Abstract
Homologous recombination provides an effective way to repair DNA double-strand breaks (DSBs) and is required for genetic recombination. During the process of homologous recombination, a heteroduplex DNA structure, or a ‘Holliday junction’ (HJ), is formed. The movement, or branch migration, of this junction is necessary for recombination to proceed correctly. In prokaryotes, the RecQ protein or the RuvA/RuvB protein complex can promote ATP-dependent branch migration of Holliday junctions. Much less is known about the processing of Holliday junctions in eukaryotes. Here, we identify RecQL1 as a predominant ATP-dependent, HJ branch migrator present in human nuclear extracts. A reduction in the level of RecQL1 induced by RNA interference in HeLa cells leads to an increase in sister chromatid exchange. We propose that RecQL1 is involved in the processing of Holliday junctions in human cells.
INTRODUCTION
DNA double-strand breaks (DSBs) arise as a result of ionizing radiation, DNA-damaging chemicals and as a product of blocked replication forks. Mechanisms that repair DNA DSBs are critical for the maintenance of genome integrity and cell viability. Homologous recombination provides an efficient way to repair DNA DSBs. Homologous recombination also occurs during meiosis providing an effective source of genetic variation. During the process of homologous recombination, complementary DNA strand exchange leads to the formation of Holliday junctions (HJs) (1). The initial strand invasion process is carried out by RecA in prokaryotes and the RecA homolog (RAD51) in conjunction with RAD54 in eukaryotes (2). In prokaryotes, the movement, or branch migration, of this junction is facilitated by the RuvA and RuvB proteins. RuvA is a Holliday junction specific binding protein that loads RuvB, an ATP-dependent unwinding protein onto the HJ. The RuvA and RuvB proteins cooperate with the highly specific HJ endonuclease, RuvC, which cuts junctions by symmetrically nicking opposing strands of like polarity. This reaction (termed resolution) produces ligatable products (3). Similar to RuvA, RuvC is a HJ structure specific binding protein. However, it cleaves HJs in a sequence-specific manner within the consensus sequence 5′-a/tTTg/c-3′ (4). Branch migration of the HJ by the RuvAB complex allows RuvC to scan for cleavable sequences (5).
The Escherichia coli RecQ protein is another prokaryotic enzyme that can facilitate ATP-dependent branch migration of HJs (6). RecQ belongs to the recF recombination pathway and has been shown to be involved in the resumption of DNA synthesis following DNA damage (7,8). RecQ has also been shown to suppress illegitimate recombination (9).
Much less is known about the processing of HJs in eukaryotes. Branch migration and resolution activities have been detected in mammalian cell free extracts. However, attempts to identify the proteins responsible were unsuccessful (10,11). Recently, it was reported that extracts prepared from cultured mammalian cell lines that have mutations in the RAD51 paralogs RAD51C and XRCC3 have reduced levels of resolution activity. Furthermore, immunodepletion of RAD51C from fractionated mammalian extracts resulted in a loss of branch migration and resolution activity. However, neither recombinant protein was found to possess branch migration or resolution activity (12).
Bloom's syndrome is a rare genetic disorder associated with a predisposition to cancer and genomic instability. Bloom's syndrome is caused by a mutation in the gene encoding the BLM protein (13). Analysis of this protein established that it is a member of the RecQ family of helicases and is capable of binding to and inducing branch migration of HJs (14). Cultured Bloom's syndrome cells exhibit a high incidence of spontaneous sister chromatid exchanges (SCEs) (15). Recent studies have demonstrated that BLM and topoisomerase III are part of a large complex in human cells, and BLM and topoisomerase III can catalyze the dissolution of Holliday junctions in vitro (16,17). Furthermore, genetic studies have revealed that the yeast homologs of BLM and topoisomerase III, Sgs1 and Top3, suppress the formation of crossover products arising from homologous recombination (18,19). The WRN protein is another member of the RecQ family that is responsible for Werner's syndrome, a rare autosomal recessive disorder (20). Individuals with Werner's syndrome manifest the clinical symptoms of premature aging and a predisposition to certain cancers. Cells cultured from Werner's syndrome patients exhibit abnormal genomic rearrangements and large chromosomal deletions (21,22). Similar to BLM, the WRN protein is also capable of branch migrating HJs (23). Both BLM and WRN are members of a larger family of RecQ-related helicases in humans that also includes RecQL1, RecQ4 and RecQ5ß (24). RecQL1 was first identified as an abundant DNA-dependent ATPase with DNA helicase activity in human cells (25) and through a fortuitous protein association (26), whereas other members of the family have been identified through homology. The precise functions of the different RecQ family proteins in maintaining the stability of the genome remain unresolved. Although both BLM and WRN are capable of branch migrating HJs, nuclear extracts prepared from BLM−/− and WRN−/− cell lines still possess comparable levels of ATP-dependent HJ branch migration activity which suggests that another protein may be performing this function (10). We therefore set out to identify the protein responsible for the predominant ATP-dependent HJ branch migration activity present in human cell lines.
MATERIALS AND METHODS
Purification of RecQL1
RecQL1 was purified from log phase growth HEK293 cell nuclei (2 × 1010 cells) cultured in DMEM supplemented with 10% fetal calf serum (FCS) (GIBCO-BRL). Cells were lysed in 4 l hypotonic lysis buffer (10 mM Tris–HCl, pH 7.5, 1.5 mM MgCl2, 0.25 M Sucrose, 1 mM DTT and 0.5 mM phenylmethylsulfonyl fluoride). Nuclei were pelleted by centrifugation and the supernatant containing the cytosol was discarded. The nuclei were extracted with 100 ml of buffer F (10 mM Tris–HCl, pH 7.5, 250 mM NaCl, 30 mM Na4P2O7, 50 mM NaF, 5 µM ZnCl2, 10% glycerol and 0.1% Triton X-100). The nuclear extract was then clarified by centrifugation for 20 min at 25 000 g and dialyzed against 4 l of BC100. BC buffers contain 20 mM Tris–HCl, pH 7.8, 0.2 mM EDTA, 1 mM MgCl2, 1 mM DTT and 10% glycerol; the number following BC denotes the mM concentration of KCl. The extract was then applied to a column containing 100 ml of phosphocellulose resin (Sigma) equilibrated in BC100. The column was washed with BC100 until no protein eluted, and then sequentially eluted with BC350, BC600 and BC1000. Holliday junction ATP-dependent branch migration activity eluted in the 0.35 M fraction, whereas the majority of Holliday junction resolution activity eluted in the 0.6 M fraction. The 0.35 M fraction was diluted with BC0 to a final concentration of 150 mM KCl and loaded onto a 50 ml column of DEAE-52 resin (Whatman) equilibrated in BC150. The flow through was collected, pooled with one column volume of BC150 wash, and the bound proteins were eluted with BC500. Branch migration activity was observed in the BC150 flow through fraction. This 0.15 M fraction was then diluted to 0.1 M with BC0 and loaded onto a 25 ml column containing SP-Sepharose (Pharmacia) equilibrated in BC100. The column was washed with 50 ml of BC100 and bound proteins were eluted with 500 ml linear gradient of BC100–BC500. Branch migration activity eluted at the KCl concentration of ∼0.2 M. Fractions containing branch migration activity were pooled, dialyzed against BC100 and loaded onto a 2 ml column containing ssDNA-Agarose (Sigma) equilibrated in BC100. The column was washed with BC100 and the proteins were eluted with a 40 ml linear gradient of BC100–BC1000. Branch migration activity eluted at a KCl concentration of ∼0.5 M. The active fractions were pooled and ∼1/4 of this fraction was subjected to gel filtration on a 24 ml Superdex-200 (Pharmacia) FPLC column equilibrated and run in BC200. ATP-dependent branch migration activity elutes with a relative molecular mass of 150 kDa.
Holliday junction substrates
Synthetic Holliday junction substrate was prepared by annealing the following oligonucleotides in PCR buffer (Roche): oligonucleotide 1, (5′CCGCTACCAGTGATCACCAATGGATTGCTAGGACATCTTTGCCCACCTGCAGGTTCACCC-3′); oligonucleotide 2, (5′TGGGTGAACCTGCAGGTGGGCAAAGATGTCCTAGCAATCCATTGTCTATGACGTCAAGCT3′); oligonucleotide 3, (5′GAGCTTGACGTCATAGACAATGGATTGCTAGGACATCTTTGCCGTCTTGTCAATATCGGC-3′) and oligonucleotide 4, (5′TGCCGATATTGACAAGACGGCAAAGATGTCCTAGCAATCCATTGGTGATCACTGGTAGCGG3′).
Annealing was carried out by heating the oligos to 95°C and then allowing them to cool to 25°C. 32P-labeled oligonucleotide 2 was annealed with a 2-fold excess of the other three oligonucleotides. Holliday junctions were then purified by gel filtration on a Sephadex G-200 column equilibrated and run in STE (10 mM Tris–HCl, pH 7.5, 1 mM EDTA and 50 mM NaCl).
Holliday junction branch migration and resolution assays
Approximately 5 ng of 32P-labeled synthetic Holliday junction was incubated in a 40 µl assay with 10 µl of each fraction (in BC50) and reaction buffer (60 mM Na2HPO4/NaH2PO4, pH 7.4, 6 mM MgCl2, 1 mM DTT and 100 µg/ml BSA) in the presence or absence of 2 mM ATP when indicated. Assays performed for the first two chromatographic steps also contained sonicated salmon sperm DNA (Stratagene) 100 µg/ml to inhibit non-specific nucleases. The reactions were incubated for 45 min at 37°C. The reactions were then stopped and deproteinized with 200 µg/ml proteinase K and 0.1% SDS for 5 min at 37°C. DNA products were resolved on a 10% polyacrylamide neutral gel and visualized using X-ray film.
Transfections and FLAG-immunoprecipitations
HEK293 cells were cultured in DMEM supplemented with 10% FCS (GIBCO-BRL). Cells were transfected via the calcium phosphate method with 10 µg of vector that expresses the indicated N-terminal FLAG-fusion protein. Nuclear extract was prepared as described above. For immunoprecipitations, nuclear extracts were incubated with anti-FLAG M2 Agarose beads (Sigma) for 4 h at 4°C. The beads were then washed extensively with buffer F and eluted with 2 bead volumes of buffer F containing 50 ng/ml of FLAG peptide (Sigma) for 12 h at 4°C.
Western blot
Proteins were resolved on 10% SDS–PAGE gels. Proteins were transferred to Immobilon-P (Millipore) probed with M2-FLAG monoclonal antibody in TBST (20 mM Tris–HCl, pH 7.5, 150 mM NaCl and 0.2% Tween-20). After extensive washing the blots were probed with anti-mouse secondary antibody coupled to horseradish peroxidase. The blots were washed and developed by chemiluminescence (Amersham Biosciences).
Mass spectrometry
The protein identification was carried out by nanoscale microcapillary LC-MS/MS analysis method. An ULTIMATE capillary HPLC system with SWITCHOS II and FAMOS autoinjector (Dionex, Sunnyvale, CA) and a LCQ DECA XP PLUS ion trap mass spectrometer (Thermo Finnigan, San Jose, CA) were used. A 10 cm length fritless 75 µm id fused-silica capillary column was packed with 5 µm, 100A′, C18 resin. The flow rate was 120 nl/min and the 0.1 M acetic acid buffer system was used. All MS/MS spectra were analyzed using SEQUEST program.
RNA interference (siRNA)
For RecQL1-siRNA, SMART pool® (siRNAs) were selected and produced by Dharmacon, Inc. HeLa cells grown in DMEM supplemented with 10% calf serum to ≈30% confluency were transfected with RecQL1 siRNA. Twenty-four hours later the medium was replaced and the cells were transfected with RecQL1 siRNA for a second time. Forty-eight hours after the second transfection the cells were harvested for the SCE assay or RNA was isolated for RT–PCR. Transfections were preformed utilizing Oligofectamine™ following procedures recommended by the manufacturer (Invitrogen).
RT–PCR
RNA was extracted from cells with Trizol® (Sigma). For RT–PCR, a Superscript™ One-Step RT–PCR with Platinum® Taq kit was used, utilizing cDNA specific primer pairs, following procedures suggested by the manufacturer (Invitrogen). Products from the reactions were separated by agarose gel electrophoresis, stained with ethidium bromide, visualized by UV-transillumination and photographed. Reactions contained the following primers: RecQL1 5′ primer 5-ATGGCGTCCGTTTCAGCTCTA-3, 3′ primer 5-TAACATTCATATCAGGCATCATCG-3; RAD54L 5′ primer 5-ATGAGGAGGAGCTTGGCTCCC-3, 3′ primer 5-ACCCAGACCAGCTGGTTATCA-3; RAD51C 5′ primer 5-ATGCGCGGGAAGACGTTCCGC-3, 3′ primer 5-TTGAGATTTGTTTCTGGGTTA-3
SCE assay
SCE assays were performed as described previously (27). Briefly, HeLa cells grown to ≈60% confluency, 24 h after the first transfection with siRNA, were grown for 48 additional hours (two cell cycles) in the presence of 20 µM BrdU. The cells were then incubated with 0.1 µg/ml Colcemid for 2 h. The cells were harvested with Trypsin, swelled for 20 min in 75 mM KCl and fixed for 30 min in methanol:acetic Acid (3:1). The cells were dropped onto moist 45°C pre-warmed glass slides and allowed to air dry. The slides were aged overnight and stained with 100 µg/ml Hoechst 33258 in dH2O for 20 min at 25°C. The slides were then bleached with a 120-watt plant light at a distance of 20 cm for 3 h. The slides were next stained with 10% Giemsa stain for 15 min. The slides were air-dried and mounted with glass coverslips in Permount® (Sigma). For MMC-induced SCE, Mitomycin C (MMC) was added to the culture at a final concentration of 60 nM for 16 h prior to harvesting. Slides were analyzed with Nikon E800 bright-field microscope equipped with a 100× objective. Pictures were taken with a digital camera.
RESULTS AND DISCUSSION
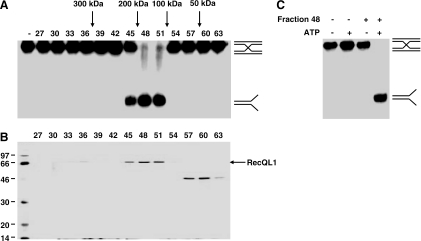
The factor(s) mediating the branch migration of HJs in human cells remains undefined since nuclear extracts prepared from BLM−/− and WRN−/− cell lines still possess comparable levels of ATP-dependent HJ branch migration activity (10). We therefore set out to identify the protein(s) responsible for the predominant ATP-dependent HJ branch migration activity present in human cell lines using biochemical purification. To accomplish this, we utilized an in vitro assay employing synthetic 32P-labelled Holliday junctions (Figure 1A). The synthetic HJs contain a 26 bp homologous core flanked by regions of non-homology, as described previously (10). This assay detects both ATP-dependent HJ branch migration and HJ cleavage activity. ATP-dependent HJ branch migration leads to the formation of splayed DNAs whereas HJ cleavage produces nicked DNA products. Employing the in vitro assay, we followed ATP-dependent HJ branch migration activity through an extensive chromatographic fractionation of HEK293 cell nuclear extracts (Figure 1B). Owing to the presence of non-specific nucleases, the assays performed on early chromatography steps contained sonicated salmon sperm DNA in addition to the synthetic 32P-labelled synthetic HJs. The nuclear extract was first subjected to phosphocellulose chromatography. The extract was loaded in a buffer containing 100 mM KCl and the bound proteins were eluted with sequential steps of buffer containing 350 mM, 600 mM and 1 M KCl. These fractions were then assayed for HJ processing activities. As depicted in Figure 1C, ATP-dependent HJ branch migration activity was detected in the 350 mM fraction, whereas the majority of HJ cleavage activity was present in the 600 mM fraction. We believe that a portion of this cleavage activity is the same as an activity previously described as Resolvase A, because after further fractionation an identical cleavage pattern was observed (data not shown) (28). The 350 mM fraction containing ATP-dependent HJ branch migration activity was then subjected to chromatographic fractionation on four more resins as summarized in Figure 1B. By the third column step (SP-Sepharose), all HJ cleavage activity was removed from the peak of branch migration activity. The final step of chromatography was a Superdex-200 gel filtration column. As depicted in Figure 2A, HJ branch migration activity eluted from the Superdex-200 column with an apparent molecular mass of 150 kDa. Silver staining of an SDS–polyacrylamide gel loaded with fractions from the Superdex-200 column revealed that HJ branch migration activity coeluted with a polypeptide of ∼70 kDa (Figure 2B). Mass spectrometry analysis identified this polypeptide as RecQL1. Owing to its gel filtration elution profile, we predict that RecQL1 is likely a homodimer, consistent with a previous report (29). The HJ branch migration activity exhibited by purified RecQL1 requires ATP (Figure 2C). Based on purification yields, RecQL1 appears to be a fairly abundant protein. Assuming a 25% yield and the protein concentration by Bradford assay, we estimate that there are at least 50 000 molecules of RecQL1 per nucleus in log phase HEK293 cells. The latter is similar to the 21 000 molecules estimated in EBV-immortalized lymphoblastoid cells (30). The same assay led to the purification of RecQL1 as the most abundant HJ branch migration activity in HeLa cells as well as HEK293 (data not shown).
Figure 1.
(A) Schematic diagram of a synthetic Holliday Junction and the products of ATP-dependent branch migration and resolution. (B) Chromatographic scheme used to purify RecQL1. (C) Holliday junction branch migration and cleavage assay with HEK293 cell nuclear extract phosphocellulose fractions. Fractions were mixed with 32P-labelled synthetic Holliday junctions, with added ATP as indicated. 32P-labelled DNA products were separated by PAGE and analyzed by autoradiography.
Figure 2.
(A) Holliday junction branch migration assay with fractions from the Superdex-200 gel filtration column. Fractions were mixed with 32P-labelled synthetic Holliday junctions, with added ATP as indicated. 32P-labelled DNA products were separated by PAGE and analyzed by autoradiography. (B) SDS–PAGE and silver staining of protein fractions from the Superdex-200 gel filtration column. Peptide sequencing by microcapillary HPLC-ion trap mass spectrometry identified the 70 kDa protein as RecQL1. Fractions were loaded as indicated and the positions of RecQL1 and molecular weight markers are indicated. (C) The purified RecQL1 protein exhibits ATP-dependent Holliday junction branch migration activity. Fraction 48 from the Superdex-200 gel filtration column was tested in the assay. The fraction and ATP were mixed with 32P-labelled synthetic Holliday junctions as indicated. 32P-labelled DNA products were separated by PAGE and analyzed by autoradiography.
We next tested immunopurified FLAG-tagged RecQL1 for ATP-dependent HJ branch migration activity, along with other immunopurified FLAG-tagged proteins expressed in HEK293 cells. As depicted in Figure 3A, only the immunopurified FLAG-tagged RecQL1 eluate exhibits ATP-dependent HJ branch migration activity. Human RAD54L was also tested because it was shown to promote both transient ATP-dependent strand separation of duplex DNA and, in conjunction with RAD51, DNA pairing that leads to the formation of recombination DNA intermediates (2). We assayed human RuvBL1, because it is thought to the human homolog of the prokaryotic RuvB by sequence homology (31). This protein was found to be a component of a chromatin modifying complex (32). We also tested the hRAD51 paralogs RAD51C and XRCC3. Nuclear extracts prepared from cell lines carrying mutations in the genes encoding these proteins were shown to have reduced levels of HJ processing activity (12). Of all the proteins examined, only the RecQL1 immunoprecipitation eluate exhibited HJ branch migration activity. None of the other proteins alone or by coimmunoprecipitation was capable of branch migrating HJs. Immunopurified FLAG-tagged eluates used in this assay were also analyzed by SDS–PAGE followed by western blotting with anti-FLAG antibody. The corresponding eluates contained FLAG-tagged proteins with the correct molecular masses (Figure 3B).
Figure 3.
(A) Holliday junction branch migration assay with eluates of anti-FLAG immunoprecipitations from HEK293 cells transfected with various vectors containing FLAG-tagged proteins. Eluates were mixed with 32P-labelled synthetic Holliday junctions and ATP as indicated. 32P-labelled DNA products were separated by PAGE and analyzed by autoradiography. (B) Western blot analysis of the FLAG-tagged protein eluates used in the branch migration assay. Gel was loaded; (lane 1) FLAG-RAD54L, (lane 2) FLAG-RuvBL1, (lane 3) FLAG-RecQL1, (lane 4) FLAG-RAD51C and (lane 5) FLAG-XRCC3. Blot was probed with anti-FLAG antibody and visualized by chemiluminescence.
RecQ homologs have been shown to suppress illegitimate and homologous recombination in bacteria, yeast and humans (9,33,34). Cultured Bloom's syndrome cells exhibit excessive rates of spontaneous SCE (15) which is reduced upon restoration of BLM expression (34). We therefore wanted to determine whether a reduction of RecQL1 levels in HeLa cells could have a similar effect on SCE. HeLa cells were chosen because they transfect well and form good chromosome spreads. Employing RNA interference (siRNA) we were able to achieve a >95% knockdown in RecQL1 mRNA levels (Figure 4A). SCEs were monitored using the SCE assay, which utilizes the differential staining of sister chromatids from cells that have replicated their DNA in the presence of bromodeoxyuridine (BrdU). The RecQL1-siRNA HeLa cells exhibited an ∼3.5-fold increase in the number of SCEs per cell when compared with wild-type HeLa cells (Figure 4B and C). To assess if RecQL1 was also critical for DNA damage-associated SCE, we treated HeLa cells with mitomycin C (MMC), which is a DNA cross-linking agent that causes DNA replication fork arrest and increases the frequency of SCE events. We observed a similar 3-fold increase in MMC-induced SCEs in the RecQL1-siRNA HeLa cells compared with wild-type HeLa cells. Transfection of a control siRNA to GFP had no effect on the frequency of SCE with or without MMC (data not shown).
Figure 4.
(A) RT-PCR with RNA prepared from wild type HeLa cells (lanes 1,3 and 5) or RecQL1-siRNA HeLa cells (lanes 2, 4 and 6). Primer pairs for RecQL1 were used for reactions loaded in lanes 1 and 2, RAD54L for lanes 3 and 4, and RAD51C for lanes 5 and 6. Products were separated by agarose gel electrophoresis and stained with ethidium bromide. (B) Table summarizing spontaneous and MMC induced SCE in wild type and RecQL1-siRNA HeLa cells. The mean number of SCEs per cell ± the standard deviation and the number of cells counted are shown. (C) Differentially stained chromosome spreads from wild type HeLa cells (left panel) and RecQL1-siRNA HeLa cells (right panel). This experiment was repeated 3 times with similar results.
Here, we report the identification of RecQL1 as the protein responsible for a predominant ATP-dependent HJ branch migration activity present in nuclear extracts prepared from proliferating HEK293 and HeLa cells. Further support for RecQL1 as a critical factor in HJ resolution came from HeLa cells with reduced levels of RecQL1 mediated by siRNA, which exhibit a substantial increase in SCE that is further stimulated by MMC-induced DNA damage. It is important to consider our findings in light of a previous report that a genetic knockout of the RecQL1 gene in the chicken lymphocyte cell line DT40 yielded viable cells with no major defect in SCE (35). RecQ family proteins are widely expressed in most cells, but their abundance is highly variable (30). In immortalized lymphocytes, the WRN protein is by far the most abundant, followed by BLM and RecQL1. We do not know the relative abundance of these proteins in HEK293 or HeLa cells. Moreover, in DT40 cells, the double knockout (RecQL1−/−, BLM−/−) cells had a slower growth rate, elevated spontaneous cell death and increased MMC-induced SCE compared with the BLM−/− cells (35), suggesting functional redundancy among the proteins. The number of SCEs per cell is in the same general range for RecQL1 siRNA treated HeLa cells (17-34 SCEs/cell) compared with DT40 knockout cells (26-64 SCEs/cell) (35).
Human RecQL1 is a member of the RecQ helicase family of proteins, which participate in maintaining genomic stability as well as double-strand DNA repair (24,36,37). RecQ helicases are also proposed to function during DNA replication in restoring stalled or broken replication forks through homologous recombination (24). Inherited mutations in the genes encoding three members of this family, BLM, WRN and RecQL4, cause Bloom's syndrome, Werner's syndrome and Rothmund-Thomson syndrome, respectively. All three display genomic instability and a predisposition to cancer in humans or mouse models (24,38). Mutations in the RecQL1 gene have not been shown to be associated with any human disorders as of yet. Since it is likely that RecQL1 is a predominant HJ branch migration protein in proliferating cells, its loss may be too detrimental for survival. Alternately, this protein may serve other critical functions in development beyond an overlapping role with other RecQ family members in genomic stability.
Acknowledgments
The authors would like to thank Jane Flint, Peter Houston, Humayra Ali, Victoria Cowling and Brenden Rickards for helpful discussions and critical reading of the manuscript. The authors would also like to thank Nathan Ellis for help with the SCE assays. This study was supported by a grant from the NIH/NCI to MDC. Funding to pay the Open Access publication charges for this article was provided by NIH RO1 CA055248.
Conflict of interest statement. None declared.
REFERENCES
- 1.West S.C. Molecular views of recombination proteins and their control. Nature Rev. Mol. Cell. Biol. 2003;4:435–445. doi: 10.1038/nrm1127. [DOI] [PubMed] [Google Scholar]
- 2.Sigurdsson S., Van Komen S., Petukhova G., Sung P. Homologous DNA pairing by human recombination factors Rad51 and Rad54. J. Biol. Chem. 2002;277:42790–42794. doi: 10.1074/jbc.M208004200. [DOI] [PubMed] [Google Scholar]
- 3.West S.C. Processing of recombination intermediates by the RuvABC proteins. Annu. Rev. Genet. 1997;31:213–244. doi: 10.1146/annurev.genet.31.1.213. [DOI] [PubMed] [Google Scholar]
- 4.Shah R., Cosstick R., West S.C. The RuvC protein dimer resolves Holliday junctions by a dual incision mechanism that involves base-specific contacts. EMBO J. 1997;16:1464–1472. doi: 10.1093/emboj/16.6.1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Gool A.J., Shah R., Mezard C., West S.C. Functional interactions between the Holliday junction resolvase and the branch migration motor of Escherichia coli. EMBO J. 1998;17:1838–1845. doi: 10.1093/emboj/17.6.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harmon F.G., Kowalczykowski S.C. RecQ helicase, in concert with RecA and SSB proteins, initiates and disrupts DNA recombination. Genes Dev. 1998;12:1134–1144. doi: 10.1101/gad.12.8.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakayama K., Irino N., Nakayama H. The recQ gene of Escherichia coli K12: molecular cloning and isolation of insertion mutants. Mol. Gen. Genet. 1985;200:266–271. doi: 10.1007/BF00425434. [DOI] [PubMed] [Google Scholar]
- 8.Courcelle J., Hanawalt P.C. RecQ and RecJ process blocked replication forks prior to the resumption of replication in UV-irradiated Escherichia coli. Mol. Gen. Genet. 1999;262:543–551. doi: 10.1007/s004380051116. [DOI] [PubMed] [Google Scholar]
- 9.Hanada K., Ukita T., Kohno Y., Kato J., Ikeda H. RecQ DNA helicase is a suppressor of illlegitamate recombination in Escherichia coli. Proc. Natl Acad. Sci. USA. 1997;94:3860–3865. doi: 10.1073/pnas.94.8.3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Constantinou A., Davies A.A., West S.C. Branch migration and Holliday junction resolution catalyzed by activities from mammalian cells. Cell. 2001;104:259–268. doi: 10.1016/s0092-8674(01)00210-0. [DOI] [PubMed] [Google Scholar]
- 11.Elborough K.M., West S.C. Resolution of synthetic Holliday junctions in DNA by an endonuclease activity from calf thymus. EMBO J. 1990;9:2931–2936. doi: 10.1002/j.1460-2075.1990.tb07484.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Y., Masson J.Y., Shah R., O'Regan P., West S.C. RAD51C is required for Holliday junction processing in mammalian cells. Science. 2004;303:243–246. doi: 10.1126/science.1093037. [DOI] [PubMed] [Google Scholar]
- 13.Ellis N.A., Groden J., Ye T.Z., Straughen J., Lennon D.J., Ciocci S., Proytcheva M., German J. The Bloom's syndrome gene product is homologous to RecQ helicases. Cell. 1995;83:655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- 14.Karow J.K., Constantinou A., Li J.L., West S.C., Hickson I.D., van Gool A.J., Shah R., Mezard C. The Bloom's syndrome gene product promotes branch migration of Holliday junctions. Proc. Natl Acad. Sci. USA. 2000;97:6504–6508. doi: 10.1073/pnas.100448097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chaganti R.S., Schonberg S., German J. A manyfold increase in sister chromatid exchanges in Bloom's syndrome lymphocytes. Proc. Natl Acad. Sci. USA. 1974;71:4508–4512. doi: 10.1073/pnas.71.11.4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meetei A.R., Sechi S., Wallisch M., Yang D., Young M.K., Joenje H., Hoatlin M.E., Wang W. A multiprotein nuclear complex connects Fanconi anemia and Bloom syndrome. Mol. Cell Biol. 2003;23:3417–3426. doi: 10.1128/MCB.23.10.3417-3426.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu L., Hickson I.D. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- 18.Gangloff S., McDonald J.P., Bendixen C., Arthur L., Rothstein R. The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: a potential eukaryotic reverse gyrase. Mol. Cell. Biol. 1994;14:8391–8398. doi: 10.1128/mcb.14.12.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ira G., Malkova A., Liberi G., Foiani M., Haber J.E. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell. 2003;115:401–411. doi: 10.1016/s0092-8674(03)00886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu C.E., Oshima J., Fu Y.H., Wijsman E.M., Hisama F., Alisch R., Matthews S., Nakura J., Miki T., Ouais S., et al. Positional cloning of the Werner's syndrome gene. Science. 1996;272:258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]
- 21.Gebhart E., Bauer R., Raub U., Schinzel M., Ruprecht K.W., Jonas J.B. Spontaneous and induced chromosomal instability in Werner syndrome. Hum. Genet. 1988;80:135–139. doi: 10.1007/BF00702855. [DOI] [PubMed] [Google Scholar]
- 22.Fukuchi K., Martin G.M., Monnat R.J., Jr Mutator phenotype of Werner syndrome is characterized by extensive deletions. Proc. Natl Acad. Sci. USA. 1989;86:5893–5897. doi: 10.1073/pnas.86.15.5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Constantinou A., Tarsounas M., Karow J.K., Brosh R.M., Bohr V.A., Hickson I.D., West S.C. Werner's syndrome protein (WRN) migrates Holliday junctions and co-localizes with RPA upon replication arrest. EMBO Rep. 2000;1:80–84. doi: 10.1093/embo-reports/kvd004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khakhar R.R., Cobb J.A., Bjergbaek L., Hickson I.D., Gasser S.M. RecQ helicases: multiple roles in genome maintenance. Trends Cell Biol. 2003;13:493–501. doi: 10.1016/s0962-8924(03)00171-5. [DOI] [PubMed] [Google Scholar]
- 25.Seki M., Miyazawa H., Tada S., Yanagisawa J., Yamaoka T., Hoshino S., Ozawa K., Eki T., Nogami M., Okumura K., et al. Molecular cloning of cDNA encoding human DNA helicase Q1 which has homology to Escherichia coli Rec Q helicase and localization of the gene at chromosome 12p12. Nucleic Acids Res. 1994;22:4566–4573. doi: 10.1093/nar/22.22.4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Puranam K.L., Blackshear P.J. Cloning and characterization of RECQL, a potential human homologue of the Escherichia coli DNA helicase RecQ. J. Biol. Chem. 1994;269:29838–29845. [PubMed] [Google Scholar]
- 27.Dracopoli N.C., Haines J.L., Korf B.R., Moir D.T., Morton C.C., Seidman C.E., Seidman J.G., Smith D.R. Current Protocols in Human Genetics. New York, NY: Wiley; 1994. [Google Scholar]
- 28.Constantinou A., Chen X.B., McGowan C.H., West S.C. Holliday junction resolution in human cells: two junction endonucleases with distinct substrate specificities. EMBO J. 2002;21:5577–5585. doi: 10.1093/emboj/cdf554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cui S., Arosio D., Doherty K.M., Brosh R.M., Jr, Falaschi A., Vindigni A. Analysis of the unwinding activity of the dimeric RECQ1 helicase in the presence of human replication protein A. Nucleic Acids Res. 2004;32:2158–2170. doi: 10.1093/nar/gkh540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kawabe T., Tsuyama N., Kitao S., Nishikawa K., Shimamoto A., Shiratori M., Matsumoto T., Anno K., Sato T., Mitsui Y., et al. Differential regulation of human RecQ family helicases in cell transformation and cell cycle. Oncogene. 2000;19:4764–4772. doi: 10.1038/sj.onc.1203841. [DOI] [PubMed] [Google Scholar]
- 31.Qiu X.B., Lin Y.L., Thome K.C., Pian P., Schlegel B.P., Weremowicz S., Parvin J.D., Dutta A. An eukaryotic RuvB-like protein (RUVBL1) essential for growth. J. Biol. Chem. 1998;273:27786–27793. doi: 10.1074/jbc.273.43.27786. [DOI] [PubMed] [Google Scholar]
- 32.Ikura T., Ogryzko V.V., Grigoriev M., Groisman R., Wang J., Horikoshi M., Scully R., Qin J., Nakatani Y. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell. 2000;102:463–473. doi: 10.1016/s0092-8674(00)00051-9. [DOI] [PubMed] [Google Scholar]
- 33.Watt P.M., Hickson I.D., Borts R.H., Louis E.J. SGS1, a homologue of the Bloom's and Werner's syndrome genes, is required for maintenance of genome stability in Saccharomyces cerevisiae. Genetics. 1996;144:935–945. doi: 10.1093/genetics/144.3.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ellis N.A., Proytcheva M., Sanz M.M., Ye T.Z., German J. Transfection of BLM into cultured bloom syndrome cells reduces the sister-chromatid exchange rate toward normal. Am. J. Hum. Genet. 1999;65:1368–1374. doi: 10.1086/302616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang W., Seki M., Narita Y., Nakagawa T., Yoshimura A., Otsuki M., Kawabe Y., Tada S., Yagi H., Ishii Y., et al. Functional relation among RecQ family helicases RecQL1, RecQL5, and BLM in cell growth and sister chromatid exchange formation. Mol. Cell. Biol. 2003;23:3527–3535. doi: 10.1128/MCB.23.10.3527-3535.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kusano K., Sunohara Y., Takahashi N., Yoshikura H., Kobayashi I. DNA double-strand break repair: genetic determinants of flanking crossing-over. Proc. Natl Acad. Sci. USA. 1994;91:1173–1177. doi: 10.1073/pnas.91.3.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adams M.D., McVey M., Sekelsky J.J. Drosophila BLM in double-strand break repair by synthesis-dependent strand annealing. Science. 2003;299:265–267. doi: 10.1126/science.1077198. [DOI] [PubMed] [Google Scholar]
- 38.Mann M.B., Hodges C.A., Barnes E., Vogel H., Hassold T.J., Luo G. Defective sister-chromatid cohesion, aneuploidy and cancer predisposition in a mouse model of type II Rothmund-Thomson syndrome. Hum. Mol. Genet. 2005;14:813–825. doi: 10.1093/hmg/ddi075. [DOI] [PubMed] [Google Scholar]