Abstract
The search for promoters has largely been confined to sequences upstream of open reading frames (ORFs) or stable RNA genes. Here we used a cloning approach to discover other potential promoters in Escherichia coli. Chromosomal fragments of ∼160 bp were fused to a promoterless lacZ reporter gene on a multi-copy plasmid. Eight clones were deliberately selected for high activity and 105 clones were selected at random. All eight of the high-activity clones carried promoters that were located upstream of an ORF. Among the randomly-selected clones, 56 had significantly elevated activity. Of these, 7 had inserts which also mapped upstream of an ORF, while 49 mapped within or downstream of ORFs. Surprisingly, the eight promoters selected for high activity matched the canonical σ70 −35 and −10 sequences no better than sequences from the randomly-selected clones. For six of the nine most active sequences with orientations opposite to that of the ORF, chromosomal expression was detected by RT–PCR, but defined transcripts were not detected by northern analysis. Our results indicate that the E.coli chromosome carries numerous −35 and −10 sequences with weak promoter activity but that most are not productively expressed because other features needed to enhance promoter activity and transcript stability are absent.
INTRODUCTION
Many Escherichia coli promoters are difficult to recognize by sequence alone. Numerous studies have shown that the optimal E.coli σ70-dependent RNA polymerase-binding site contains −35 (TTGACA) and −10 (TATAAT) consensus hexamers spaced 17 bp apart (1–6), though much deviation from this consensus is encountered with bona fide promoters, especially if specific DNA-binding proteins activate transcription from the promoters. Historically, most promoters were sought upstream of open reading frames (ORFs) or stable RNA genes, but their presence elsewhere has not been systematically investigated. In general, promoters were not expected to be found within ORFs though some internal promoters do exist (7–10).
However, the possibility of promoters not associated with ORFs or even contained within ORFs needs to be considered. Based on the degeneracy of the RNA polymerase-binding site in bona fide promoters, many more than 4000 RNA polymerase-binding sites are predicted to be encoded by the E.coli genome (11) some of which might act as promoters. Furthermore, whole genome transcription analyses indicate that >3000 genes show expression from the antisense strand (12). In addition, recent screens for small, noncoding RNAs based on inter-species sequence conservation have indicated that >60 transcripts are expressed from intergenic regions (13–15). These findings led us to consider whether promoters also exist at sites other than the sequences immediately upstream of ORFs or stable RNA genes.
We therefore undertook a direct experimental approach for identifying chromosomal sequences with promoter activity. Random fragments of the E.coli chromosome were cloned upstream of a promoterless lacZ gene on a plasmid, and the β-galactosidase activities of these constructs were assayed. The inserts were sequenced to determine their locations on the chromosome. Many sequences derived from within ORFs showed promoter activity in our multi-copy tester plasmid; a significant fraction with strong activity. The promoter activities of these sequences, like those of bona fide promoters, correlated with homology to optimal −35 and −10 sequences. However, while transcripts expressed from the corresponding regions of the chromosome were detected by RT–PCR, we did not observe defined transcripts by northern analysis indicating that these promoters do not give rise to stable transcripts. We show that the activity of a potential promoter is enhanced by the insertion of a tyrT promoter UP element or lacZ translational signals and suggest that transcripts from many potential promoters are not observed because of low expression and message instability.
MATERIALS AND METHODS
Promoter expression library
E.coli DNA was partially digested with Sau3A and Tsp509I and separated by gel electrophoresis. Fragments of ∼160 bp were purified using the DNA Purification System (Promega, Madison, WI). The fragments were then ligated upstream of the promoterless lacZ gene in plasmid pRS551 (16), digested previously with EcoRI and BamHI and treated with alkaline phosphatase. DH5α cells (Life Technologies, Rockville, MD) transformed by the plasmid pool were selected as intensely blue colonies on Luria broth (LB) plates containing 100 µg/ml of ampicillin and 40 µg/ml of the chromogen, 5-bromo-4-chloro-3-indolyl β-d-galactopyranoside (Xgal) or isolated at random from LB plates containing 100 µg/ml of ampicillin. The inserts were sequenced on an ABI 310 Prism Analyzer using the Biosystems DNA sequencing kit (Warrington, England) with primers: 5′-GCC ATA AAC TGC CAG GAA TTG GGG-3′ and 5′-GCG GAA CTG GCG GCT GTG GG-3′ corresponding to upstream and downstream sequences of the plasmid pRS551 site of insertion.
β-Galactosidase assays
Plasmid- or prophage-containing cells were grown overnight, diluted 1:1000 and grown to A600 = 0.1–0.2 in LB at 32°C. Most expression under these growth conditions is likely to involve σ70-dependent RNA polymerase. β-Galactosidase activities were measured (17) in duplicate at least twice, with SDs of <10%, and are expressed in Miller units (MU).
RT–PCR
For total RNA used in RT–PCR analysis, the wild-type MG1655 strain was grown in LB at 37°C to A600 ≈ 0.46, and RNA was isolated by acid hot-phenol extraction (18). Residual genomic DNA was removed with TURBO DNA-free DNase (Ambion Inc., Austin, TX). The absence of DNA was verified by PCR using 1 µg of total RNA as the template. cDNA synthesis was performed by using 2 µg of total RNA together with 1 µl of 10 mM dNTP Mix (Invitrogen Corp., Carlsbad, CA) and 1 µl of the specific (reverse) primers (2 µM concentration) listed in Supplementary Table S1 in a final volume of 15 µl. The samples were heated for 5 min at 65°C and then placed on ice for 5 min. Subsequently, 4 µl of 5× first-strand buffer and 1 µl of 0.1 M DTT were added, the reaction mixture was incubated for 2 min at 55°C, and 0.5 µl of SuperScript III RT (Invitrogen Corp.) was added to the RTase plus samples. The reaction mixture was incubated for 30 min at 50°C and then for 20 min at 55°C and for 15 min at 70°C. PCRs were carried out using 0.25 µM forward and reverse primers listed in Supplementary Table S1, 1 µl of the cDNA products, and the AccuPrime SuperMix I (Invitrogen Corp.) in a final volume of 20 µl. The cycling condition was as follows: 95°C for 2 min, 30 cycles of (95°C for 30 s, 52°C for 15 s, 68°C for 15 s), and 68°C for 1 min. The RT–PCR products were stored at 4°C and then analyzed by electrophoresis on 2% agarose gels using UltraPure Agarose-1000 (Invitrogen Corp.) and ethidium bromide staining.
Northern analysis
Total RNA isolated from MG1655 and used for RT–PCR analysis was separated on 1% agarose gels alongside RNA Millennium size markers (Ambion Inc.). The RNA was transferred to NYTRAN nylon transfer membranes (Schleicher and Schuell Inc., Keene NH) by capillary action. Membranes were UV-crosslinked and probed with 32P-labeled oligonucleotide probes (listed in Supplementary Table S1) in ULTRAhyb-Oligo buffer (Ambion Inc.) at 45°C. The membranes were then washed as described in the manual for NYTRAN nylon transfer membranes. The marker lanes were detected by probing with 32P-labeled Millennium markers.
Construction of specific lysogens
To construct the longer antisense yfjN-lacZ transcriptional and translational fusion plasmids, the yfjN gene fragment of E.coli MG1655 was amplified by PCR using primers yfjN-Bam (5′-TTT GGA TCC GGG GAC GGT TCG ACT ACA ATT CAA TAT CTC-3′) and yfjN-Eco (5′-ATA GAA TTC CAC ACT ACG GTA TGA GCG-3′). The fragment was then digested with BamHI and EcoRI and cloned into the corresponding sites of plasmids pRS551 and pRS552. The term ‘antisense yfjN-lacZ’ indicates that the antisense message of the yfjN ORF is being monitored by lacZ.
To make single-copy fusions, we recombined each construct with λRS45 (16) and integrated the phage into the chromosome of the ΔlacZ strain DJ480 (19). Multiple lysogens were selected and assayed for β-galactosidase activity. For most constructs we found presumptive single and multiple lysogens which differed from one another in activity by factors of 2 and 3. Those with the lowest activity were chosen and many were verified to contain single prophage by PCR (20).
Primer extension analysis
For total RNA used in primer extension analysis, the host strain DJ480, DJ480 harboring the antisense yfjN-lacZ fusion prophage, and DJ480 harboring the antisense yfjN-lacZ fusion plasmids were grown in LB at 37°C to A600 ≈ 0.4. Kanamycin (20 µg/ml) was added when appropriate. RNA was isolated by acid hot-phenol extraction (18). The reverse transcription reactions were carried out using 100 U of SuperScript II RT (Invitrogen Corp.) and oligonucleotide yfjN-F4 (5′-CAC TGG AGC CAA TCG TTC TCT GGG CCA AG-3′) labeled at the 5′ end with 32P and 30 µg of total RNA from strains without a plasmid and 5 µg of total RNA from strains with a plasmid. The samples were incubated for 1 h at 42°C. The cDNA products were fractionated in 8% polyacrylamide-urea gels together with a sequencing ladder obtained using labeled YfjN-F4 and the Thermo Sequenase Cycle Sequencing Kit (USB; Cleveland, OH). The template for the sequencing reaction was a DNA fragment generated by PCR using yfjN-5U (5′-CCG GTA GAC GAT CCT GCC CTA TAG-3′) and yfjN-Eco as primers and E.coli K-12 genomic DNA.
Promoter homology scores
Homology scores were calculated using the pftools 2.2 program (Swiss Bioinformatics Institute site: http://www.isrec.isb-sib.ch/ftp-server/pftools/) (5,13).
RESULTS
Cloning strategy
To detect regions of the E.coli chromosome with promoter activity, total genomic DNA was partially digested to give fragments of ∼160 bp in length and cloned into the plasmid pRS551 (16) to generate lacZ transcriptional fusions. These plasmids were used to transform the E.coli strain DH5α. One set of transformants was selected on plates containing Xgal, a chromogenic indicator of β-galactosidase activity, and eight colonies exhibiting the most intense color were chosen. Intensely colored colonies represented ∼0.5% of the population. Another 105 colonies were selected at random in the absence of the chromogen. For both the colonies selected for high activity and the colonies selected at random, the levels of β-galactosidase activity were determined in early log phase cultures in LB broth and compared with the activity of a strain carrying the control plasmid lacking an insert [which exhibited 3.6 ± 0.16 MU (17) of β-galactosidase specific activity in 24 assays performed in duplicate]. Each of the inserts was sequenced and its position on the chromosome was identified (21).
Fragments selected for high activity
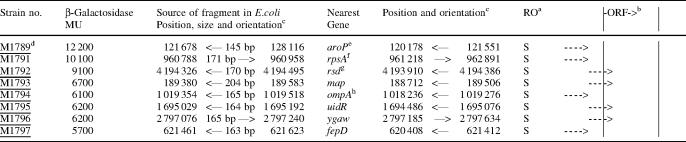
The eight clones that were selected on the basis of high activity are listed in Table 1. The average size of the corresponding inserts was 168 ± 17 bp, and the average β-galactosidase activity was 7800 MU. All of the sequences are <200 bp upstream of, and oriented in the same direction as, the nearest ORF (Table 1), except for rpsA where the insert is >260 bp upstream (Table 1, footnotes d and f). In the four cases where the 5′ ends of the cognate mRNAs have been mapped, the inserts of the strains selected for high promoter activity contain bona fide promoters: M1789 contains the P1 promoter of aroP (22,23); M1791 contains the P1 promoter of rpsA (24); M1793 contains the strong promoter for map which lies within 70 bp of the ORF (25); and M1797 contains the P1 promoter of fepD (26). We infer that all of the eight inserts selected on the basis of high activity contain bona fide promoters.
Table 1.
Activities and genomic locations of eight fragments selected for high activity
aRO refers to the relative orientation of fragment and the nearest gene. O = opposite, S = same.
bOrientation of potential promoter relative to the ORF.
cArrows indicate orientations of promoter or nearest gene. Forward arrows represent the clockwise or Watson strand and reverse arrows represent the counterclockwise or Crick strand. Numbers refer to chromosomal positions (21).
dUnderlined strain numbers indicate those with ‘immediately upstream’ promoters, arbitrarily defined as having the 3′ end of the sequence no farther than 200 bp upstream of the ORF and the 5′ end no closer than 50 bp from the ORF, assuming that a minimal promoter plus leader would be ∼50 bp long.
eThe sequence ∼80 bp upstream of the aroP promoter in our strain and in MG1655 (21) differs from the strain used by Pittard and coworkers (22,23). The sequence in our strain is TTGAGAGGGGTTGAGGCTGAGCTTTACA which matches (in bold) the optimal hexamers in 8 of 12 positions (separated by 16 bp), whereas the sequence in their strain is TACGGGGATCTGTTGAGGCTGAGCTTTACA which matches in only 5 positions.
fThe insert upstream of rpsA does not fit the definition of ‘immediately upstream’ but contains the known transcription start site of rpsA at 960 940 (24).
gOnly the P2 promoter of rsd is contained in the fragment.
hA possible alternative ORF start codon is GTG at 1 019 345.
Fragments selected at random
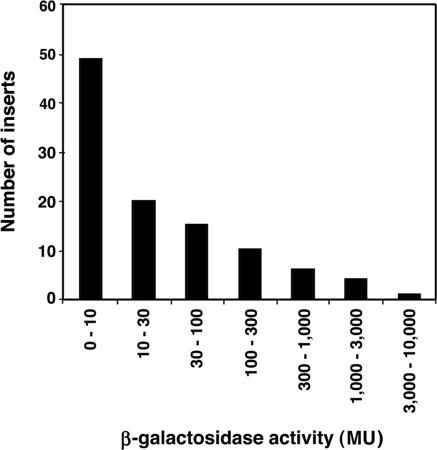
The 105 clones isolated at random were shown to have single inserts with an average length of 163 ± 24 bp, statistically the same size as the inserts present in the clones selected for high activity. The β-galactosidase activities of the strains selected at random varied in activity from 1.6 to 3220 MU and their distribution is shown in Figure 1. The average activity of the eight most active random clones (1600 MU) was nearly 5-fold lower than the average activity of the clones selected on the basis of high activity (7800 MU).
Figure 1.
The distribution of promoter (β-galactosidase) activities of 105 random fragments of E.coli as assayed in the promoterless lacZ tester plasmid, pRS551. We defined a sequence as having potential promoter activity when the β-galactosidase activity of the strain was increased by 3-fold or more over that of the strain with the parental plasmid pRS551 (i.e. to >10 MU). We found that 56 (53%) of the inserts (tested in only one orientation) exhibited promoter activity by this criterion. However, some of the inserts may contain more than one promoter. Applying the Poisson distibution, P(0) = e−λ, where P(0) is the probability of finding no activity (i.e. the null class of plasmids expressing 0–10 MU = 47%), and solving for λ (the average number of potential promoters per insert), we calculate there are on average 0.76 promoters per fragment (in one of the two possible orientations). We therefore conclude that there are ∼1.52 (twice 0.76) promoters per 163 bp or one full promoter in either direction per ∼107 bp.
We defined a sequence as having ‘potential promoter’ activity when it increased the β-galactosidase activity of the plasmid by 3-fold over that of the parental plasmid pRS551, i.e, to >10 MU. We found that 56 (53%) of the inserts from the random clones carried at least one such potential promoter. Given the two possible orientations of each fragment, this would correspond to one potential promoter per ∼107 bp or the ∼40 000 potential promoters per chromosome (for calculations see legend to Figure 1).
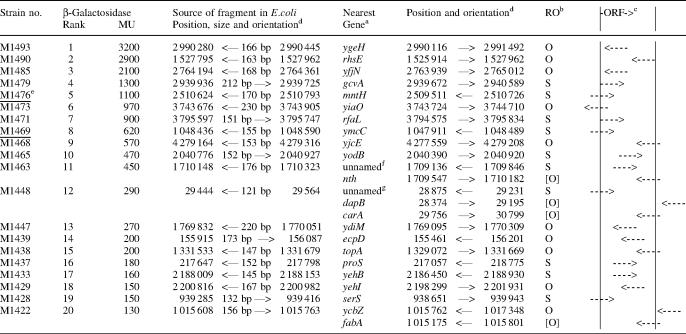
The mapping of the pRS551 inserts revealed that the potential promoters are apparently randomly distributed on the chromosome. Some properties of the 20 most active fragments are presented in Table 2. Only 7 of the 56 sequences with promoter activity (13%) were located immediately upstream of, and in the same orientation as, an ORF such that they might contain a bona fide promoter. The remaining 49 fragments with promoter activity >10 MU correspond to internal segments of ORFs or extend downstream from or into ORF sequences. About half of these sequences are oriented in the same direction as the ORF and conceivably could serve as internal promoters for a downstream ORF. Those potential promoters oriented in the opposite direction of the ORF, if functional in situ, might act as convergent promoters to modulate the activity of the bona fide promoter for the ORF or might direct the synthesis of antisense RNAs that could regulate mRNA stability or translation or might have no physiological consequences.
Table 2.
Activities and genomic locations of 20 most active fragments selected at random
aIn some cases, alternate possibilities are listed.
bRO refers to the relative orientation of fragment and the nearest gene. O = opposite, S = same. Alternate possibilities are indicated in brackets.
cOrientation of potential promoter relative to the ORF.
dArrows indicate orientations of promoter or nearest gene. Forward arrows represent the clockwise or Watson strand and reverse arrows represent the counterclockwise or Crick strand. Numbers refer to chromosomal positions (21).
eUnderlined strain numbers indicate those with ‘immediately upstream’ promoters, arbitrarily defined as having the 3′ end of the sequence no farther than 200 bp upstream of the ORF and the 5′ end no closer than 50 bp from the ORF, assuming that a minimal promoter plus leader would be ∼50 bp long.
fInsert is 302 bp upstream of an unannotated ORF of 237 codons.
gInsert is 213 bp upstream of an unannotated ORF of 119 codons.
Expression from potential promoters
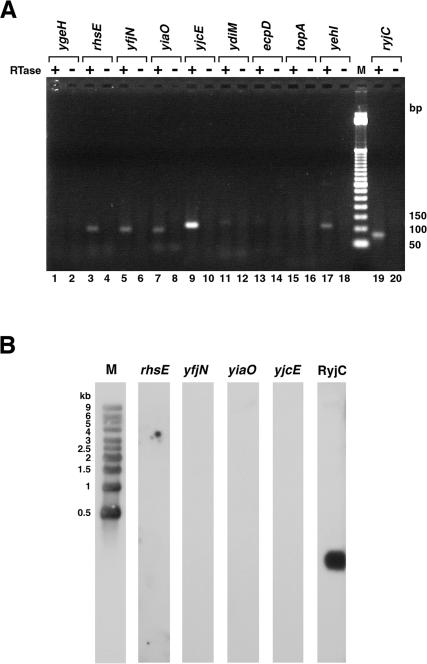
To determine whether the potential promoters within ORFs are transcriptionally active on the chromosome, we carried out RT–PCR and northern analyses on total RNA isolated from exponentially-growing MG1655 cells and probed for transcripts that could be expressed from the potential promoters. We chose to probe for transcripts from the potential promoters within the presumptive ORFs of ygeH, rhsE, yfjN, yiaO, yjcE, ydiM, ecpD, topA and yehI, since they showed the highest β-galactosidase activities when fused to the lacZ reporter gene and were oriented opposite to the ORF so that we could readily distinguish such antisense transcripts from transcripts expressed from the normal ORF promoter. For five of the loci (rhsE, yfjN, yiaO, yjcE and yehI), we detected clear RT–PCR products of the expected size, and for another locus (ydiM) a faint signal was observed (Figure 2A). No product was seen for three of the regions (ygeH, ecpD and topA). These results showed that antisense transcripts were in fact expressed for some of the regions for which we detected promoter activity. However, the RT–PCR signal did not necessarily correlate with the amount of β-galactosidase activity measured for the fusions; no expression was observed in the ygeH region, which showed high expression when fused to lacZ, and clear expression was detected for the yehI region, which showed low expression when fused to lacZ.
Figure 2.
Detection of antisense transcripts by RT–PCR and northern analysis. (A) RT–PCR was performed with primer sets listed in Supplementary Table S1 and with (plus) and without (minus) RT. The products were analyzed by electrophoresis on 2% agarose gels. Lanes 1 and 2, ygeH antisense transcript (expected size, 124 bp); lanes 3 and 4, rhsE antisense transcript (expected size, 95 bp); lanes 5 and 6, yfjN antisense transcript (expected size, 92 bp); lanes 7 and 8, yiaO antisense transcript (expected size, 87 bp); lanes 9 and 10, yjcE antisense transcript (expected size, 103 bp); lanes 11 and 12, ydiM antisense transcript (expected size, 114 bp); lanes 13 and 14, ecpD antisense transcript (expected size, 119 bp); lanes 15 and 16, topA antisense transcript (expected size, 101 bp); lanes 17 and 18, yehI antisense transcript (expected size, 108 bp); lane M for 50 bp ladder size marker and lanes 19 and 20, RyjC RNA (expected size, 77 bp). The lack of correlation between the RT–PCR signal and β-galactosidase activity may be due to differences in the hybridization efficiencies for the primer pairs used. (B) Total RNA separated on 1% agarose gels and transferred to nylon membranes was probed with primers to the antisense strands of rhsE, yfjN, yiaO and yjcE as well as to the 77 nt antisense RNA RyjC. RNA molecular weight markers were run with each set of samples for direct estimation of RNA transcript length, but the RNA marker lane for only one of the panels is shown.
We next tried to detect distinct RNA transcripts from these nine regions by northern analysis. The conditions used for the northern analysis were similar to conditions successfully used to detect the RyjC antisense RNA (27). However we were not able to detect the expected antisense transcripts (Figure 2B and data not shown). The failure to detect significant expression for ygeH, ecpD and topA by RT–PCR and the lack of distinct transcripts for rhsE, yfjN, yiaO, yjcE, yehI and ydiM by northern analysis suggested that these potential promoters have little or no activity when in their normal chromosomal location and/or that the corresponding transcripts are very unstable under the assay conditions and/or the length of the RNAs is extremely heterogeneous.
Increased expression from the potential promoter internal to yfjN
To evaluate possible explanations for the inability to detect distinct RNAs by northern analysis, we examined transcription from the potential promoter antisense to the yfjN ORF in more detail. One explanation for the low transcript levels could be ‘promoter occlusion’ (28), whereby expression of the antisense promoter internal to yfjN would be silenced owing to interference by RNA polymerase transcribing from the bona fide yfjN promoter. However, we found expression of the yfjN transcript to be low (data not shown) suggesting that transcriptional interference from the yfjN promoter did not contribute to the low expression from the potential antisense promoter. In addition, strong simultaneous expression of both an mRNA and the antisense transcript has been observed for probable antisense RNA regulators (18) suggesting that transcription across both strands of DNA does not necessarily lead to promoter occlusion and gene silencing.
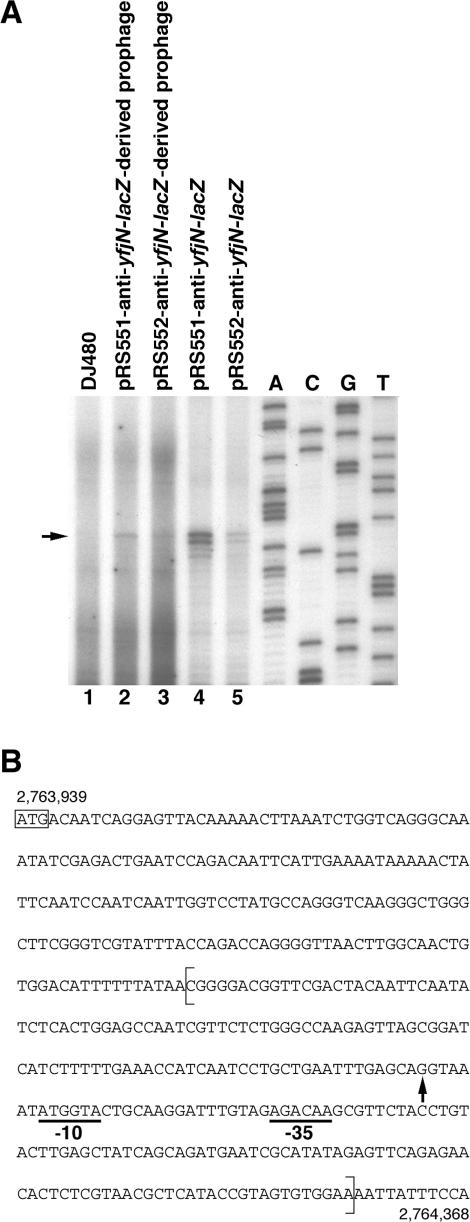
The DNA context of the putative promoter also might influence expression. Therefore, we compared the activity of the potential antisense promoter internal to the yfjN promoter in three situations: (i) its normal chromosomal context, (ii) fused to lacZ on a multi-copy plasmid or (iii) fused to lacZ on a single-copy λ prophage integrated at attλ. To do this, a 231 bp fragment containing the potential promoter internal and antisense to yfjN was cloned upstream of lacZ in the transcriptional fusion plasmid pRS551 (generating pRS551-anti-yfjN-lacZ). The lacZ-deletion strain DJ480 (containing the normal yfjN chromosomal copy) carrying the plasmid or a single prophage copy (integrated at attλ) derived from the plasmid was grown to exponential phase, and expression from the endogenous, plasmid-located and prophage-located potential antisense promoter was compared in primer extension assays (Figure 3). A strong transcription initiation signal was detected from the fragment on the plasmid, and the start site of the transcript was mapped downstream of the predicted −10 and −35 sequences (Figure 3A, lane 4 and Figure 3B). The transcription initiation signal for the strain with the prophage was very faint (lane 2) compared with the signal for the strain with the plasmid, even though 6-fold more total RNA was used in the primer extension assay. The ∼30-fold difference in signal between the plasmid strain and the prophage strain is somewhat larger than the difference expected from an ∼20-copy pRS551 plasmid (Robert Simons, personal communication) compared with a single-copy prophage. In addition, no signal was detected in the absence of the prophage or the plasmid (lane 1), indicating that the normal chromosomal copy of the potential promoter is even less active than the copy on the prophage.
Figure 3.
Identification of the transcriptional start site of the antisense RNA from yfjN by primer extension analysis. (A) Lane 1, control host strain (DJ480); lane 2, DJ480 carrying single-copy λ prophage with the pRS551-derived anti-yfjN–lacZ fusion; lane 3, DJ480 carrying single-copy λ prophage with the pRS552-derived anti-yfjN–lacZ fusion; lane 4, DJ480 harboring the pRS551-anti-yfjN-lacZ plasmid; lane 5, DJ480 harboring the pRS552-anti-yfjN –lacZ plasmid. Reverse transcription reactions were performed by using 30 µg of total RNA from the parent and prophage-containing strains (lanes 1–3) and 5 µg of total RNA from plasmid-containing strains (lanes 4 and 5); The sequence ladder was generated using the primer used in the primer extension reactions. Although the levels of the primer extension products for the prophage strains are extremely low, the same relative levels were seen in all three repetitions of the experiment. (B) Top strand sequence from nt 2 763 939 to 2 764 368 of the E.coli chromosome (1–430 of yfjN). The yfjN ORF extends to 2 765 012. The ATG start codon of yfjN is boxed. The sequence cloned in pRS551 and pRS552 is surrounded by brackets (between 2 764 127 and 2 764 357). The +1 transcriptional start site is shown by an arrow. The −10 and −35 sequences of the yfjN antisense promoter are underlined.
We compared the β-galactosidase activities of the plasmid and single-copy prophage strains carrying transcriptional fusions to the eight selected bona fide promoters with the activities of 10 potential antisense promoters (Table 3). As expected on the basis of the copy number of the plasmid, the activities for the lacZ fusions of the bona fide promoters were, on average, 16-fold greater when present on the plasmid than when present as single-copy prophage. However, this ratio increased to 30-fold for the activities of the potential promoter fusions. This observation suggests that the promoter fragments from bona fide promoters may carry additional sequence information that allows for more efficient transcription in the context of the bacterial chromosome.
Table 3.
Expression from multi-copy and single-copy fusions to potential antisense promoters and to known promoters
| Promoter | Plasmid | Prophage | Plasmid/prophagea |
|---|---|---|---|
| Potential antisense promoters | |||
| ygeH | 2900 | 120 | 25 |
| rhsE | 2400 | 84 | 29 |
| yfjN | 1700 | 56 | 30 |
| yiaO | 670 | 33 | 20 |
| yjcE | 680 | 25 | 28 |
| ydiM | 200 | 6 | 35 |
| ecpD | 170 | 5 | 34 |
| topA | 170 | 6 | 28 |
| yehI | 120 | 4 | 35 |
| fabA | 150 | 4 | 39 |
| Average (SD) | 30 (5.3) | ||
| Known promoters | |||
| ompA | 11 000 | 830 | 16 |
| rpsA | 9700 | 630 | 15 |
| aroP | 9700 | 860 | 11 |
| uidR | 7400 | 400 | 19 |
| map | 7100 | 340 | 21 |
| rsd | 6400 | 530 | 12 |
| fepD | 5800 | 350 | 17 |
| ygaW | 4700 | 260 | 18 |
| Average (SD) | 16 (3.2) | ||
aThe ratios were calculated prior to rounding of the assay results.
Specific sequence elements that increase promoter activity, such as UP elements that are bound by the C-terminal domain of the α subunit of RNA polymerase, also could be lacking from the potential promoter internal to yfjN. Thus we tested whether an UP element would increase potential promoter activity by replacing the 90 bp immediately upstream of the anti-yfjN-lacZ −35 hexamer with the 102 bp sequence found upstream of the −35 hexamer of the tyrT promoter (29). When cells with this construct present on a single-copy prophage derived from pRS551 were assayed, they had 9.9 times the activity of the same construct without the UP element (890 MU compared with 90 MU). Thus, the absence of such elements seems to be another reason for the low-level activity of the anti-yfjN and, presumably, other potential promoters.
Transcript instability could be another reason for our inability to detect defined transcripts. With this possibility in mind, we postulated that base-pairing between the yfjN mRNA and the antisense transcript could result in a double-stranded RNA that might be degraded by the RNase III endoribonuclease. Thus we tried to detect defined transcripts in an RNase III -deficient (rnc) strain. However, the levels of the antisense transcripts were not significantly increased in the mutant strain (data not shown).
An additional determinant of RNA stability might be translation of the message; the chromosomal yfjN antisense sequence does not contain a plausible translation start signal. We therefore cloned the same anti-yfjN fragment into the plasmid pRS552, which lacks a ribosome-binding site preceding the lacZ structural gene (generating pRS552-anti-yfjN-lacZ). Expression from this plasmid and from a corresponding integrated λ prophage were compared with the expression from the pRS551 constructs carrying the same anti-yfjN fragment cloned upstream of lacZ which has a ribosome-binding site (Figure 3A). The steady-state level of the RNA expressed from the plasmid pRS552-derivative (lane 5) was ∼4-fold lower than the level expressed from the pRS551-derivative (lane 4). Expression of RNA from the pRS552-derived prophage (lane 3) was similarly lower than the expression from the pRS551-derived prophage (lane 2). Together these assays of the modified anti-yfjN–lacZ fusions suggest that the inability to detect defined chromosomal transcripts from the potential promoters is probable to be due to the lack of features needed to enhance promoter activity as well as transcript stability.
Comparison of potential promoter sequences with the canonical σ70 sequence
To evaluate the sequences of the inserts selected for high transcriptional activity and of those isolated at random, we analyzed each of the cloned sequences with the computer program pftools2.2 (5,13). This program is designed to identify promoter sequences by using weighted matches to the optimal −35, −10 and spacer regions and then to compute a homology score. A homology score of 45.0 or more was considered a significant ‘hit’ (5).
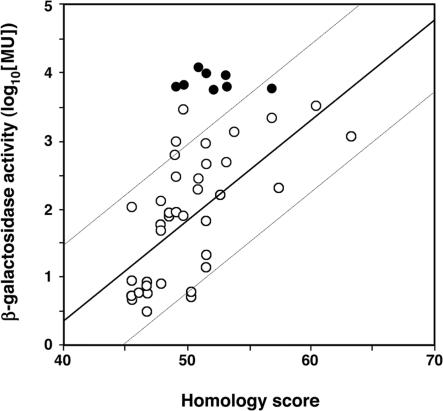
Two general conclusions can be drawn from this analysis. First, for both the sequences isolated at random and the eight clones selected for high activity, there is a correlation between the β-galactosidase activities and the frequency of computer ‘hits’ (Supplementary Table S2) as well as between the logarithm of β-galactosidase activity and the homology scores (Figure 4), analogous to that observed with bona fide promoters (5). Second, the fragments selected for high activity fit the optimal promoter sequence no better than the most active of the fragments selected at random: an average homology score of 52.1 ± 2.43 was calculated for the eight promoters selected for high expression (average ∼7800 MU, Table 1) and an average homology score of 54.2 ± 5.45 was calculated for the eight most active of the randomly isolated promoters (average ∼1700 MU; Table 2). Thus the 4.5-fold greater activity in the eight bona fide promoters does not correlate with greater homology to the optimally configured promoter. This is clearly seen in Figure 4 where the activities of seven of the eight selected promoters are two SDs greater than that found for the promoters isolated at random. These results confirm the importance of factors other than appropriately spaced −35 and −10 hexamers for determining promoter activity and suggest that a very large number of E.coli chromosomal sites with near-perfect homology to the optimal −35 and −10 hexamers are inconsequential because they lack additional critical sequence information.
Figure 4.
Correlation between match to optimal σ70 promoter (−35 and −10 sequences) measured by the pftools 2.2 program (homology score, X) and log10 of the promoter (β-galactosidase, Y) activity. The regression line for the 39 randomly isolated promoters that have homology scores of 45 or greater (open circles) has the formula Y = −5.5496 + 0.14772X and an R2 of 0.417. Two SDs are indicated by the dashed lines. The eight inserts selected on the basis of high activity are shown by filled circles.
DISCUSSION
By assaying random ∼160 bp chromosomal fragments in a system designed to detect promoter activity on plasmids, we identified a large number of segments internal to ORF sequences that had sufficient activity to warrant examination as potential promoters. Systems similar to that used here have been used repeatedly and successfully to analyze documented functional promoters in intergenic regions (16,30). We found that many of the potential promoters located within ORFs have homology scores (a measure of fit to the optimal RNA polymerase-binding site hexamers and spacing) that are very similar to the scores of documented promoters. However, despite the high levels of expression found when these segments were on plasmids and despite the close matches to documented promoters, we were not able to detect distinct transcripts from the corresponding chromosomal sequences. Although it is possible that some of these potential antisense transcripts are expressed at higher levels under growth conditions that we did not assay, a more probable explanation is that these segments carry functional −35 and −10 sequences but not other elements of bona fide promoters that are needed for productive expression.
Based on our evaluation of expression of the putative antisense promoter internal to yfjN, we suggest that multiple factors contribute to productive promoter activity. First, the yfjN antisense promoter fusion constructed in plasmid pRS551, which contains the translation signals of lacZ, gave significantly higher levels of mRNA than the fusion constructed in plasmid pRS552, which lacks these translational signals. The simplest explanation is that translation has a significant impact on whether or not a transcript can be detected, most probably by affecting the stability of the mRNA. It is also possible that other sequence differences between the two fusions affect RNA stability. Similarly, terminator sequences and other stability elements such as those present on noncoding RNAs probably affect whether a transcript can be detected. Second, relative to their expression on the multi-copy plasmid, the yfjN antisense promoter fusion, as well as those of other potential promoters within ORFs, showed significantly lower expression when present in single copy on the chromosome compared with bona fide promoters. Thus bona fide promoters together with the surrounding region also appear to contain sequence information that protects against possible negative effects of supercoiling or other structural constraints associated with the bacterial chromosome.
The results suggest that bona fide promoters may have evolved for greater activity not so much by optimizing their σ70-binding signals but by improving other promoter features. These include decreasing the binding of repressors or enhancing the binding of activators; enhancing the binding of RNA polymerase via UP elements; and lowering the energy required for open complex formation and/or polymerase clearance. Indeed, of the six sequences selected for high activity that others have studied in greater depth, five are believed to be regulated by transcriptional activators (25,26,31–34). Additional computational and genetic studies of bona fide promoters should allow further definition of the sequences required for the expression of detectable transcripts.
Our observations are similar to recent findings in eukaryotic organisms. There is increasing evidence for the expression of RNAs outside of and on the opposite strand of known and predicted coding genes based on clones present in cDNA libraries derived from mouse and human cell lines (35–37). However, few of these have been shown to correspond to defined RNAs. In fact, transcripts encoded in several intergenic regions in Saccharomyces cerevisiae could only be detected as heterogeneous bands by northern analysis and only in strains lacking the Rrp6p exonuclease indicating that there are specific quality control mechanisms to degrade the transcripts made from cryptic promoters (38). Based on these observations and our own results, we suggest caution before attributing biological significance to antisense transcripts that have not been shown to be distinct.
Nevertheless, our results that many functional −35 and −10 sites are present throughout the chromosome and within genes are consistent with recent experiments in which E.coli RNA polymerase-binding sites in rifampicin-treated cells were mapped by chromatin immunoprecipitation and microarrays (39). These studies showed that a significant fraction of the RNA polymerase was bound within ORFs. An open question is whether these RNA polymerase-binding sites and the accompanying low level of intragenic transcription have a function. The possibilities that the binding sites serve as docks for RNA polymerase or that the low level of transcription helps to maintain a more accessible state of the chromosome warrant further investigation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
We thank Thomas Brodigan for excellent technical assistance and Virgil Rhodius and Robert Weisberg for comments on the manuscript. M.K. was supported by a research fellowship from the Japan Society for the Promotion of Science. This work was supported by the Intramural Research Program of the NIH (NIDDK and NICHD). Funding to pay the Open Access publication charges for this article was provided by the Intramural Research Program of NIDDK.
Conflict of interest statement. None declared.
REFERENCES
- 1.Gross C.A., Chan C., Dombroski A., Gruber T., Sharp M., Tupy J., Young B. The functional and regulatory roles of sigma factors in transcription. Cold Spring Harb. Symp. Quant. Biol. 1998;63:141–155. doi: 10.1101/sqb.1998.63.141. [DOI] [PubMed] [Google Scholar]
- 2.Harley C.B., Reynolds R.P. Analysis of E.coli promoter sequences. Nucleic Acids Res. 1987;15:2343–2361. doi: 10.1093/nar/15.5.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hawley D.K., McClure W.R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983;11:2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mulligan M.E., McClure W.R. Analysis of the occurrence of promoter-sites in DNA. Nucleic Acid Res. 1986;14:109–126. doi: 10.1093/nar/14.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mulligan M.E., Hawley D.K., Entriken R., McClure W.R. Escherichia coli promoter sequences predict in vitro RNA polymerase selectivity. Nucleic Acids Res. 1984;12:789–800. doi: 10.1093/nar/12.1part2.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu. Rev. Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- 7.Alifano P., Piscitelli C., Blasi V., Rivellini F., Nappo A.G., Bruni C.B., Carlomagno M.S. Processing of a polycistronic mRNA requires a 5′ cis element and active translation. Mol. Microbiol. 1992;6:787–798. doi: 10.1111/j.1365-2958.1992.tb01529.x. [DOI] [PubMed] [Google Scholar]
- 8.Bauerle R.H., Margolin P. Evidence for two sites for initiation of gene expression in the tryptophan operon of Salmonella typhimurium. J. Mol. Biol. 1967;26:423–436. doi: 10.1016/0022-2836(67)90313-0. [DOI] [PubMed] [Google Scholar]
- 9.Jackson E.N., Yanofsky C. Internal promoter of the tryptophan operon of Escherichia coli is located in a structural gene. J. Mol. Biol. 1972;69:307–313. doi: 10.1016/0022-2836(72)90232-x. [DOI] [PubMed] [Google Scholar]
- 10.Wek R.C., Hatfield G.W. Examination of the internal promoter, PE, in the ilvGMEDA operon of E.coli K-12. Nucleic Acids Res. 1986;14:2763–2777. doi: 10.1093/nar/14.6.2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huerta A.M., Collado-Vides J. Sigma70 promoters in Escherichia coli: specific transcription in dense regions of overlapping promoter-like signals. J. Mol. Biol. 2003;333:261–278. doi: 10.1016/j.jmb.2003.07.017. [DOI] [PubMed] [Google Scholar]
- 12.Selinger D.W., Cheung K.J., Mei R., Johanson E.M., Richmond C., Blattner F.R., Lockhart D.J., Church G.M. RNA expression analysis using a 30 base pair resolution Escherichia coli genome array. Nat. Biotechnol. 2000;18:1262–1268. doi: 10.1038/82367. [DOI] [PubMed] [Google Scholar]
- 13.Chen S., Lesnik E.A., Hall A.T., Sampath R., Griffey R.H., Ecker D.J., Blyn L.B. A bioinformatics based approach to discover small RNA genes in the Escherichia coli genome. BioSystems. 2002;65:157–177. doi: 10.1016/s0303-2647(02)00013-8. [DOI] [PubMed] [Google Scholar]
- 14.Rivas E., Klein R.J., Jones T.A., Eddy S.R. Computational identification of noncoding RNAs in E.coli by comparative genomics. Curr. Biol. 2001;11:1369–1373. doi: 10.1016/s0960-9822(01)00401-8. [DOI] [PubMed] [Google Scholar]
- 15.Wassarman K.M., Repoila F., Rosenow C., Storz G., Gottesman S. Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev. 2001;15:1637–1651. doi: 10.1101/gad.901001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simons R.W., Houman F., Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- 17.Miller J.H. Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 18.Kawano M., Oshima T., Kasai H., Mori H. Molecular characterization of long direct repeat (LDR) sequences expressing a stable mRNA encoding for a 35-amino-acid cell-killing peptide and a cis-encoded small antisense RNA in Escherichia coli. Mol. Microbiol. 2002;45:333–349. doi: 10.1046/j.1365-2958.2002.03042.x. [DOI] [PubMed] [Google Scholar]
- 19.Cabrera J.E., Jin D.J. Growth phase and growth rate regulation of the rapA gene, encoding the RNA polymerase-associated protein RapA in Escherichia coli. J. Bacteriol. 2001;183:6126–6134. doi: 10.1128/JB.183.20.6126-6134.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Powell B.S., Rivas M.P., Court D.L., Nakamura Y., Turnbough C.L., Jr Rapid confirmation of single copy lambda prophage integration by PCR. Nucleic Acid Res. 1994;22:5765–5766. doi: 10.1093/nar/22.25.5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blattner F.R., Plunkett G., III, Bloch C.A., Perna N.T., Burland V., Riley M., Collado-Vides J., Glasner J.D., Rode C.K., Mayhew G.F., et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 22.Wang P., Yang J., Pittard A.J. Promoters and transcripts associated with the aroP gene of Escherichia coli. 1997;179:4206–4212. doi: 10.1128/jb.179.13.4206-4212.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang J., Wang P., Pittard A.J. Mechanism of repression of the aroP P2 promoter by the TyrR protein of Escherichia coli. J. Bacteriol. 1999;181:6411–6418. doi: 10.1128/jb.181.20.6411-6418.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pedersen S., Skouv J., Kajitani M., Ishihama A. Transcriptional organization of the rpsA operon of Escherichia coli. Mol. Gen. Genet. 1984;196:135–140. doi: 10.1007/BF00334105. [DOI] [PubMed] [Google Scholar]
- 25.Ben-Bassat A., Bauer K., Chang S.Y., Myambo K., Boosman A., Chang S. Processing of the initiation methionine from proteins: properties of the Escherichia coli methionine aminopeptidase and its gene structure. J. Bacteriol. 1987;169:751–757. doi: 10.1128/jb.169.2.751-757.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Christoffersen C.A., Brickman T.J., Hook-Barnard I., McIntosh M.A. Regulatory architecture of the iron-regulated fepD-ybdA bidirectional promoter region in Escherichia coli. J. Bacteriol. 2001;183:2059–2070. doi: 10.1128/JB.183.6.2059-2070.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kawano M., Reynolds A.A., Miranda-Rios J., Storz G. Detection of 5′ and 3′ UTR-derived small RNAs and cis-encoded antisense RNAs in Escherichia coli. Nucleic Acid Res. 2005;33:1040–1050. doi: 10.1093/nar/gki256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adhya S., Gottesman M. Promoter occlusion: transcription through a promoter may inhibit its activity. Cell. 1982;29:939–944. doi: 10.1016/0092-8674(82)90456-1. [DOI] [PubMed] [Google Scholar]
- 29.Lamond A.I., Travers A.A. Requirement for an upstream element for optimal transcription of a bacterial tRNA gene. Nature. 1983;305:248–250. doi: 10.1038/305248a0. [DOI] [PubMed] [Google Scholar]
- 30.Silhavy T.J., Berman M.L., Enquist L.W. Experiments with Gene Fusions. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1984. [Google Scholar]
- 31.Gibert I., Barbe J. Cyclic AMP stimulates transcription of the structural gene of the outer-membrane protein OmpA of Escherichia coli. FEMS Microbiol. Lett. 1990;56:307–311. doi: 10.1111/j.1574-6968.1988.tb03197.x. [DOI] [PubMed] [Google Scholar]
- 32.Kajitani M., Ishihama A. Promoter selectivity of Escherichia coli RNA polymerase. Differential stringent control of the multiple promoters from. J. Biol. Chem. 1984;259:1951–1957. [PubMed] [Google Scholar]
- 33.Jishage M., Ishihama A. Transcriptional organization and in vivo role of the Escherichia coli rsd gene, encoding the regulator of RNA polymerase. J. Bacteriol. 1999;181:3768–3776. doi: 10.1128/jb.181.12.3768-3776.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blanco C., Mata-Gilsinger M., Ritzenthaler P. The use of gene fusions to study the expression of uidR, a negative regulatory gene of Escherichia coli K-12. Gene. 1985;36:159–167. doi: 10.1016/0378-1119(85)90080-0. [DOI] [PubMed] [Google Scholar]
- 35.Lavorgna G., Dahary D., Lehner B., Sorek R., Sanderson C.M., Casari G. In search of antisense. Trends Biochem Sci. 2004;29:88–94. doi: 10.1016/j.tibs.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 36.Numata K., Kanai A., Saito R., Kondo S., Adachi J., Wilming L.G., Hume D.A. RIKEN GER Group, GSL Members. Hayashizaki Y., et al. Identification of putative noncoding RNAs among the RIKEN mouse full-length cDNA collection. Genome Res. 2003;13:1301–1306. doi: 10.1101/gr.1011603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kiyosawa H., Yamanaka I., Osato N., Kondo S. RIKEN GER Group, GSL Members. Hayashizaki Y. Antisense transcripts with FANTOM2 clone set and their implications for gene regulation. Genome Res. 2003;13:1324–1334. doi: 10.1101/gr.982903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wyers F., Rougemaille M., Badis G., Rousselle J.C., Dufour M.E., Boulay J., Regnault B., Devaux F., Namane A., Seraphin B., et al. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 39.Herring C.D., Raffaelle M., Allen T.E., Kanin E.I., Landick R., Ansari A.Z., Palsson B.O. Immobilization of Escherichia coli RNA polymerase and location of binding sites by use of chromatin immunoprecipitation and microarrays. J. Bacteriol. 2005;187:6166–6174. doi: 10.1128/JB.187.17.6166-6174.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.