Abstract
The primary function of the HIV-1 regulatory protein Tat, activation of transcription from the viral LTR, is highly regulated by complex interactions between Tat and a number of host cell proteins. Tat nuclear import, a process mediated by importin β, is a prerequisite for its activity. Here, we report and characterize the interaction of the human inhibitor of MyoD family domain-containing protein (I-mfa), HIC, with Tat at a biochemical and a functional level. This interaction was shown to occur in vivo and in vitro and to involve the nuclear localization signal and the transactivation responsive element-binding domains of Tat and the I-mfa domain of HIC. Coexpression of HIC and Tat resulted in the down-regulation of transactivation of the HIV-1 LTR, and colocalization studies revealed the cytoplasmic sequestration of Tat by HIC. Functionally this sequestration appears to be the underlying mechanism of LTR transcriptional repression by HIC and represents a unique mechanism for the control of Tat activity and regulation of HIV-1 replication.
Keywords: NLS, protein-protein interaction, nuclear import
HIV type I (HIV-1) encodes the transactivator protein, Tat, a polypeptide of 86-101 aa in size, which is essential for efficient transcription of the provirus and for HIV-1 replication (1, 2). The primary function of Tat is the activation of transcription from the HIV-1 LTR where it binds specifically to the transactivation responsive element region (3). Although Tat may enhance the rate of transcription initiation, its primary function involves promoter clearance and transcriptional elongation (4). Protein-protein interactions are essential for Tat activity as exemplified by the interaction and recruitment of the positive transcription elongation factor b complex (P-TEFb) to the viral promoter via a specific interaction with Tat and cyclin T1, which is part of a complex with the cyclin-dependent kinase 9 (CDK9) kinase (4, 5). Once recruited to the transactivation responsive element region, the complex phosphorylates the RNA polymerase II C-terminal domain, promoting its processivity (4-6).
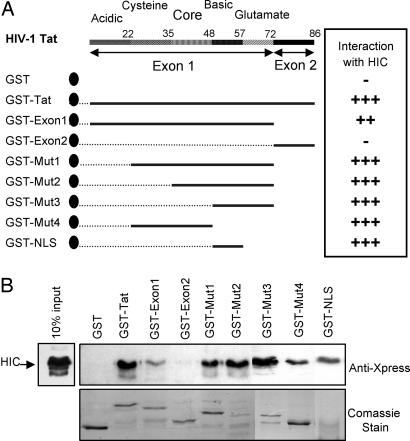
Encoded by two exons, Tat contains several distinct regions based on its amino acid composition (Fig. 1 A). The core, acidic, and cysteine regions correspond to the minimal activation domain whereas the basic region together with the core and glutamine-rich regions confers RNA-binding activity. In addition, the basic domain contains a nuclear localization signal (NLS), which mediates the nuclear transport of Tat. Tat is primarily localized in the nucleus/nucleolus, which is a prerequisite for its transactivation function (7-11).
Fig. 1.
Direct interaction between HIV-1 Tat and HIC in vitro by using GST pull-down assays. (A) Schematic representation of HIV-1 Tat and deletion mutants. (B) Detection of HIC, Tat, and its deletion mutants by Western Blot analysis. Shown are Coomassie-stained polyacrylamide gels documenting the quantity and the quality of GST-Tat and GST.
The trafficking of proteins in and out the nucleus is mediated by members of the importin β family of nuclear receptors via specific recognition of their localization signal such as NLS and/or nuclear export signal (12, 13). Nuclear import of Tat is an energy-dependant process and is selectively mediated by the soluble import factor importin β (14). This involves the molecular recognition of the NLS by importin β, with no apparent need for the adapter protein importin α (14). Subsequently, importin β interacts with nuclear pore complexes and mediates the translocation of Tat into the nucleus.
The identification of key interactions of Tat with host proteins is essential to our understanding of HIV replication and of the potential of the virus to disrupt normal cellular activities (15, 16). In this report, we describe the interaction between Tat and the human inhibitor of MyoD family (I-mfa) domain-containing protein, HIC (17). HIC belongs to the I-mfa domain-containing protein family and shares with I-mfa a highly conserved cysteine rich C-terminal domain. HIC and I-mfa have also been reported to function as a transcriptional regulator and a modulator of different pathways including the Wnt and c-jun N-terminal kinase (JNK) pathways (18-20). Although one previous study (17) has reported that HIC can both down-regulate Tatdependant transcription of the HIV-1 LTR and stimulate the activation of the human T-lymphotrophic virus-1 LTR in the presence of the viral regulatory protein Tax, the underlying mechanisms involved, direct or indirect, were not investigated. Subsequently, a study (21) demonstrated that HIC interacts with Tat in vivo via its I-mfa domain and colocalizes with Tat in the nucleolus. In addition, it was shown that ectopic expression of HIC stimulates Tat transactivation of the HIV-1 LTR in HeLa, COS7, and NIH 3T3 cells (21). Here, we clearly demonstrate the direct and specific interaction of Tat and HIC, both in vitro and in vivo, and describe the domains within the proteins involved. In contrast to this report (21), we clearly show that HIC-Tat complex formation impairs the nuclear import of Tat and results in the cytoplasmic accumulation of Tat. This cytoplasmic sequestration of Tat by HIC is associated with the down-regulation of Tat transactivation of the HIV-1 LTR in 293T and COS7 cells. Overall, our data suggest that HIC regulates Tat subcellular distribution, which in turn modulates its transactivation function.
Materials and Methods
Yeast Two-Hybrid Screening (YTHS). By using the Tat-encoding vector PCV1 as a template (22), a PCR product encoding the full length of Tat(1-86) was cloned in PAST2-1 (Clontech) and used as bait in the YTHS. The screening was performed by using the Human Leukocyte MATCHMAKER cDNA library (Clontech) containing 3.5 × 106 independent clones. The Clontech MATCHMAKER GAL4 two-hybrid system 2 (K1604-1) was used according to the manufacturer's instructions.
Expression and Purification of Recombinant Proteins. The full length of Tat was cloned in pGEX-5X (Amersham Pharmacia). Deletion mutants GST-exon1 (first exon of Tat), GST-exon2 (second exon of Tat), GST-Mut-1(22-72), GST-Mut-2(36-72), GST-Mut-3(47-72), and GST-Mut-4(22-47) were generated by cloning the respective PCR products into pGEX-5X. Additionally, a PCR fragment encoding the deletion mutant GST-NLS was generated by annealing two complementary oligonucleotides encoding the amino acid sequence 49-RKKRRQRRRPPQ-60 and cloned into pGEX-5X. GST-fusion proteins were purified from transformed BL21 cell lysates by using gluthatione Sepharose beads (Amersham Pharmacia). Full-length HIC(2-246) was amplified by PCR from clone 254 and cloned into pBAD/His B (Invitrogen). Recombinant proteins were expressed in Top 10 F′ (Invitrogen) cells and purified by using nickel resin (Qiagen) according to the manufacturer's instructions.
In Vitro “Pull-Down” Assays. After immobilization of equivalent amounts of GST, GST-Tat, and deletion mutants onto Gluthatione-agarose beads, purified recombinant protein HIC was added. Bound proteins were eluted twice in 25 μl of glutathione elution buffer (Amersham Pharmacia). One-half of the eluted complexes was analyzed by Western blot by using the anti-Xpress Ab (Invitrogen) to reveal the presence of HIC. The other half was used for Coomassie blue staining to verify the quality and quantity of GST-fused proteins used in the assay.
Coimmunoprecipitation Assays. Tat was cloned in pCAGGS (23). HIC was cloned into pFLAG-CMV-2 (Sigma). A HIC(2-144) was subcloned into pFLAG-CMV-6a (Sigma), and HIC(144-246) was subcloned into pFLAG-CMV6c (Sigma). By using FuGENE 6 (Roche), 293T cells were transfected with 3 μg of pCAGGS or pCAGGS-Tat and 2 μg of pFLAG, pFLAG-HIC, pFLAG-HIC(2-144), or pFLAG-HIC(144-246). After transfection (48 h), cells were lysed and incubated with ANTI-FLAG M2-agarose affinity resin (Sigma). Bound complexes were eluted twice in 20 μl of 0.1 M glycine at pH 3.5. Western blotting was carried out with ANTI-FLAG M5 (Sigma) or with a mAb to HIV-1 Tat (NT3 2D1) (3).
Transient Transfections and Luciferase Assays. The HIV-1 promoter region from -644 bp to +78 bp was cloned upstream to the firefly luciferase gene in pGL3 (pGL3-LTR) (Promega). The 293T cells were transfected by using FuGENE 6 (Roche) with pGL3-LTR and pRL-TK (Promega) or pRL-β-actin to control transfection efficiency, pCAGGS or pCAGGS-Tat, and pFLAG, pFLAG-HIC, pFLAG-HIC(2-144), or pFLAG-HIC(144-246). Total amounts of DNA were equilibrated by addition of parent plasmid. Dual-luciferase assays (Promega) were performed according to the manufacturer's instructions. Luciferase activity was measured 24 or 48 h posttransfection and normalized against pRL-TK or pRL-β-actin activity.
Indirect Immunofluorescence and Confocal Microscopy. By using FuGENE 6 (Roche), Cos7 cells were cotransfected with 0.5 μg of pCAGGS or pCAGGS-Tat and 0.5 μg of pFLAG, pFLAG-HIC or pFLAG-HIC(2-144). At 48 h posttransfection, cells were fixed in 4% paraformaldehyde and permeabilized with 0.2% Triton X-100. Indirect immunofluorescence was performed with rabbit polyclonal Ab against HIV-1 Tat (HIV-1 BH10 Tat antiserum) (24) and mouse monoclonal ANTI-FLAG M2 (Sigma) Ab as primary Abs, and with Texas red-conjugated goat anti-rabbit Ig (Molecular Probes) and FITC-conjugated rabbit anti-mouse Ig (DAKO) as secondary Abs. Stained cells were visualized by using a Leica inverted scanning confocal microscope, and images were captured by using lcs lite software (Leica).
Subcellular Fractionation and Western Blot Analysis. By using Fu-GENE 6 (Roche), Cos7 cells were cotransfected with 1 or 2 μg of pCAGGS-Tat and 2 μg of pFLAG, pFLAG-HIC. Whole-cell protein extracts were prepared from one-tenth of transfected Cos7 lysed in Leammli sample buffer. Nuclear cytoplasmic fractionation was carried out on the rest of the cells by using the NE-PER kit (Pierce) according to the manufacturer's instructions. The protein concentrations of each fraction were determined with the bicinchoninic acid protein assay (Pierce). Twenty micrograms of total protein from each fraction was subjected to Western blot analysis by using ANTI-FLAG M2 (Sigma) and a mAb to HIV-1 Tat (NT3 2D1) (3). The same membrane was stripped and reprobed with Sp-1 mAb (Santa Cruz Biotechnology) as fractionation control.
Results
YTHS. To identify cellular proteins interacting with Tat, we performed the screening of a Human Leukocyte cDNA library (Clontech) with the YTHS approach, using Tat as a bait. From 80 positive clones resulting from nutritional and colorimetric selection, two distinct clones (clone 208 and clone 254) were found to contain a cDNA of 879 bp (GenBank accession no. AY196485) encompassing the full-length open-reading frame of the HIC protein.
Direct Interaction of Tat and HIC in Vitro. To validate the interaction between Tat and HIC, we used in vitro GST-based pull-down assays. GST-Tat fusion protein and GST alone were immobilized on glutathione-Sepharose beads and incubated with purified recombinant 6× His-Xpress-tagged HIC protein. As shown in Fig. 1B, HIC was specifically retained by GST-Tat, whereas there was no significant association between HIC and beads containing GST alone. Thus, HIC interacts directly and physically with Tat without the involvement of a cellular intermediate.
To delineate the HIC-interaction domains of Tat, we carried out deletion studies by using the GST pull-down approach with truncated forms of Tat (Fig. 1 A and C). The first exon of Tat (Mut-1) was found to retain its binding capacity while exhibiting a relatively weaker affinity for HIC. In contrast, the second exon (Mut-2) had lost this activity, suggesting a minimal binding site in the first exon. A series of deletion mutants Mut-1, Mut-2, and Mut-3, lacking the acidic, the cysteine, and the core domains, respectively, exhibited no significant differences in HIC-Tat complex formation. Finally, a mutant encoding the NLS domain of Tat located between residues 48 and 60, GST-NLS, was sufficient to interact with HIC. In addition, Mut-4, which includes the cysteine and the core domains, also showed significant binding. Thus, it would appear that the interaction involves a number of regions within the protein, specifically the first exon, and to different degrees the cysteine, the core, and the NLS regions. The NLS region contains six arginine residues, which are essential for Tat nuclear localization and for its binding to the transactivation responsive element region in the HIV-1 LTR.
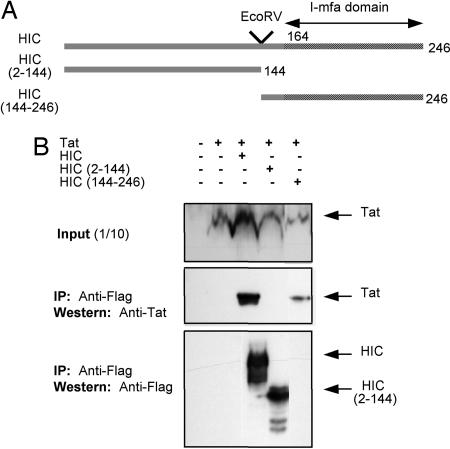
Tat Interacts with HIC in Vivo. After cotransfection (48 h) of 293T cells, immunoprecipitation of HIC was performed, and Tat was specifically detected in the eluted complex when HIC and Tat were coexpressed (Fig. 2B), providing additional evidence of the Tat-HIC complex formation in vivo. To further determine the specificity of the Tat-HIC complex formation, Tat was coexpressed with deletion mutants HIC(2-144) and HIC(144-246) (Fig. 3 A and C). Only HIC(144-246), containing the I-mfa domain, retained its binding property, whereas HIC(2-144) did not. However, for undetermined reasons HIC(2-144) could not be detected by Western blot. Thus, Tat-HIC complex formation is specific and depends on the presence of the C-terminal cysteine-rich I-mfa domain. In addition, it could be noted that when HIC and Tat were coexpressed, the level of Tat expression was increased.
Fig. 2.
HIV-1 Tat and HIC interact in vivo.(A) Schematic representation of HIC and deletion mutants. (B) Immunoprecipitates were assayed by Western blotting to detect coimmunoprecipitation of HIV-1 Tat and to show the corresponding expression levels of HIC, HIC(2-144), and HIC(144-246).
Fig. 3.
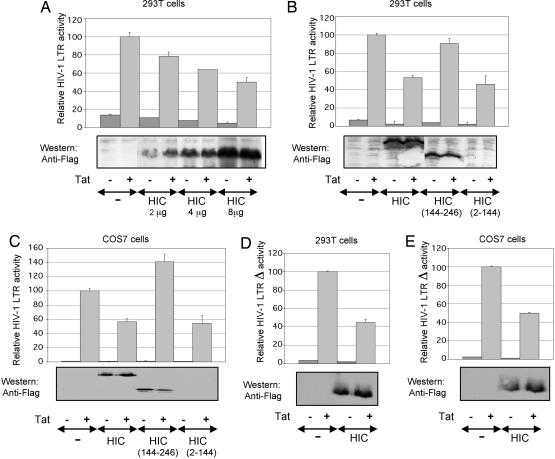
Down-regulation of Tat-mediated transactivation of the HIV-1 LTR by HIC. (A) The 293T cells were transfected with 0.5 μg of reporter pGL3-LTR and 0.05 μg of p-RL-TK in combination with 0.05 μg of pCAGGS-Tat and 0, 2, 4, and 8 μg of pFLAG-HIC. The relative luciferase activity is compared with 100% for Tat transactivation of pGL3-LTR. Error bars indicate the SD of the mean of triplicate samples. (A-E Lower) Western blot shows the corresponding levels of HIC expression. (B) The I-mfa domain is involved in the down-regulation of HIV-1 LTR by HIC. Conditions were as above, but 293T cells were transiently transfected with 0.3 μg of reporter pGL3-LTR and 0.03 μg of p-TK in combination with 0.03 μg of pCAGGS-Tat and 4 μg of pFLAG-HIC, pFLAG-HIC(2-144), or pFLAG-HIC(144-246). (C) As above, but Cos7 cells were transiently transfected with 0.1 μg of reporter pGL3-LTR and 0.03 μgof p-RL-bactin in combination with 0.005 μg of pCAGGS-Tat and 2 μg of pFLAG-HIC, pFLAG-HIC(2-144), or pFLAG-HIC(144-246). (D and E) As described in B and C, but 293T and Cos7 cells were transiently transfected with pGL3-LTRΔ instead of pGL3-LTR.
HIC Down-Regulates Tat-Dependent Transcription from the HIV-1 LTR. To assess the functional consequences of Tat-HIC complex formation on Tat-mediated transactivation of the HIV-1 LTR, 293T cells were transfected with a luciferase-reporter plasmid under the control of HIV-1 LTR and increasing amounts of HIC with or without Tat (Fig. 3A). In corroboration of the results of Thebault et al. (17), who performed this experiment in human T-lymphoblastoid (CEM) cells, HIC decreased LTR transactivation by Tat in a dose-dependant manner. A maximal down-regulation of transactivation was achieved with 8 μg of HIC corresponding to 50% inhibition. The inhibitory effect is dependant on the presence of the I-mfa domain because HIC(2-144) deletion mutant showed no effect on the LTR transactivation by Tat, whereas HIC(144-246) deletion mutant down-regulated transactivation in a similar fashion to the full-length HIC protein (Fig. 3B). Accordingly, the same experiments were performed in Cos7 cells and similar results were obtained (Fig. 3C).
To narrow the number of potential cellular transcription factors with binding sites on the HIV-1 LTR, which could be influenced by the ectopic expression of HIC, transactivation studies were performed by using pGL3-LTRΔ, a luciferase-reporter plasmid under the control of a truncated HIV-1 LTR lacking the modulatory region (-450 to -105). Under this conditions, Tat-mediated transactivation of HIV-1 LTRΔ in 293T and Cos7 cells was down-regulated by HIC in a similar fashion as was the full-length HIV-1 LTR (Fig. 3 D and E). Hence, the cis-acting sequences within the modulatory region and their corresponding transcription factors were not involved in the ability of HIC to repress the HIV-1 promoter.
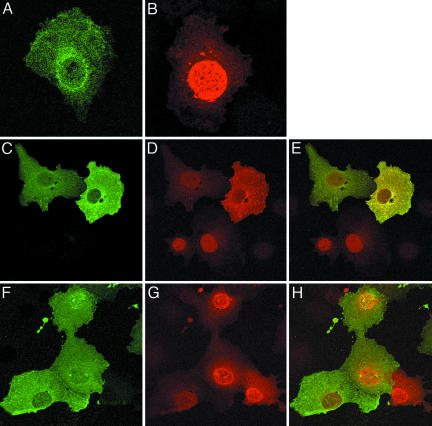
Intracellular Localization of HIV-1 Tat and HIC. Indirect immunofluorescence staining and confocal microscopy were used to confirm the colocalization of Tat and HIC and to determine whether HIC may alter the expected nuclear localization of Tat. Cos7 cells expressing Tat exhibited two patterns of expression; in >80% of transfected cells, Tat was primarily localized in the nucleus and distributed in a speckled pattern (Fig. 4A); in the remainder of the cells, the protein was distributed diffusely in both the nucleus and cytoplasm. Cos7 cells expressing HIC exhibited a diffuse cytoplasmic and nuclear staining pattern, which was more intense in the perinuclear region (Fig. 4B). In cells coexpressing HIC and Tat, the merge image shows that the interacting proteins were colocalized (yellow) in the cytoplasm (Fig. 4E). In 80% of cells, coexpression of both proteins resulted in a distinct redistribution of Tat to the cytoplasm with only weak expression of Tat in the nucleus, demonstrating that HIC sequesters Tat in the cytoplasm. In the additional 20% of cells expressing the two proteins, Tat was equally distributed in both the cytoplasm and the nucleus. As expected, cells in the same microscopic field expressing Tat alone displayed a clear intranuclear distribution pattern (Fig. 4E, arrows). To confirm that the sequestration of Tat in the cytoplasm is the result of HIC-Tat complex formation, HIC(2-144), lacking the I-mfa domain responsible for the interaction of the two proteins, and Tat were coexpressed in Cos7 cells. As is shown in Fig. 4 F-H, HIC(2-144) did not alter the pattern of Tat expression. There was no cytoplasmic sequestration of Tat, which remained primarily localized in the nucleus.
Fig. 4.
Intracellular localization of Tat and HIC. (A) Cos7 cells expressing Tat. (B) Cos7 cells expressing HIC. (C-E) Colocalization of HIC and Tat. (C) HIC (green) remains localized in the cytoplasm. (D) Redistribution of Tat (red) in the cytoplasm. (E) Merged images showing colocalization of HIC and Tat (yellow). (F-H) The I-mfa domain is involved in the sequestration of Tat. When HIC(2-144) and Tat are coexpressed, HIC localizes in the cytoplasm (F) and Tat localizes in the nucleus (G). Merged images (H) confirmed the absence of colocalization. Arrows indicate cells expressing Tat only.
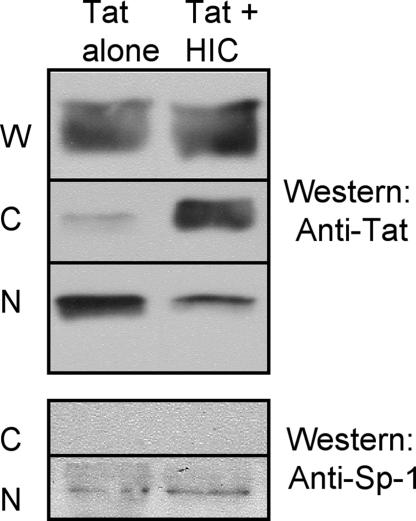
As a complementary approach, the subcellular distribution of Tat was monitored by subcellular fractionation of transfected Cos7 cells, followed by Western blot analysis (Fig. 5). Expression levels of HIC and Tat were first determined in the whole-cell lysate (one-tenth of the transfected cells), and it could be noted that Tat cellular abundance increased when coexpressed with HIC. Therefore, it was necessary to increase the amount of transfected pCAGGS-Tat vector from 1 to 2 μg to obtain a comparable level of Tat expression. As shown in the nuclear and cytoplasmic fractions, Tat was mainly observed in the nuclear fraction when expressed on its own. However, HIC and Tat coexpression resulted in a shift in the steady-state localization of Tat from the nucleus to the cytoplasm confirming our previous results.
Fig. 5.
Subcellular fractionation analysis. Cos7 cells were transfected with the indicated plasmids. Whole-cell lysate (W), cytoplasmic (C), and nuclear (N) fractions were prepared as described in Materials and Methods. Then 20 mg of protein from each fraction was subjected to Western blot analysis by using the indicated Abs. In all of the extracts prepared, Sp1 was present exclusively in the nuclear fractions, which demonstrated the accuracy of the fractionation (Lower).
Discussion
A better understanding of protein-protein interactions underlying the dynamics of virus-host interactions is critical to our understanding of the molecular pathogenesis of HIV-1 infection. In this report, we describe a binary interaction between HIV-1 Tat and the cellular protein HIC, identified while screening a human leukocyte cDNA library with the YTHS approach by using Tat as a bait and validated by relevant biochemical and biological assays. First, in vitro GST pull-down assays demonstrated that HIC and Tat physically and directly interacted. Furthermore, by using the same approach, deletion mutants studies delineated accurately and precisely two functional domains present within Tat first exon, the arginine-rich and cysteine domains, as being independently sufficient for HIC and Tat interaction. Subsequently, coimmunoprecipitation experiments confirmed that HIC-Tat complex formation was taking place within the cellular environment and that this interaction depended on the presence of the I-mfa domain of HIC. HIC-Tat interaction was previously reported; however, the distinct interaction sites within Tat and the direct nature of this interaction remained to be determined (21).
Because HIV transcriptional regulation is primarily orchestrated by Tat, the functional consequences of the HIC-Tat interaction on Tat-dependent activation of the HIV-1 LTR were investigated in vivo. In this report, transactivation assays in 293T and Cos7 cells demonstrated that HIC down-regulated Tatdependant activation of the HIV-1 LTR in a dose-dependant manner. Furthermore, it was shown that this inhibition depended on the I-mfa domain of HIC, which is necessary for HIC-Tat complex formation. These results are consistent with the transactivation studies performed in CEM cells reported by Thebault et al. (17). The HIV-1 LTR is divided into three functional regions according to their influence on LTR activity: a basal promoter, an enhancer region, and a modulatory region. They encompass numerous target sequences for a variety of cellular transcription factors, which positively and/or negatively regulate LTR-driven transcription (25-27). Although it is clear that Tat is the main orchestrator of LTR activation, other cellular factors contribute to the regulation of this activity. Therefore, one cannot rule out a possible interaction between HIC and another cellular transcription factor(s), which could produce the effects described above. Illustrating this possibility is LEF-1, which has three binding sites located in the regulatory region of the LTR and has been shown to be an activator of the HIV-1 LTR (28, 29). In a recent report, HIC and I-mfa have been shown to bind to LEF-1 through their I-mfa domain (19); consequently this additional interaction could influence the overall effect of HIC on LTR transactivation. Therefore, to narrow the number of potential cofactors, which could be influenced by the ectopic expression of HIC, transactivation assays were performed in 293T and Cos7 cells in which a reporter gene under the control of a truncated version of the HIV-1 LTR selectively harboring the basal and enhancer regions. These conditions did not affect HIC effect on Tat-dependent activation of the LTR, thus excluding the involvement of cofactors such as LEF-1 and further establishing the HIC-Tat complex formation as the underlying mechanism involved in HIC inhibition of transactivation. Of note, it is interesting that HIC inhibits Tat-mediated LTR transactivation because the first observed consequence of HIC and Tat coexpression was the increased cellular accumulation of Tat protein, which might be expected to result in an enhancement of LTR transactivation.
Previous studies have suggested that interactions involving HIC or I-mfa result in the inhibition of the activities of their partner proteins in a similar fashion, either by preventing nuclear localization or DNA binding or both (18, 20, 30). This is directly supported by our subcellular colocalization studies where it could be shown at a single-cell level that although Tat was predominantly nuclear when expressed on its own, coexpression of HIC and Tat resulted in a distinctive localization of Tat in the cytoplasm. In parallel, our subcellular fractionation studies, which broaden the observation to the whole population of transfected cells, confirmed the redistribution of Tat to the cytoplasm together with a dramatic decrease in nuclear localization.
Tat nucleocytoplasmic transport is an active and signal-mediated process orchestrated by the soluble import factor importin β, which selectively binds its NLS and mediates the translocation of Tat to the nucleus (14). The initial step of NLS molecular recognition by specific import factors is critical in directing proteins to their correct subcellular localization, and enhancing or preventing this step is central to the control of the flux of proteins between the cytoplasm and nucleus (17). Consistent with this are our findings that one of Tat-interacting regions with HIC includes the NLS domain. Together our data suggest that HIC binding to Tat would mask the NLS domain of Tat and consequently sterically interfere with its molecular recognition by importin β and its subsequent nuclear translocation. Although it is clear that HIC reduces the rate of nuclear import, it does not impose a complete block. It remains to be determined whether the net nuclear amounts of Tat depend on the relative abundance of HIC and importin β. Notably, in our systems, HIC is expressed under ectopic conditions whereas importin β is expressed at endogenous levels, and as such, the observed cyoplasmic localization of Tat would appear to be the result of an overwhelming competition of HIC over importin β for the Tat NLS. Alternatively, in response to specific stimuli, some regulatory mechanism could govern the relative affinity of Tat for HIC and importin β, and mediate the release of Tat from sites of sequestration in the cytoplasm. Possible regulatory mechanisms could involve posttranslational modifications of the binding partners such as phosphorylation or acetylation, which could enhance or disrupt complex formation.
Based on the interaction sites of HIC and Tat, additional mechanisms could mediate the inhibition of Tat transactivation function by HIC. First, HIC interaction via the basic domain could interfere with the binding of Tat with the transactivation responsive element region of the LTR. Secondly, HIC interaction with the cysteine-rich domain could disrupt the binding of Tat with its cellular cofactors such as cyclin T1. At present, the relative contribution of the amino acid sequence and overall charge of the Tat NLS to the HIC interaction and inhibition of Tat function is unclear. This is currently under investigation by using scrambled peptides and related NLS sequences (HIV-1 Rev and human T-lymphotrophic virus-1 Rex) to define both the sequence specificity and charge requirements for such interactions.
In a recent study, Young et al. (21) assessed the functional consequences of HIC-Tat complex formation and reported conflicting results regarding the effect of HIC on Tat transactivation function and Tat subcellular distribution. Their transactivation studies performed in Cos7 resulted in an enhancement of Tat activation of the LTR when coexpressed with HIC. Furthermore, they have suggested that Tat causes the redistribution of HIC in the nucleoli, an effect also reported to depend on the presence of the I-mfa domain. This apparent discrepancy presumably reflects the inherent differences in the setting of each colocalization studies. Young et al. (21) used a truncated mutant of Tat, which only encompassed the first exon (Tat 72 aa) and with HIC and Tat tagged with large fusion proteins, the EGFP and the red fluorescent protein from Discosoma (DsRed), respectively, which could disturb their respective structures and/or complex formation. Although the reasons for the conflicting results are not clear, two scenarios remain possible. First, although both studies agree that the I-mfa domain is crucial for the binding of HIC to Tat, it is not known whether sequestration of Tat to the cytoplasm requires the integrity of HIC N terminus, which could be impaired by the fusion of HIC to GFP. Alternatively or additionally, because they form obligate tetramers, DsRed used as a fusion protein could results in the undesirable formation of multimers of Tat. In accordance with our sequestration model, this could be especially problematic in the situation where HIC proteins would mask all but one NLS domain, which by itself should be sufficient to redirect the large complex to the nucleus and or the nucleoli.
Restricting the access of transcription factors to their target genes is an important aspect of transcriptional regulation. Overall, this study has provided a comprehensive set of biochemical and functional data supporting a role for HIC in the repression of Tat-mediated transactivation of the HIV-1 LTR via a unique mechanism involving protein-protein interaction whereby HIC can physically restrict the distribution of the HIV-1 transactivator protein Tat to the cytoplasm. Ultimately, this could have important implications for our understanding of HIV-1 postintegration latency.
Acknowledgments
The mAb to HIV-I Tat (NT3 2D1) from Dr. J. Karn was obtained through the National Institute for Biological Standards and Control Centralised Facility for AIDS Reagents supported by European Union Program European Vaccine Against AIDS and the U.K. Medical Research Council. The rabbit antiserum to HIV-1 Tat (HIV-1 BH10 Tat antiserum) from Bryan Cullen was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health. This work was supported by the Japanese Foundation for AIDS Prevention.
Author contributions: V.W.G. and W.W.H. designed research; V.W.G. performed research; V.W.G., M.D., and K.H. contributed new reagents/analytical tools; V.W.G., N.S., and W.W.H. analyzed data; and V.W.G. and W.W.H. wrote the paper.
Confict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: I-mfa, inhibitor of MyoD family; HIC, human I-mfa domain-containing protein; NLS, nuclear localization signal; YTHS, yeast two-hybrid screening.
Data deposition: The sequence reported in this paper was deposited in the GenBank database (accession no. AY196485).
References
- 1.Dayton, A. I., Sodroski, J. G., Rosen, C. A., Goh, W. C. & Haseltine, W. A. (1986) Cell 44, 941-947. [DOI] [PubMed] [Google Scholar]
- 2.Fisher, A. G., Feinberg, M. B., Josephs, S. F., Harper, M. E., Marselle, L. M., Reyes, G., Gonda, M. A., Aldovini, A., Debouk, C. & Gallo, R. C. (1986) Nature 320, 367-371. [DOI] [PubMed] [Google Scholar]
- 3.Dingwall, C., Ernberg, I., Gait, M. J., Green, S. M., Heaphy, S., Karn, J., Lowe, A. D., Singh, M., Skinner, M. A. & Valerio, R. (1989) Proc. Natl. Acad. Sci. USA 86, 6925-6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones, K. A. (1997) Genes Dev. 11, 2593-2599. [DOI] [PubMed] [Google Scholar]
- 5.Wei, P., Garber, M. E., Fang, S. M., Fischer, W. H. & Jones, K. A. (1998) Cell 92, 451-462. [DOI] [PubMed] [Google Scholar]
- 6.Cujec, T. P., Okamoto, H., Fujinaga, K., Meyer, J., Chamberlin, H., Morgan, D. O. & Peterlin, B. M. (1997) Genes Dev. 11, 2645-2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ruben, S., Perkins, A., Purcell, R., Joung, K., Sia, R., Burghoff, R., Haseltine, W. A & Rosen, C. A. (1989) J. Virol. 63, 1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siomi, H., Shida, H., Maki, M. & Hatanaka, M. (1990) J. Virol. 64, 1803-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luznik, L., Martone, M. E., Kraus, G., Zhang, Y., Xu, Y., Ellisman, M. H. & Wong-Staal, F. (1995) AIDS Res. Hum. Retroviruses 11, 795-804. [DOI] [PubMed] [Google Scholar]
- 10.Li, Y. P. (1997) J. Virol. 71, 4098-4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stauber, R. H. & Pavlavis, G. N. (1998) Virology 252, 126-136. [DOI] [PubMed] [Google Scholar]
- 12.Strom, A. & Weis, K. (2001) Genome Biol. 2, 1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mattaj, I. W. & Englmeier, L. (1998) Annu. Rev. Biochem. 67, 265-306. [DOI] [PubMed] [Google Scholar]
- 14.Truant, T. & Cullen, B. R. (1999) Mol. Cell. Biol. 19, 1210-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De la Fuente, C., Santiago, F., Deng, L., Eadie, C., Zilberman, I., Kehn, K., Maddukuri, A., Baylor, S., Wu, K., Lee, C. G., et al. (2002) BMC Biochem. 3, 14-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jeang, K. T., Xiao, H. & Rich, E. A. (1999) J. Biol. Chem. 274, 28837-28840. [DOI] [PubMed] [Google Scholar]
- 17.Thebault, S., Gachon, F., Lemasson, I., Devaux, C. & Mesnard, J. M. (2000) J. Biol. Chem. 275, 4848-4857. [DOI] [PubMed] [Google Scholar]
- 18.Chen, C. M., Kraut, N., Groudine, M. & Weintraub, H. (1996) Cell 86, 731-741. [DOI] [PubMed] [Google Scholar]
- 19.Kusano, S. & Raab-Traub, N. (2002) Mol. Cell. Biol. 22, 6393-6405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Snider, L., Thirlwell, H., Miller, J. R., Moon, R. T., Groudine, M. & Tapscott S. J. (2001) Mol. Cell. Biol. 21, 1866-1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Young, T. M., Wang, Q., Peery, T. & Mathews, M. B. 2003. Mol. Cell. Biol. 23, 6373-6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arya, S. K., Guo, C., Josephs, S. F. & Wong-Stall, F. (1985) Science 22, 69-73. [DOI] [PubMed] [Google Scholar]
- 23.Niwa, H., Yamamura, K. & Miyazaki, S. A. (1991) Gene 108, 193-199. [DOI] [PubMed] [Google Scholar]
- 24.Hauber, J., Perkins, A., Heimer, E. P. & Cullen B. R. (1987) Proc. Natl. Acad. Sci. USA 84, 6364-6368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones, K. A. & Perterlin, M. B. (1994) Annu. Rev. Biochem. 63, 717-743. [DOI] [PubMed] [Google Scholar]
- 26.Pereira, L. A., Bentley, K., Peeters, A., Churchill, M. J. & Deacon, N. J. (2000) Nucleic Acids Res. 28, 663-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang, H. (1999) Annu. Rev. Genet. 33, 905-912. [DOI] [PubMed] [Google Scholar]
- 28.Sheridan, P. L., Sheline, C. T., Cannon, K. & Jones, K. A. (1995) Genes Dev. 9, 2090-2104. [DOI] [PubMed] [Google Scholar]
- 29.Waterman, M. L. & Jones, K. A. (1990) New Biol. 2, 621-636. [PubMed] [Google Scholar]
- 30.Mizugishi, K., Hatayama, M., Tohmonda, T., Ogawa, M., Inoue, T., Mikoshiba, K. & Aruga, J. (2004) Biochem. Biophys. Res. Commun. 320, 233-240. [DOI] [PubMed] [Google Scholar]