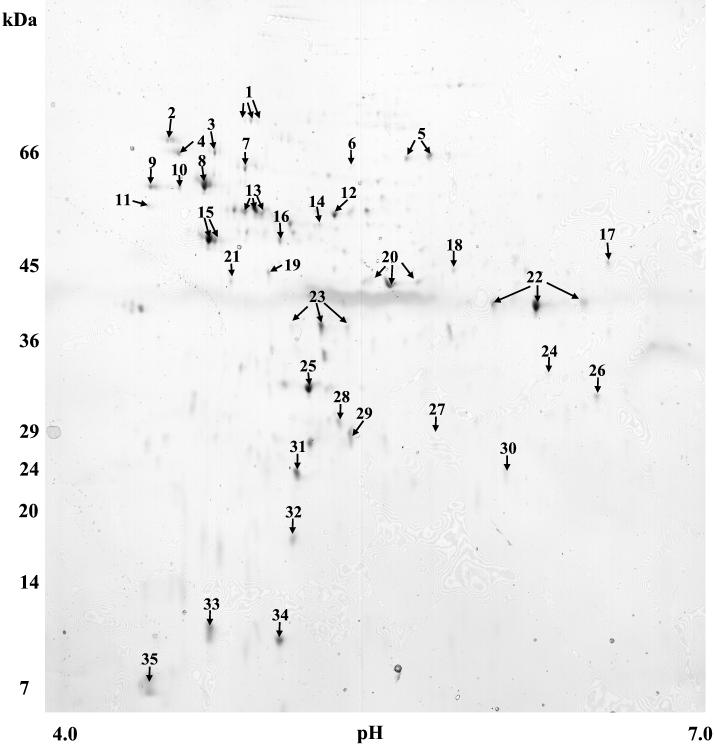
Cellular proteins extracted from mid-exponential-phase cells of S. mutans were resolved by 2-D PAGE and visualized following silver staining. Preliminary analysis of expressed proteins using IPG strips in the ranges 3 to 6, 4 to 7, and 5 to 8 demonstrated that greater than 95% of proteins from bacteria grown in either culture condition focused in the pI 4 to 7 region (data not shown). Subsequent experiments designed to monitor modulated expression of proteins at low pH were therefore carried out using IPG strips in the pI range of 4 to 7 only, because this facilitated analysis of the major part of the proteome. For 2-D gels that were prepared for peptide mass fingerprinting, colloidal Coomassie brilliant blue staining was used because this yielded better MALDI-TOF mass spectra than silver staining. Examination of 2-D maps of colloidal Coomassie brilliant blue-stained preparations demonstrated that 188 (±28) and 163 (±44) proteins were detected when bacteria were cultured initially at pH 7.0 and 5.2, respectively. The results of the resolution of cellular proteins extracted from pH 5.2-grown cultures are shown in Fig. 2; all of the proteins marked were also expressed by pH 7.0-grown cells and were either among the 10 most abundantly expressed or were differentially expressed, depending on the protein under consideration (data not shown).
FIG. 2.
Resolution of cellular proteins of S. mutans cultured at pH 5.2 by 2-D PAGE. S. mutans was cultured in BHI adjusted to pH 5.2. Extracted proteins were resolved in the pI range of 4 to 7 in the first dimension and by SDS-gradient (12 to 14%) PAGE in the second dimension. Proteins were visualized following staining with colloidal Coomassie brilliant blue. Proteins with distinct isoforms are given a single identifying number, and isoforms are indicated by arrows. The proteins numbered account for the 10 most abundantly expressed proteins and those with differential expression following growth at low pH.
The most abundant S. mutans proteins.
The 10 most abundant proteins for each growth condition comprised approximately 30% of the total protein observed (Table 1). These proteins were excised from gels and subjected to in-gel digestion to demonstrate that polypeptides resolved in this manner were amenable to identification when the peptide mass fingerprints were searched against the S. mutans genomic sequence data. Of the 20 proteins analyzed, 19 were unambiguously identified, although fructose bisphosphate aldolase was from the S. pneumoniae genomic data. The unidentified protein (spot 34 in Fig. 2) had an observed molecular mass of <15 kDa. Lactate dehydrogenase was represented in pH 7.0-grown cells, and fructose bisphosphate aldolase and phosphoglycerate kinase ranked among the most abundant proteins only in cells cultured at pH 5.2 (Table 1). Protein translation elongation factor Tu (EF-Tu), EF-G, and small-subunit (SSU) ribosomal protein S1P were detected as abundant proteins in pH 7.0-derived S. mutans, while only EF-Tu ranked among the most highly expressed proteins from bacteria grown at low pH. The 60-kDa molecular chaperonin (GroEL homolog) was one of the most abundant proteins extracted from pH 5.2- but not pH 7.0-grown cells. Spot 34 was abundant in the pH 5.2-grown cells but could not be identified.
TABLE 1.
Most abundant proteins expressed by S. mutans cultured at pH 7.0 or 5.2
| Rankinga |
S. mutans cultured at pH 7.0
|
S. mutans cultured at pH 5.2
|
||||
|---|---|---|---|---|---|---|
| Spotb | Protein identityc | % of total proteind (SD) | Spot | Protein identity | % of total protein (SD) | |
| 1 | 22 | Glyceraldehyde-3-phosphate dehydrogenase | 6.45 (2.68) | 22 | Glyceraldehyde-3-phosphate dehydrogenase | 4.95 (1.01) |
| 2 | 13 | EF-Tu | 5.01 (1.43) | 15 | Enolase | 4.89 (0.23) |
| 3 | 15 | Enolase | 2.87 (0.71) | 25 | Fructose-bisphosphate aldolasee | 4.52 (1.35) |
| 4 | 29 | Phosphoglycerate mutase | 2.85 (0.71) | 8 | 60-kDa chaperonin | 4.38 (2.08) |
| 5 | 16 | SSU ribosomal protein S1P | 2.30 (0.55) | 13 | EF-Tu | 3.32 (0.81) |
| 6 | 1 | EF-G | 2.23 (0.70) | 35 | LSU ribosomal protein L12P | 2.99 (0.64) |
| 7 | 2 | DnaK | 2.16 (0.89) | 20 | Phosphoglycerate kinase | 2.55 (0.54) |
| 8 | 35 | LSU ribosomal protein L12P | 2.15 (0.10) | 29 | Phosphoglycerate mutase | 2.53 (0.60) |
| 9 | 12 | Pyruvate kinase | 2.09 (0.62) | 12 | Pyruvate kinase | 2.51 (0.54) |
| 10 | 23 | l-Lactate dehydrogenase | 1.94 (0.64) | 34 | No matchf | 1.19 (0.20) |
Proteins are ranked according to abundance (greatest mean IOD).
Numbers refer to the proteins labeled in Fig. 2.
Putative functions for proteins were assigned on the basis of the presence of homologs in the database for S. mutans. LSU, large subunit.
Percentage of total protein detected on gels as measured by mean IOD (three replicates).
The putative function was assigned on the basis of the presence of homologs in the database for S. pneumoniae.
Peptide mass fingerprint data failed to attain match criteria, and no function could be assigned to the protein.
Identification of modulated proteins.
Thirty proteins in the mass and pI ranges investigated here were expressed in different amounts (1.5-fold or greater up- or down-regulation) when S. mutans was cultured at pH 5.2 compared with cells grown at pH 7.0. Expression of 18 proteins was up-regulated (10 of these significantly), and that of 12 proteins was down-regulated (7 of these significantly). Homologs were identified for 26 of the 30 proteins with altered expression by using the S. mutans genomic database (Table 2), and one other protein, fructose bisphosphate aldolase, was identified using S. pneumoniae genomic database. All of the proteins identified matched the stringent search criteria, and there was usually good agreement between theoretical and observed mass and pI values (usually less than 10% variation). Enolase, glutamate dehydrogenase, lactate dehydrogenase, and EF-Tu were exceptions, as the observed masses were higher than the theoretical masses since the gene sequences in the databases were apparently truncated. Three modulated proteins (spots 10, 33, and 34 in Fig. 2), with observed mass/pI values of 56.0/4.6, 15.4/5.0, and 14.1/4.6, respectively, could not be assigned putative functions, although all yielded good mass spectra.
TABLE 2.
S. mutans whole-cell proteins up- or down-regulated following growth at pH 5.2
| Functional category | Spota | Protein identityb | RMNc | Coverage (%)d | Observed migratione
|
Theoretical migrationf
|
Peptides matchedg | MOWSE scoreh | Mean spot vol ratio, pH 5.2/7.0i | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| pI | Mr | pI | Mr | ||||||||
| Cellular processes and stress proteins | 2 | DnaK protein | 00169 | 24 | 4.6 | 67.0 | 4.6 | 65.3 | 9 | 5.4 × 106 | 0.53 |
| 8 | 60-kDa chaperonin | 01340 | 26 | 4.7 | 57.6 | 4.7 | 57.1 | 8 | 3.6 × 106 | 2.53j | |
| 9 | Trigger factor, Ppiase | 01099 | 16 | 4.5 | 56.0 | 4.5 | 47.5 | 6 | 5.6 × 104 | 0.50 | |
| 11 | Cell division protein FtsZ | 01544 | 14 | 4.5 | 52.8 | 4.4 | 45.7 | 4 | 7.4 × 103 | 3.07 | |
| 14 | Cell division protein FtsA | 01545 | 14 | 5.2 | 51.5 | 5.0 | 48.7 | 4 | 9.6 × 103 | 2.35 | |
| 31 | Superoxide dismutase (Mn) | 00054 | 39 | 5.1 | 23.5 | 5.0 | 22.6 | 6 | 4.1 × 105 | 4.18j | |
| Central and intermediary metabolism | 3 | Neutral endopeptidase | 01555 | 29 | 4.7 | 65.9 | 4.7 | 71.8 | 13 | 1.8 × 1012 | 4.74j |
| 6 | Glucan-1,6-α-glucosidase | 01417 | 11 | 5.3 | 60.0 | 5.0 | 62.0 | 4 | 2.4 × 102 | 0.24 | |
| 7 | Phosphoglucomutase/phoshomannomutase | 02078 | 26 | 4.9 | 60.0 | 4.9 | 63.1 | 13 | 1.8 × 108 | 5.37j | |
| 15 | Enolase | 02125 | 19 | 4.7 | 48.6 | 4.7 | 39.5 | 6 | 6.8 × 104 | 1.70j | |
| 18 | NADP-specific glutamate dehydrogenase | 00335 | 46 | 5.8 | 47.3 | 5.5 | 46.8 | 15 | 8.4 × 1013 | 0.11j | |
| 23 | l-Lactate dehydrogenase | 00108 | 25 | 5.2 | 37.0 | 5.0 | 35.2 | 7 | 2.3 × 106 | 1.71 | |
| 24 | Enoyl-[acyl-carrier protein] reductase (NADH) | 01983 | 37 | 6.1 | 32.9 | 5.7 | 33.5 | 9 | 7.7 × 107 | 0.15j | |
| 25 | Fructose bisphosphate aldolase | 16 | 5.1 | 31.7 | 5.0 | 31.4 | 4 | 1.1 × 103 | 2.28j | ||
| 26 | Acetoin reductase | 01801 | 30 | 6.3 | 30.0 | 5.5 | 22.4 | 4 | 1.8 × 104 | 6.69j | |
| 28 | Metal dependent hydrolase | 01483 | 22 | 5.2 | 28.2 | 5.1 | 28.8 | 4 | 1.4 × 104 | 4.71 | |
| 32 | Lactoylglutathione lyase | 01881 | 33 | 5.1 | 20.5 | 4.9 | 15.3 | 5 | 4.4 × 104 | 50.63j | |
| Transport/binding proteins | 4 | Phosphoenolpyruvate: protein phosphotransferase | 00388 | 18 | 4.6 | 65.5 | 4.6 | 63.4 | 10 | 1.8 × 1011 | 0.53j |
| 17 | Multiple sugar binding transport ATP-binding transport protein MsmK | 00117 | 22 | 6.3 | 44.5 | 5.9 | 42.0 | 6 | 1.4 × 103 | 0.41 | |
| Translation | 1 | EF-G | 02018 | 28 | 4.9 | 70.5 | 4.8 | 76.7 | 15 | 2.2 × 1012 | 0.49j |
| 13 | EF-Tu | 00164 | 34 | 5.0 | 52.0 | 4.8 | 43.9 | 13 | 4.9 × 1011 | 0.66 | |
| 16 | SSU ribosomal protein S1P | 00334 | 19 | 5.1 | 48.1 | 5.0 | 43.7 | 7 | 1.6 × 105 | 0.22j | |
| 19 | EF-Ts | 01267 | 9 | 5.0 | 41.5 | 4.9 | 37.7 | 5 | 7.9 × 102 | 0.25j | |
| 21 | DNA-directed RNA polymerase alpha chain | 00821 | 27 | 4.8 | 40.1 | 4.8 | 34.6 | 7 | 6.4 × 105 | 0.03j | |
| 30 | Ribosome recycling factor | 01607 | 28 | 5.9 | 24.0 | 5.6 | 20.6 | 4 | 5.7 × 103 | 2.51 | |
| Unknown function | 5 | Hypothetical protein | 00557 | 22 | 5.8 | 65.8 | 5.7 | 67.5 | 7 | 1.8 × 107 | 4.06 |
| 27 | ORF 1749 | 01749 | 24 | 5.6 | 30.6 | 5.4 | 29.4 | 5 | 4.0 × 104 | 4.77j | |
Numbers refer to the proteins labeled in Fig. 2.
Putative functions for proteins were assigned on the basis of the presence of homologs in the database for S. mutans.
RMN, number assigned to each ORF in the database for S. mutans.
Percent amino acid coverage (amino acids in peptides observed in peptide mass fingerprint/total amino acids in translated gene sequence in the database for S. mutans).
Calculated from data shown in Fig. 2.
As given in the database for S. mutans.
Number of peptides in peptide mass fingerprint contributing to MOWSE score.
As calculated by MS-Fit software using peptide mass fingerprint data.
Ratio of IOD for each protein when extracted from S. mutans cultured at pH 5.2 or 7.0 (mean of three replicates for each growth condition).
Significantly up- or down-regulated (P < 0.05) as calculated using Student's t test.
The cell division proteins FtsZ and FtsA, the ORFs of which are located adjacently on the same contig, were both up-regulated during growth at low pH (Table 2), as were the stress proteins 60-kDa chaperonin and Mn superoxide dismutase. However, DnaK and trigger factor were down-regulated under these same conditions. Examination of the expression of proteins involved in central and intermediary metabolism showed that key glycolytic enzymes and expression of a number of proteins of intermediary metabolism increased. In addition to effects on enzymes of intermediary metabolism, expression of components of two independent transport systems, the multiple sugar transport ATP-binding protein MsmK and a component of a phosphoenolpyruvate:sugar phosphotransferase system (PTS), were down-regulated.
Key proteins involved in protein synthesis, i.e., EF-Tu, EF-Ts, EF-G, SSU ribosomal protein S1P, and the DNA-directed RNA polymerase alpha chain, were down-regulated under acidic conditions, while ribosome recycling factor was up-regulated. Two other modulated proteins (RMN00557 and RMN01749) were matched to ORFs in the S. mutans genomic database, but no putative functions were identifiable. RMN00557 exhibited sequence homology (p score = 2.89 × 10−17) with RMN00580, a lysozyme M1 precursor, while RMN01749 exhibited very low homology (p score = 8.1 × 10−2) with a Pyrococcus abyssii protein, SUA5.
Protein isoforms at low pH.
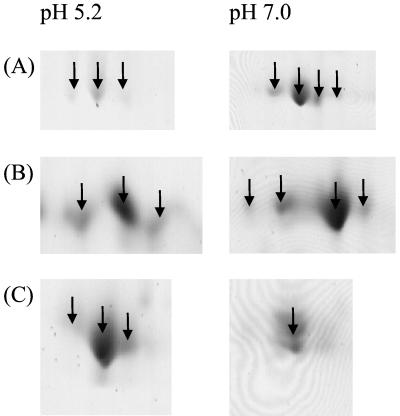
A number of S. mutans proteins, including EF-G, EF-Tu, enolase, phosphoglycerate kinase, glyceraldehyde-3-phosphate dehydrogenase, and lactate dehydrogenase (spots 1, 13, 15, 20, 22, and 23, respectively, in Fig. 2), were present as isoforms on the gels. Comparison of these data with those for pH 7.0-cultured S. mutans indicated that the expression of isoforms, in terms of both the number of isoforms present and their relative abundances, was altered by growth of the organism under the two different conditions. Figure 3 shows the expanded regions of the pH 7.0 and pH 5.2 2-D gels in which EF-G, EF-Tu, and the 60-kDa chaperonin are resolved. The number of isoforms of EF-G and EF-Tu decreased from four polypeptides detected in pH 7.0-grown cells to three forms detected in cells cultured at pH 5.2, and the relative abundance of each isoform was altered, as judged by the IOD. The 60-kDa chaperonin that was up-regulated in low-pH-grown cells (Table 2) existed as a single form of the protein when extracted from cultures adjusted to pH 7.0, but three isoforms were observed in the aciduric cultures.
FIG. 3.
Isoforms of representative S. mutans proteins differentially expressed following growth at low pH. Cellular proteins extracted from cells of S. mutans cultured at pH 5.2 or 7.0 were resolved by 2-D PAGE; relevant regions of gels are shown expanded. Proteins were identified following peptide mass fingerprinting and comparison with the S. mutans genomic database. (A) EF-G; (B) EF-Tu; (C) 60-kDa chaperonin.
DISCUSSION
In this study we have investigated the response of S. mutans to growth in a nutrient-rich medium adjusted initially to pH 5.2 or 7.0. Thirty proteins were modulated during growth under these conditions, but it should also be borne in mind that the pH of the medium was not constant during the growth experiments. However, it has been demonstrated that the initial pH determines the rate of glucose consumption and acidic end product formation (14), and the acid tolerance of S. mutans may be modified by the nature of the carbohydrate provided in pH challenge assays (6). The modulation of protein expression may also be strain specific, and it may also be that an organism growing in batch culture will express different proteins when grown as a biofilm (10, 19, 50). However, we clearly demonstrated that the S. mutans strain investigated is aciduric, as no appreciable lag period was observed when it was grown at pH 5.2. In addition an acid tolerance response was induced, since cells grown in medium initially at pH 5.2 were more resistant to killing at pH 4.0 than were cells grown in medium initially at pH 7.0. This study of the S. mutans proteins with modulated expression during growth at low pH has relevance to the pathogenesis of dental caries, especially as increased protein synthesis due to instantaneous acid shock is predominantly transient (28).
The growth rate in the acidic medium was reduced, which may explain the observed down-regulation of EF-G, EF-Ts, EF-Tu, and SSU ribosomal protein S1P; however, we found that FtsZ and FtsA, proteins essential for cell division (22), were up-regulated. FtsZ is a bacterial GTPase (20), and ftsZ and ftsA occur as consecutive ORFs of the same contig of the S. mutans genomic data and in S. pneumoniae. It is unclear why these proteins are up-regulated in cells growing at the reduced rate observed in this study.
The predominant S. mutans proteins visualized from pH 7.0-grown cells in the pI range of 4 to 7 were primarily enzymes involved in glycolysis, and many of these were up-regulated following growth at low pH. S. mutans is more resistant than other oral streptococci to the acid shock-induced inactivation of glycolytic enzymes (51), but our data indicate that an increase in the amounts of these proteins may also be an important factor for survival at low pH, perhaps resulting in an increase in ATP production and consequently increased proton extrusion via the H+-ATPase. In addition to the observed effects on glycolytic proteins, the expression of components msmK and dexB of the multiple sugar metabolism (41, 42) operon was also down-regulated at pH 5.2, as was the PTS enzyme I. This effect might explain the down-regulation of components of the multiple sugar metabolism operon, since it is regulated via the PTS as evident by the observation that mutants defective in PTS enzyme I were less efficient at transporting and utilizing raffinose (16). The PTS also exerts a control effect on sugar metabolism generally, facilitating the transport of available sugars to yield the greatest net gain in energy (52, 53).
The enzymes that form key branch points in pathways involved in the formation of end products of carbohydrate fermentation were differentially expressed. Acetoin reductase, which was up-regulated at low pH, is involved in the formation of acetoin from fermentable carbohydrates, and S. mutans produces up to 6 mM acetoin under aerobic conditions, although its production was increased in lactate dehydrogenase-negative mutants (30). Lactate dehydrogenase was also up-regulated at low pH, confirming the observations of Iwami et al. (31). Our data suggest the possibility that both lactate and acetoin production are likely to be increased as a result of carbohydrate fermentation by S. mutans at low pH.
Many studies have investigated the active efflux of protons via the H+-ATPase by S. mutans as a means of surviving at low pH; this membrane-associated system extrudes protons, regulating the pH of the cytoplasm (40). Further, it has been shown that as part of an adaptive response to low pH, S. mutans is able to increase H+-ATPase levels as a result of environmental acidification. Studies on the genes for the structural components of this complex indicate that the genes are organized in an ATPase operon and that the transcriptional start site does not alter as a function of pH (46). The subunits of the H+-ATPase were not identified among the 30 polypeptides differentially expressed by S. mutans. No change in the expression of the components of the H+-ATPase at low pH was detected, possibly because there is a hyperbolic relationship between ATPase levels and acid tolerance in glycolysis (5) and above a minimum pH (3.5), relatively small changes in ATPase levels were associated with significant changes in the acid tolerance of glycolysis. Therefore, in these acid-grown cells the acid-adaptive response had already occurred, since the adaptive response of S. mutans is rapid (48). The response of the H+-ATPase in S. mutans is different from that observed in Streptococcus oralis, where components of the ATPase system were up-regulated at low pH (55).
Expression of a number of general shock proteins, including the 60-kDa chaperonin, lactoylglutathione lyase (a homolog of the enzyme glyoxalase I), and superoxide dismutase, was up-regulated in S. mutans at low pH. Chaperonins are a class of proteins expressed in response to a range of environmental stresses, playing a role in refolding or degradation of denatured proteins, and it may be that the 60-kDa chaperonin (the GroEL homolog) is produced in increased amounts to counteract the effects of environmental acidification. S. mutans, unlike S. pneumoniae, possesses only a single superoxide dismutase gene, for the Mn type of the enzyme, which is involved in the response to oxidative stress (37), but it is the same Mn form of the superoxide dismutase that is also expressed in response to oxidative stress, and is a putative virulence determinant, in S. pneumoniae (56). Glyoxalase I is involved in the tolerance of oxidative stress in mammals (36), nematodes (47), and yeasts (1). The genes for glyoxalase I have been cloned from Salmonella and identified in sequences of several pathogenic microorganisms (12, 13). Although lactoylglutathione lyase was not identified as differentially expressed in S. oralis, another enzyme, NAD(P)H nitroreductase/dihydropteridine reductase, the gene for which is located upstream on the same contig as the lactoylglutathione lyase gene in S. pneumoniae, was up-regulated at low pH. The function of lactoylglutathione lyase has yet to be determined, but enzymes within this family of NADH oxidases have been shown to be produced as a response of S. mutans to oxidative stress (29). The heat shock protein DnaK is usually produced by S. mutans in the stationary phase and in response to thermal stress (32). Additionally, steady-state levels of both dnaK mRNA and the DnaK protein were increased in response to acid shock and were elevated in acid-adapted cells (33). In this study DnaK was down-regulated during growth at low pH. The previous data apparently conflict with our study, but they may reflect the differences between the responses of cells exposed instantaneously to low pH and those grown at low pH.
A number of proteins, including the 60-kDa chaperonin, were present as isoforms, and these were expressed in different amounts, in terms of both the number of isoforms and their relative amounts, under the two different growth conditions. We have previously made the same observations with S. oralis grown at low pH (55). The isoforms most likely arise as a result of posttranslational modifications, including phosphorylation, which have been described for eukaryotic and, more recently, prokaryotic systems. The nature of the modifications that give rise to the different isoforms are unknown and their biological relevance has yet to be clarified, but it is unlikely, given the relatively small differences in mass between the isoforms, that these arise solely as a result of processing by endogenous bacterial proteases.
We have used 2-D PAGE in conjunction with peptide mass fingerprinting to identify S. mutans proteins that have altered expression as a result of culture of the organism at pH 5.2. While 2-D technologies have the advantage that expression of a large number of proteins can be monitored simultaneously, not all proteins are detected, and it has been shown that some classes of proteins (for example, very large, low-abundance, or hydrophobic proteins) may not be detected. Other investigators have implicated other proteins in the acid stress response of S. mutans, including a GTP-binding protein, proteins involved in the synthesis of d-alanyl-lipoteichoic acid, an acid-inducible apyrimidinic-apurinic endonuclease, and Ffh, which may be involved in the maintenance of a functional membrane protein composition (3, 9, 25, 26). These proteins were not detected by us among those differentially expressed by S. mutans, perhaps because we monitored protein expression during growth and not during the immediate physiological response to acid stress conditions or because different methodologies may have a bias towards the detection of a particular subset of proteins. Nonetheless, we have shown that this proteomics-based approach has identified new target genes for further mutagenesis studies, which will be required to assess their physiological significance. A wealth of data has accumulated from conventional physiological and genetics studies, and this is now being extended from planktonic-phase organisms to the investigation of gene expression within biofilms of dental plaque bacteria (10, 50).
Acknowledgments
J. J. Ferretti (University of Oklahoma) kindly provided the S. mutans isolate used during this study. We thank the Streptococcus mutans Genome Sequencing Project and B. A. Roe, R. Y. Tian, H. G. Jia, Y. D. Qian, S. P. Lin, S. Li, S. Kenton, H. Lai, J. D. White, R. E. McLaughlin, M. McShan, D. Ajdic, and J. Ferretti for the genomic data so freely accessed.
The Pathological Society of Great Britain and Ireland is acknowledged for funding the Ph.D. Studentship that provided support for J. C. Wilkins throughout this research. The Streptococcus mutans Genome Sequencing Project is funded by a Public Health Service grant from the National Dental Institute.
REFERENCES
- 1.Aguilera, J., and J. A. Prieto. 2001. The Saccharomyces cerevisiae aldose reductase is implied in the metabolism of methylglyoxal in response to stress conditions. Curr. Genet. 39:273-283. [DOI] [PubMed] [Google Scholar]
- 2.Alam, S., S. R. Brailsford, S. Adams, C. Allison, E. Sheehy, L. Zoitopoulos, E. A. Kidd, and D. Beighton. 2000. Genotypic heterogeneity of Streptococcus oralis and distinct aciduric subpopulations in human dental plaque. Appl. Environ. Microbiol. 66:3330-3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baev, D., R. England, and H. K. Kuramitsu. 1999. Stress-induced membrane association of the Streptococcus mutans GTP-binding protein, an essential G protein, and investigation of its physiological role by utilizing an antisense RNA strategy. Infect. Immun. 67:4510-4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baev, D., S. H. Ohk, and H. K. Kuramitsu. 2000. Protein interactions of SGP, an essential Streptococcus mutans GTPase, revealed by biochemical and yeast two-hybrid system analyses. FEMS Microbiol. Lett. 184:149-153. [DOI] [PubMed] [Google Scholar]
- 5.Belli, W. A., and R. E. Marquis. 1991. Adaptation of Streptococcus mutans and Enterococcus hirae to acid stress in continuous culture. Appl. Environ. Microbiol. 57:1134-1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belli, W. A., and R. E. Marquis. 1994. Catabolite modification of acid tolerance of Streptococcus mutans GS-5. Oral Microbiol. Immunol. 9:29-34. [DOI] [PubMed] [Google Scholar]
- 7.Bowden, G. H., and I. R. Hamilton. 1998. Survival of oral bacteria. Crit. Rev. Oral Biol. Med. 9:54-85. [DOI] [PubMed] [Google Scholar]
- 8.Bowden, G. H. 1990. Microbiology of root surface caries in humans. J. Dent. Res. 69:1205-1210. [DOI] [PubMed] [Google Scholar]
- 9.Boyd, D. A., D. G. Cvitkovitch, A. S. Bleiweis, M. Y. Kiriukhin, D. V. Debabov, F. C. Neuhaus, and I. R. Hamilton. 2000. Defects in d-alanyl-lipoteichoic acid synthesis in Streptococcus mutans results in acid sensitivity. J. Bacteriol. 182:6055-6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burne, R. A., R. G. Quivey, Jr., and R. E. Marquis. 1999. Physiologic homeostasis and stress responses in oral biofilms. Methods Enzymol. 310:441-460. [DOI] [PubMed] [Google Scholar]
- 11.Chia, J. S., Y. Y. Lee, P. T. Huang, and J. Y. Chen. 2001. Identification of stress-responsive genes in Streptococcus mutans by differential display reverse transcription-PCR. Infect. Immun. 69:2492-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clugston, S. L., E. Daub, R. Kinach, D. Miedema, J. F. Barnard, and J. F. Honek. 1997. Isolation and sequencing of a gene coding for glyoxalase I activity from Salmonella typhimurium and comparison with other glyoxalase I sequences. Gene 186:103-111. [DOI] [PubMed] [Google Scholar]
- 13.Clugston, S. L., and J. F. Honek. 2000. Identification of sequences encoding the detoxification metalloisomerase glyoxalase I in microbial genomes from several pathogenic organisms. J. Mol. Evol. 50:491-495. [DOI] [PubMed] [Google Scholar]
- 14.Concha, M. L., A. Castillo, J. Liebana, J. Gutierrez, and A. Garcia-Mendoza. 1996. Initial pH as a determining factor of glucose consumption and lactic and acetic acid production in oral streptococci. Microbios 87:207-216. [PubMed] [Google Scholar]
- 15.Crowley, P. J., L. J. Brady, S. M. Michalek, and A. S. Bleiweis. 1999. Virulence of a spaP mutant of Streptococcus mutans in a gnotobiotic rat model. Infect. Immun. 67:1201-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cvitkovitch, D. G., D. A. Boyd, and I. R. Hamilton. 1995. Regulation of sugar transport via the multiple sugar metabolism operon of Streptococcus mutans by the phosphoenolpyruvate phosphotransferase system. J. Bacteriol. 177:5704-5706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cvitkovitch, D. G., J. A. Gutierrez, J. Behari, P. J. Youngman, J. E. Wetz, P. J. Crowley, J. D. Hillman, L. J. Brady, and A. S. Bleiweis. 2000. Tn917-lac mutagenesis of Streptococcus mutans to identify environmentally regulated genes. FEMS Microbiol. Lett. 182:149-154. [DOI] [PubMed] [Google Scholar]
- 18.Dashper, S. G., and E. C. Reynolds. 1992. pH regulation by Streptococcus mutans. J. Dent. Res. 71:1159-1165. [DOI] [PubMed] [Google Scholar]
- 19.Davey, M. E., and G. A. O'Toole. 2000. Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 64:847-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Boer, P., R. Crossley, and L. Rothfield. 1992. The essential bacterial cell-division protein FtsZ is a GTPase. Nature 359:254-256. [DOI] [PubMed] [Google Scholar]
- 21.de Soet, J. J., B. Nyvad, and M. Kilian. 2000. Strain-related acid production by oral streptococci. Caries Res. 34:486-490. [DOI] [PubMed] [Google Scholar]
- 22.Feucht, A., I. Lucet, M. D. Yudkin, and J. Errington. 2001. Cytological and biochemical characterization of the FtsA cell division protein of Bacillus subtilis. Mol. Microbiol. 40:115-125. [DOI] [PubMed] [Google Scholar]
- 23.Giard, J.-C., A. Hartke, S. Flahaut, P. Boutibonnes, and Y. Auffray. 1997. Glucose starvation response in Enterococcus faecalis JH2-2: survival and protein analysis. Res. Microbiol. 148:27-35. [DOI] [PubMed] [Google Scholar]
- 24.Gutierrez, J. A., P. J. Crowley, D. P. Brown, J. D. Hillman, P. Youngman, and A. S. Bleiweis. 1996. Insertional mutagenesis and recovery of interrupted genes of Streptococcus mutans by using transposon Tn917: preliminary characterization of mutants displaying acid sensitivity and nutritional requirements. J. Bacteriol. 178:4166-4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gutierrez, J. A., P. J. Crowley, D. G. Cvitkovitch, L. J. Brady, I. R. Hamilton, J. D. Hillman, and A. S. Bleiweis. 1999. Streptococcus mutans ffh, a gene encoding a homologue of the 54 kDa subunit of the signal recognition particle, is involved in resistance to acid stress. Microbiology 145:357-366. [DOI] [PubMed] [Google Scholar]
- 26.Hahn, K., R. C. Faustoferri, and R. G. Quivey. 1999. Induction of an AP endonuclease activity in Streptococcus mutans during growth at low pH. Mol. Microbiol. 31:1489-1498. [DOI] [PubMed] [Google Scholar]
- 27.Hamada, S., and H. D. Slade. 1980. Biology, immunology, and cariogenicity of Streptococcus mutans. Microbiol. Rev. 44:331-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hamilton, I. R., and G. Svensäter. 1998. Acid-regulated proteins induced by Streptococcus mutans and other oral bacteria during acid shock. Oral Microbiol. Immunol. 13:292-300. [DOI] [PubMed] [Google Scholar]
- 29.Higuchi, M., Y. Yamamoto, L. B. Poole, M. Shimada, Y. Sato, N. Takahashi, and Y. Kamio. 1999. Functions of two types of NADH oxidases in energy metabolism and oxidative stress of Streptococcus mutans. J. Bacteriol. 181:5940-5947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hillman, J. D., S. W. Andrews, and A. L. Dzuback. 1987. Acetoin production by wild-type strains and a lactate dehydrogenase-deficient mutant of Streptococcus mutans. Infect. Immun. 55:1399-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iwami, Y., K. Abbe, S. Takahashi-Abbe, and T. Yamada. 1992. Acid production by streptococci growing at low pH in a chemostat under anaerobic conditions. Oral Microbiol. Immunol. 7:304-308. [DOI] [PubMed] [Google Scholar]
- 32.Jayaraman, G. C., and R. A. Burne. 1995. DnaK expression in response to heat shock of Streptococcus mutans. FEMS Microbiol. Lett. 131:255-261. [DOI] [PubMed] [Google Scholar]
- 33.Jayaraman, G. C., J. E. Penders, and R. A. Burne. 1997. Transcriptional analysis of the Streptococcus mutans hrcA, grpE and dnaK genes and regulation of expression in response to heat shock and environmental acidification. Mol. Microbiol. 25:329-341. [DOI] [PubMed] [Google Scholar]
- 34.Kuramitsu, H. K. 1993. Virulence factors of mutans streptococci: role of molecular genetics. Crit. Rev. Oral Biol. Med. 4:159-176. [DOI] [PubMed] [Google Scholar]
- 35.Loesche, W. J. 1986. Role of Streptococcus mutans in human dental decay. Microbiol. Rev. 50:353-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mitsumoto, A., K. R. Kim, G. Oshima, M. Kunimoto, K. Okawa, A. Iwamatsu, and Y. Nakagawa. 1999. Glyoxalase I is a novel nitric-oxide-responsive protein. Biochem. J. 344:837-844. [PMC free article] [PubMed] [Google Scholar]
- 37.Nakayama, K. 1992. Nucleotide sequence of Streptococcus mutans superoxide dismutase gene and isolation of insertion mutants. J. Bacteriol. 174:4928-4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neuhoff, V., N. Arold, D. Taube, and W. Ehrardt. 1988. Improved staining of proteins in polyacrylamide gels including isoelectric-focusing gels with clear background at nanogram sensitivity using Coomassie brilliant blue G250 and R250. Electrophoresis 9:255-262. [DOI] [PubMed] [Google Scholar]
- 39.Pappin, D. J. C., P. Hojrup, and A. J. Bleasby. 1993. Rapid identification of proteins by peptide-mass fingerprinting. Curr. Biol. 3:327-332. [DOI] [PubMed] [Google Scholar]
- 40.Quivey, R. G., W. L. Kuhnert, and K. Hahn. 2000. Adaptation of oral streptococci to low pH. Adv. Microb. Physiol. 42:239-274. [DOI] [PubMed] [Google Scholar]
- 41.Russell, R. R., J. Aduse-Opoku, I. C. Sutcliffe, L. Tao, and J. J. Ferretti. 1992. A binding protein-dependent transport system in Streptococcus mutans responsible for multiple sugar metabolism. J. Biol. Chem. 267:4631-4637. [PubMed] [Google Scholar]
- 42.Russell, R. R., and J. J. Ferretti. 1990. Nucleotide sequence of the dextran glucosidase (dexB) gene of Streptococcus mutans. J. Gen. Microbiol. 136:803-810. [DOI] [PubMed] [Google Scholar]
- 43.Sansone, C., J. van Houte, K. Joshipura, R. Kent, and H. C. Margolis. 1993. The association of mutans streptococci and non-mutans streptococci capable of acidogenesis at a low pH with dental caries on enamel and root surfaces. J. Dent. Res. 72:508-516. [DOI] [PubMed] [Google Scholar]
- 44.Sato, T., J. Wu, and H. Kuramitsu. 1998. The sgp gene modulates stress responses of Streptococcus mutans: utilization of an antisense RNA strategy to investigate essential gene functions. FEMS Microbiol. Lett. 159:241-245. [DOI] [PubMed] [Google Scholar]
- 45.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal. Chem. 68:850-858. [DOI] [PubMed] [Google Scholar]
- 46.Smith, A. J., R. G. Quivey, and R. C. Faustoferri. 1996. Cloning and nucleotide sequence analysis of the Streptococcus mutans membrane-bound, proton-translocating ATPase operon. Gene 183:87-96. [DOI] [PubMed] [Google Scholar]
- 47.Sommer, A., P. Fischer, K. Krause, K. Boettcher, P. P. M. Brophy, R. D. Walter, and E. Liebau. 2001. A stress-responsive glyoxalase I from the parasitic nematode Onchocerca volvulus. Biochem. J. 353:445-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Svensäter, G., U. B. Larsson, E. C. Greif, D. G. Cvitkovitch, and I. R. Hamilton. 1997. Acid tolerance response and survival by oral bacteria. Oral Microbiol. Immunol. 12:266-273. [DOI] [PubMed] [Google Scholar]
- 49.Svensäter, G., B. Sjogreen, and I. R. Hamilton. 2000. Multiple stress responses in Streptococcus mutans and the induction of general and stress-specific proteins. Microbiology 146:107-117. [DOI] [PubMed] [Google Scholar]
- 50.Svensäter, G., J. Welin, J. C. Wilkins, D. Beighton, and I. R. Hamilton. 2001. Protein expression by planktonic and biofilm cells of Streptococcus mutans. FEMS Microbiol. Lett. 205:139-146. [DOI] [PubMed] [Google Scholar]
- 51.Takahashi, N., M. Horiuchi, and T. Yamada. 1997. Effects of acidification on growth and glycolysis of Streptococcus sanguis and Streptococcus mutans. Oral Microbiol. Immunol. 12:72-76. [DOI] [PubMed] [Google Scholar]
- 52.Vadeboncoeur, C., and M. Pelletier. 1997. The phosphoenolpyruvate:sugar phosphotransferase system of oral streptococci and its role in the control of sugar metabolism. FEMS Microbiol. Rev. 19:187-207. [DOI] [PubMed] [Google Scholar]
- 53.Vadeboncoeur, C., S. St. Martin, D. Brochu, and I. R. Hamilton. 1991. Effect of growth rate and pH on intracellular levels and activities of the components of the phosphoenolpyruvate:sugar phosphotransferase system in Streptococcus mutans Ingbritt. Infect. Immun. 59:900-906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van Houte, J. 1994. Role of micro-organisms in caries etiology. J. Dent. Res. 73:672-681. [DOI] [PubMed] [Google Scholar]
- 55.Wilkins, J. C., K. A. Homer, and D. Beighton. 2001. Altered protein expression of Streptococcus oralis cultured at low pH revealed by two-dimensional gel electrophoresis. Appl. Environ. Microbiol. 67:3396-3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yesilkaya, H., A. Kadioglu, N. Gingles, J. E. Alexander, T. J. Mitchell, and P. W. Paton. 2000. Role of manganese-containing superoxide dismutase in oxidative stress and virulence of Streptococcus pneumoniae. Infect. Immun. 68:2819-2826. [DOI] [PMC free article] [PubMed] [Google Scholar]