Abstract
Functional analysis of the Plasmodium falciparum genome is restricted because of the limited ability to genetically manipulate this important human pathogen. We have developed an efficient transposon-mediated insertional mutagenesis method much needed for high-throughput functional genomics of malaria parasites. A drug-selectable marker, human dihydrofolate reductase, added to the lepidopteran transposon piggyBac, transformed parasites by integration into the P. falciparum genome in the presence of a transposase-expressing helper plasmid. Multiple integrations occurred at the expected TTAA target sites throughout the genome of the parasite. We were able to transform P. falciparum with this piggyBac element at high frequencies, in the range of 10-3, and obtain stable clones of insertional mutants in a few weeks instead of 6–12 months. Our results show that the piggyBac transposition system can be used as an efficient, random integration tool needed for large-scale, whole-genome mutagenesis of malaria parasites. The availability of such an adaptable genetic tool opens the way for much needed forward genetic approaches to study this lethal human parasite.
Keywords: malaria, mutagenesis, transposon
Malaria is a deadly infectious disease, annually causing clinical illness in 400–600 million people and killing 2–3 million (1). Caused by several different Plasmodium species, malaria remains endemic in many tropical and temperate climates. The most severe effect results from infections of P. falciparum in the children of subSaharan Africa. Traditional measures to control malaria are becoming increasingly ineffective because of widespread resistance to many of the available antimalarial drugs and insecticide resistance in the mosquito vectors of the parasite (2–5). There is an urgent need for the development of new drugs and vaccines to reverse a progressive resurgence in malaria morbidity and mortality. Better understanding of the biology of malaria parasites is essential for the development of new interdiction therapies and their efficient use for long-lasting control of this insidious disease.
Application of new technologies has produced a wealth of information in recent years about the genomes, proteomes, and other aspects of the basic composition of the malaria parasites. Many aspects of the parasites' biology can be inferred through these approaches, and, yet, our ability to use this new information to reveal the complex biology of Plasmodium has been painstakingly slow because of the lack of robust and user-friendly molecular genetic tools. Manipulating the Plasmodium genome has been a great challenge because of the very low efficiency of transfection of this parasite, estimated to be ≈10-6 (6). Gene-targeting to identify gene functions is a cumbersome process hindered by the need to build individual targeting plasmids for each homologous recombination and requires a lengthy (6–12 months) selection process to isolate clones of transformed parasites with the desired genome integrants (7–10). Further complicating this process in P. falciparum is the tendency of the parasite to maintain extracellular plasmid DNA as stable self-replicating episomal concatamers (11).
Transposable elements have been widely used as tools to manipulate genomes ranging from different microbes to higher invertebrates like Drosophila, mammals, and even plants. Transposable elements do not occur naturally in many lower eukaryotes, including Plasmodium (12). So far, efficient transposon-mediated random mutagenesis in parasitic protozoa has been reported only in Leishmania (13). There has been one report of transposition in Plasmodium using the Drosophila mariner transposable element, but the transposition events occurred at a very low frequency independent of transposase and with only two integrations in the same locus (14).
The piggyBac transposable element is derived from the cabbage looper moth Trichoplusia ni and is a member of the TTAA-target site-specific class of transposable elements (15–18). piggyBac is a class II transposable element that exclusively targets the tetra nucleotide target site TTAA and always inserts and excises in a precise manner. piggyBac-based transposon vectors have been widely used to manipulate genomes of various invertebrate species, and piggyBac is currently the preferred vector of choice for enhancer trapping, gene discovery, and identifying gene function in Drosophila, other insects, and mammals (19–24). The attribute of piggyBac to nonpreferentially integrate into the genome of Drosophila has made it more attractive than the popular P-element, which seems to have preferential hot spots for insertion in untranslated 5′ regulatory sequences (20). In this report, we tested piggyBac's ability to integrate into the P. falciparum genome to assess its potential for use as a high-throughput tool to manipulate a genetic system that has been otherwise difficult to manipulate.
Methods
Genomic DNA Extraction. P. falciparum genomic DNA was isolated from blood-stage parasites by using a standard phenol/chloroform method. Briefly, parasite cultures were lysed in 0.15% saponin in TSE (50 mM Tris, pH 8.0/50 mM EDTA/100 mM NaCl), incubated for 30 min at 37°C, and washed twice in TSE. The DNA-containing pellet was resuspended in TSE containing 2% SDS and proteinase K (100 μg/ml) and incubated at 37°C overnight. Extraction was performed twice in phenol/chloroform (1:1) and once in chloroform. DNA was precipitated from the aqueous phase with 3 M sodium acetate (1/10 vol) and ice-cold 100% ethanol (2 vol), allowed to incubate (5 min at -20°C), centrifuged (12,000 × g with no brake), and washed twice with 70% ethanol. The pellet was air-dried and resuspended in TE (10 mM Tris·HCl, pH 8.0/1 mM EDTA).
PCR Amplification and Cloning. A 1.8-kb fragment 5′ to the coding sequence of hsp 86 was amplified from P. falciparum 3D7 clone by using primers 5′-taaatggtaccT TGATATATTTTTAGATATATGGAT-3′ and 5′-TCCGTATGCATTTTATTCGAAATGTG-3′. The PCRs using standard reagents were subjected to 35 cycles of 15 s at 94°C, 30 s at 45°C, and 2 min at 65°C. A 1-kb fragment 3′ to the coding sequence of hsp 86 was amplified from P. falciparum 3D7 by using primers 5′-ataggtaccGGATTTATATAATATATTTATG-3′ and 5′-ataaagcttATTAAGGAAACAAAATGAAAG-3′, and the PCR conditions were 35 cycles of 15 s at 94°C, 30 s at 41°C, and 1 min at 65°C. Both PCR products were then cloned into the vector pCR2.1 by using the TOPO TA-cloning kit (Invitrogen).
Plasmid Constructs. pXL-BACII-DHFR contained the human dihydrofolate reductase (hdhfr) coding sequence under the control of 5′ calmodulin and 3′ histidine-rich protein-2 was excised as a 2.2-kb BglII-EcoRI fragment from the plasmid pHH1 (25) and cloned between the inverted terminal repeats of the piggyBac element in the vector pXL-BacII (26).
pHTH, the piggyBac transposase coding sequence, was cut from the vector p32 as a BamHI fragment, cloned downstream of 5′ hsp86 in pCR2.1, and screened for the right orientation. 3′ hsp86 was cut out as a HindIII fragment from pCR2.1, cloned downstream to the transposase coding sequence, and screened for the right orientation to complete the helper plasmid.
Parasite Culture, Transfection, and Selection of Transformed Parasites. The NF54 clone of P. falciparum was obtained from the Naval Medical Research Center and maintained in culture according to standard methods at 37°C and gassing (5% O2 and 5% CO2, nitrogen balanced) with 5% hematocrit in RPMI medium 1640 supplemented with 0.5% Albumax I (Invitrogen), 0.25% sodium bicarbonate, and 0.01 mg/ml gentamicin. Human RBCs were obtained from the Indiana Blood Bank and were washed three times with RPMI medium 1640, resuspended to 50% hematocrit, and stored at 4°C.
Transfection of P. falciparum NF54 was achieved by parasite invasion of plasmid DNA-loaded RBCs (27). For plasmid DNA loading, 250 μl of processed RBCs were washed once with 250 μl of incomplete cytomix and combined with the desired amount of plasmid DNA and incomplete cytomix to a final volume of 400 μl. The mixture was transferred to a 0.2-cm cuvette (Bio-Rad), chilled on ice, and electroporated by using a Bio-Rad Gene Pulser II and standard conditions of 0.31 kV and 950 μF capacitance. The RBCs were loaded with either 100 μg each of plasmids pXL-BacII-DHFR and pHTH (experiment 1) or with 100 μg of pXL-BacII-DHFR and 50 μg of pHTH (experiments 2–8).
To ensure parasite invasion of only loaded RBCs, late-stage parasites were purified from culture by passage through a MACS magnetic column (Miltenyi Biotec). In brief, 20 ml of parasite culture at 5–10% late-stage parasitemia was passed through the MACS column with a magnet attached. The column was washed with 30 ml of incomplete RPMI medium 1640 to remove uninfected RBCs and early-stage parasites. The column was removed from the magnet, and the late-stage parasites still bound to the column were eluted in 20 ml of incomplete RPMI medium 1640 and pelleted by centrifugation at 1,250 × g for 5 min without brakes. The pellet was washed once with complete media and resuspended in 1 ml of complete media. In experiments 1 and 2, transfections were initiated in a 5-ml culture volume with 2 × 105 parasites, and, after 48-h posttransfection, 2.5 nM WR99210 was added to the culture, and the parasites were maintained in the drug until the reappearance of parasites in Giemsa-stained smears. Experiments 3–6 were initiated in a 200-μl culture volume of a 96-well microtiter plate with 2 × 103 parasites. Experiments 7 and 8 were initiated in a 5-ml culture volume with 1 × 105 parasites. For experiments 3–8, 2.5 nM WR99210 was added to the culture after four generations of growth. After the parasites reappeared in Giemsa-stained smears, the concentration of WR99210 was increased to 5 nM, and genomic DNA was extracted from drug-resistant parasites for Southern blot hybridizations.
Limiting Dilution of Parasite Clones and Parasite Lactate Dehydrogenase Assay. Drug-resistant parasites were cloned by limiting dilution at 0.5 and 0.25 parasites per well in a 96-well plate. Parasitemia was counted by Giemsa-stained smears, and parasites were diluted in RPMI medium 1640, such that there were 10 parasites per μl. For each plate, 500 parasites were mixed with 19.2 ml of RPMI medium 1640 and 0.8 ml of 50% hematocrit. 200 μl of this mixture were added to each well in a 96-well plate to obtain a final dilution of 0.5 parasite per well at 2% hematocrit. Similarly, 250 parasites were used as above to obtain a dilution of 0.25 parasite per well. The culture medium was changed and 0.4% hematocrit added on day 7 and day 14. On day 17, the presence of parasites was detected by a parasite lactate dehydrogenase assay (28). Briefly, 20 μl of parasite culture was lysed by freeze-thawing three times, and 100 μl of Malstat reagent (Flow Laboratories) was added. Ten microliters of 1 mg/ml nitroblue tetrazolium (Sigma) and 10 μl of 2 mg/ml diaphorase (Sigma) were added to the mixture. The mixture was incubated at room temperature for 20 min, and wells positive for parasites were identified by colorimetric analysis.
Southern Blot Hybridization. Genomic DNA (2 μg) extracted from transformed parasites was digested with 10 units of EcoRI overnight and separated on a 0.8% agarose gel. The DNA was depurinated in 0.25 M HCl, denatured in 0.5 M NaOH/1.5 M NaCl, neutralized in 500 mM Tris·HCl, pH 7.6/1.5 M NaCl and blotted overnight to a nylon membrane. The blot was then hybridized to 32P-labeled hdhfr, washed three times in 2× SSC/0.5% SDS (1× SSC/0.15 M sodium chloride/0.015 M sodium citrate, pH 7.0) for 15 min, and exposed to a Kodak photographic film at -80°C to visualize the hybridized fragments.
Inverse PCR and Sequencing. piggyBac insertion sites in transformed parasites were identified by using an inverse PCR technique (29). Genomic DNA (1 μg) from drug-resistant parasite populations was digested overnight with 10 units of Sau3AI or RsaI, precipitated with 2 vol of ethanol and 1/10 vol of 3 M sodium acetate, and self-ligated in a 100-μl reaction. The ligation reaction was precipitated as above and resuspended in 20 μl of ddH20. The Sau3AI-digested, self-ligated fragments were digested with 10 units of TseI to remove the episomal fragments. The inverted terminal repeat (ITR) 2 insertion sites of piggyBac element were identified by using 1 μl of the Sau3AI/TseI-digested or RsaI-digested ligation reaction as a template for the inverse PCR (35 cycles of 94°C for 30 s, 45°C for 30 s, and 65°C for 2 min) with one of the sense primers 5′-AGATGTCCTA A ATGCACAGCGAC-3′, 5′-CTCCAAGCGGCGACTGAG-3′, or 5′-CATTGACAAGCACGCCTCAC-3′ and one of the antisense primers 5′-GTCAATGCGGTAAGTGTCACTGA-3′, 5′-GTAAGTGTCACTGATTTTGAACTATAACG-3′, or 5′-GACGCATGATTATCTTTTACGTGAC. The PCR products were cloned into pGEM T-Easy vector (Promega) and sequenced by the dideoxy nucleotide chain termination method by using M13 forward and reverse primers in a Beckman CEQ 8000 sequencer.
Sequence Analysis and Identification of Insertion Sites. The sequences obtained by sequencing inverse PCR products were analyzed by using macvector software (Accelrys, San Diego), and the insertion sites in the genome were identified by performing a blast search using the PlasmoDB database (30).
PCR for Confirmation of ITR1 Insertion. To confirm the complete insertion of the piggyBac element into the P. falciparum genome, locus-specific primer for each insertion was used in a PCR reaction with one of the antisense primers 5′-AGATCTCCTAAATGCACAGCGAC-3′, 5′-CCTCGATATACAGACCGATAAAAC-3′, or 5′-GTTTGTTGAATTTATTATTAGTATGTAAGT-3′. The locus-specific primers for the insertions were “a,” 5′-GTTTGTATGTATGTGTGTTTGTTTC-3′; “c,” 5′-GGGAAATTATAAAATGGATTATAGG-3′; “d,” 5′-CCTTTATGAATGCCGCAAC-3′; “e,” 5′-ATGGGATCCACCATATGTATAACC-3′; “f,” 5′-GGACGGGCTAATATCCTTACG-3′; and “i,” 5′-CTTGATGGAAAAATGATAGGATC-3′. The conditions for the PCR were 35 cycles of 94°C for 15 s, 45°C for 30 s, and 65°C for 1 min. The PCR products were cloned into the pGEM T-Easy vector and sequenced to confirm the presence of ITR1 and duplication of the target site TTAA.
Results
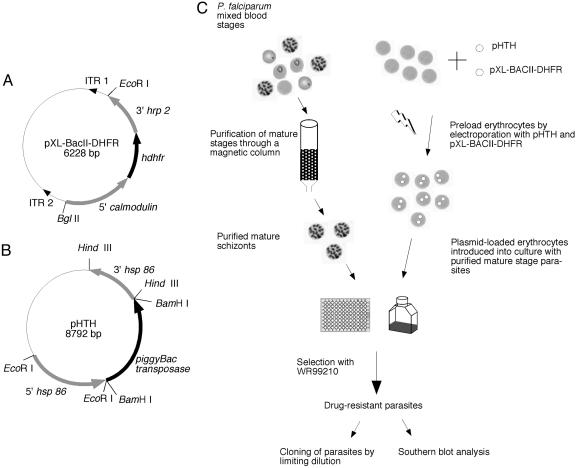
Design of the piggyBac Transposon and Helper Plasmids for Use in P. falciparum. A minimal piggyBac transposon vector, pXL-BACII-DHFR, was created by cloning the hdhfr coding sequence under the control of Plasmodium 5′ and 3′ regulatory elements of calmodulin and histidine-rich protein-2, respectively, in the plasmid vector pXL-BACII (26). This drug-resistance cassette, widely used for malaria parasite transformation by targeted homologous recombination, was flanked by the 3′ ITR1 and the 5′ ITR2 of the piggyBac element (Fig. 1A). The ITRs are oriented such that, upon transposition, they will carry the drug-resistant cassette into the Plasmodium genome without any of the plasmid backbone. A helper plasmid, pHTH, was created by cloning the piggyBac transposase coding sequence under the control of hsp 86 regulatory elements to mobilize the piggyBac element in the erythrocytic stages of P. falciparum (Fig. 1B). Intended only for transient transfection, this helper plasmid contained no selectable marker.
Fig. 1.
Plasmid design and procedure for piggyBac transformation of P. falciparum. (A) pXL-BACII-DHFR, a piggyBac transposon vector for transformation of P. falciparum, was created by cloning the hdhfr coding sequence under the control of 5′ calmodulin and 3′ histidine-rich protein-2 into the minimal piggyBac vector pXL-BACII. The hdhfr cassette was excised from the vector pHH1 as a 2.2-kb EcoRI/BglII fragment and cloned into pXL-BacII, such that it is flanked by the piggyBac ITR 1 and ITR 2. (B) The pHTH helper plasmid was constructed by cloning the piggyBac transposase coding sequence under the control of P. falciparum 5′ and 3′ hsp 86 sequences. piggyBac transposase expressed in the blood stages of P. falciparum will catalyze the transposition of the piggyBac element from the vector pXL-BACII-DHFR into the P. falciparum genome. (C) Schematic representation of the procedure used to transform P. falciparum. Mature blood-stage parasites were purified by passage through a magnetic column (Miltenyi Biotec) and reintroduced into culture with erythrocytes preloaded by electroporation with plasmids pXL-BacII-DHFR and pHTH. After one to four generations of growth in the preloaded erythrocytes, parasites were selected with 2.5 nM WR99210 until drug-resistant parasites emerged in culture.
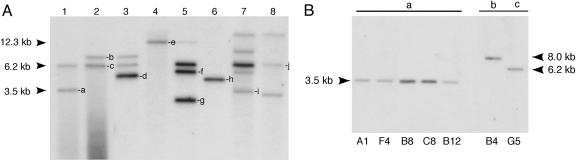
piggyBac Transformation of P. falciparum Is Rapid and Efficient. Mature blood-stage P. falciparum NF54 parasites were purified by isolation on a magnetic column (Miltenyi Biotec) (31). The purified parasitized erythrocytes were cultured in RBCs loaded with plasmids pXL-BACII-DHFR and pHTH (Fig. 1C), whereupon parasites spontaneously acquired plasmids from the erythrocytes (27). Parasites were transfected by using three different protocols to optimize the conditions required for efficient transformation. In protocol A (experiments 1 and 2), parasites were grown in 5-ml culture volume for one generation before selection with WR99210. In protocol B (experiments 7 and 8), parasites were grown in 5-ml culture volume for four generations before drug selection. In protocol C (experiments 3–6), parasites were grown in a 96-well plate in a low culture volume for four generations before drug selection. Southern blot hybridizations were performed on the drug-resistant populations obtained from these eight different experiments by using a hdhfr probe. Novel hybridization bands were detected in the parasite populations, in addition to the episomal band, indicating multiple unique integrations of the piggyBac element into the genome (Fig. 2A). The average transformation efficiency of piggyBac was highest for protocol C, in the range of 10-3 (Table 1). As expected, there was no evidence for piggyBac insertions in the absence of the helper plasmid (data not shown).
Fig. 2.
Confirmation of piggyBac integration in the P. falciparum genome. (A) Parasite genomic DNA was digested with EcoRI, and Southern blot hybridization using a hdhfr probe identified unique piggyBac hybridization bands from parasites cotransfected with pXL-BACII-DHFR and pHTH. In experiments 1 and 2, parasites were transfected with plasmids pXL-BACII-DHFR and pHTH, in 1:1 and 2:1 ratios, respectively, and the parasites were maintained for one generation in plasmid-loaded erythrocytes before selection with WR99210. In the other experiments (3–8), parasites were transfected with the plasmids in a 2:1 ratio and maintained in culture for four generations before selection with WR99210. Transfections for experiments 1, 2, 7, and 8 were initiated in a 5-ml culture volume, and, in experiments 3–6, transfections were performed in 200-μl cultures in a 96-well plate. After Southern blot hybridization, the episomal band was seen as a 6.2-kb fragment. The identified piggyBac integrations are indicated by letters. (B) Southern blot hybridization analysis of individual clones obtained from populations 1 and 2 identified clones with different sites of integrations. Clones A1, B8, B12, C8, and F4 appear to have the common insertion “a” and are likely to be of the same origin. Clone B4 and G5 have dissimilar sites of integration, “b” and “c.”
Table 1. piggyBac transformation efficiency in P. falciparum.
| Protocol | Parasite number | Generations in loaded RBCs | Insertions obtained | Transformation frequency |
|---|---|---|---|---|
| A | 4 × 105 | 1 | 3 | 7.5 × 10-6 |
| B | 2 × 105 | 4 | 8 | 4 × 10-5 |
| C | 8 × 103 | 4 | 9 | 1.1 × 10-3 |
piggyBac transformation frequency in P. falciparum was calculated as the number of insertions obtained in the genome divided by the number of parasites in the starting culture used for transfection. These results are summarized from eight different experiments. Protocols A and B include two experiments each using 5-ml cultures in flasks, whereas protocol C summarizes results from four experiments using 200-μl cultures in 96-well microtiter plates. The parasites in protocol A were grown for one generation in plasmid-loaded RBCs before applying drug, whereas in protocols B and C transfected parasites were grown for four generations. As anticipated, these preliminary results indicate that higher rates of transformation were achieved by longer exposures to the transposase (protocols B and C). Unexpectedly, fewer parasites in a transfected culture yielded higher transformation efficiency.
Insertions Are Stable. To test the stability of piggyBac integrations in the genome, parasites from populations 1 and 2 were cloned by a limiting-dilution method (32). Southern blot hybridizations with a hdhfr probe identified clones with integrations into the genome. Clones A1, B8, B12, C12, and F4, derived from population 1, appeared to have the same integration “a,” whereas clones B4 and G5, derived from population 2, had two different integrations, “b” and “c” (Fig. 2B). These clones were maintained in culture for >20 generations in the absence of the helper plasmid. The integrated piggyBac cassette was stable in all of the clones (data not shown), thereby ruling out any endogenous transposase activity as reported for mariner (14).
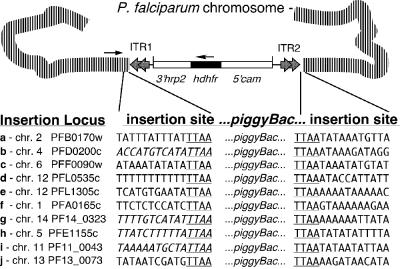
piggyBac Integrations Are TTAA-Site-Specific. The sites of integration in the transformed populations were identified by inverse PCR analyses performed using inverted oligonucleotide primers within the ITR2 arm of the transposed piggyBac element (29). From the multiple integrations obtained in the transformed populations, we isolated and identified 10 different insertion sites. These insertions were identified with ease because they represented the predominant population in each transfection experiment (Fig. 2A). Sequence analysis of these 10 insertions confirmed a consensus TTAA-site-specific integration of the piggyBac element into the parasite genome, as expected for authentic transposition (Fig. 3). Integration of the ITR1 of the piggyBac element was confirmed in separate PCR reactions using locus-specific primers and a primer in the ITR1 of piggyBac. Sequence analysis confirmed the expected TTAA duplication at the ITR1 end of the insertion for all integrations, except for integrations b, g, h, and i, because of the AT-rich repeat regions in those sequences. Instead, we confirmed the complete integration of the DHFR cassette in these populations by Southern hybridization restriction fragment length polymorphism analysis (data not shown).
Fig. 3.
Identification of piggyBac integration sites in the P. falciparum genome. Inverse PCR analysis was used to identify the piggyBac 5′ TR insertion sites. Briefly, genomic DNA from drug-resistant populations were digested with either Sau3AI or RsaI and self-ligated in a dilute reaction. Sau3AI self-ligated fragments were digested with TseI to remove the episomal fragment. The remaining self-ligated fragments were used as templates in an inverse PCR to identify sites of integration into the genome. Sequence analysis identified nine different sites of integration in eight different chromosomes, suggesting a genome-wide insertion of piggyBac. As expected, the piggyBac element had inserted in a TTAA target sequence in all of the analyzed clones. PCR analysis was then performed by using a genomic primer at each insertion site and a primer in ITR1 to confirm that the insertion of the piggyBac element was complete. Further sequence analysis confirmed the insertion of piggyBac ITRs into a TTAA target sequence that resulted in the duplication of the target site in the genome. The italicized sequences in insertions b, g, h, and i were confirmed by Southern blot hybridization analyses (data not shown).
Six insertions (c, f, g, h, i, and j) were in the 5′ region of the closest ORFs. Insertions c, i, and j were located ≈1,000 bp 5′ to the nearby ORF, whereas insertions f, g, and h were ≈300 bp, 350 bp, and 500 bp upstream, respectively. Insertions a, d, and e were 100 bp, 150 bp, and 465 bp downstream of the closest ORFs, respectively. Insertion b was ≈100 bp downstream of the start codon of a hypothetical asparagine-rich protein (PFD0200c), thereby disrupting the putative ORF of this gene. Further analysis is needed to characterize the effects of these insertions on gene expression in these transgenic parasite lines. The 10 identified insertions were widely dispersed on different chromosomes throughout the genome of the parasite, consistent with random selection of the TTAA-target insertion sites (Fig. 3).
Discussion
Genome sequencing of P. falciparum is almost complete, but many genes have unknown functions with no homologue in other species. Therefore, it is imperative that functional genomic studies are carried out with this important pathogen. Overcoming our limited ability to genetically manipulate this organism is vital so that progress can be made in understanding the parasite's unique biology. Sophisticated genetic procedures similar to what is available for model organisms are needed to employ forward genetic approaches to characterize phenotypes by producing random gene mutations. Genome-wide insertion of transposable elements will provide one such powerful tool to study gene function by allowing large-scale mutagenesis experiments. The high efficiency of the piggyBac transposition system in other organisms (20, 22, 33–35) and the lack of an efficient transposon-based mutagenesis system for Plasmodium led us to test the efficacy of this system in P. falciparum.
Parasites were transformed by using three different protocols to identify optimum conditions for piggyBac transformation. As expected, piggyBac transformation efficiency was higher when the parasites were grown longer in plasmid-loaded RBCs, probably because of more efficient uptake of plasmids. Unexpectedly, the transformation frequency was highest when using fewer parasites in low culture volume. Insertions of hdhfr-tagged piggyBac transposons were obtained throughout the genome at high transformation efficiencies in multiple experiments. These insertions occurred only in the presence of the piggyBac transposase and were stable for many generations, confirming the utility of piggyBac mutagenesis for functional genomic analyses of P. falciparum and other malaria parasites. Our strategy adapted targeted gene-disruption techniques that have evolved for targeted gene disruption in malaria parasites. The established approaches confer drug resistance to otherwise lethal antimalarial compounds through the introduction into the parasites of selectable markers such as hdhfr (36–38). Gene-specific targeting sequences that normally flank the drug-selection cassette were replaced by the minimal inverted terminal repeats of piggyBac, which are necessary for mobilization of the transposable element (26, 39). Because mobility of the piggyBac transposon depends on the presence of its transposase, another critical component needed for transposon-mediated transformation was the development of transposase-expressing helper plasmid. The coding sequence for piggyBac transposase was placed in a separate plasmid under the control of Plasmodium regulatory sequences to facilitate stable expression during blood-stage parasite development. By using this system, transformed parasites emerge <3 weeks after transfection, and the efficiency was significantly better than for methods currently in use. Furthermore, our piggyBac-mediated transformation protocols were easily adapted to a 96-well microtiter plate with conditions compatible with large-scale genetic screening, corroborating the ability of the piggyBac element to insert in the P. falciparum genome with relative ease and high efficiency.
The P element is widely used for transposon-mediated insertional mutagenesis of the D. melanogaster genome. Although this transposon preferentially inserts in the 5′ regulatory regions of genes, it effectively disrupts expression where it inserts and has been an extremely valuable genetic tool for functional characterization of the fruitfly genome (40). However, because of this bias in insertion-site preference, it is estimated that 150,000 insertions may be required to achieve ≈90% disruption of all fly genes by using the P element (41). Nevertheless, a similar rate of targeting efficiency is calculated for Agrobacterium T-DNA insertions in the Arabidopsis genome, in which insertion is totally random. Strong AT bias in noncoding regions of the P. falciparum genome (42) may affect where piggyBac insertion occurs, so we analyzed the distribution of the TTAA target sites in the P. falciparum genome to determine the likelihood and placement of piggyBac insertions within the transcribed portion of the parasite's genes. We estimate the P. falciparum genome to have 311,155 TTAA sites with 124,733 (40.5%) appearing in EST sequence databases of P. falciparum, potentially giving us ≈20 targets in each gene (42, 43). Because not all integrations will be equal in effect, implementation of high-throughput screening methods will likely be needed to achieve optimal gene identification when using piggyBac-mediated mutagenesis in forward genetic applications, such as phenotype screens. Clearly, data from our current study demonstrates that piggyBac-mediated mutagenesis can be used readily in large-scale genetic screens needed to identify interesting phenotypes.
A potential limitation of this initial methodology in relying on mobilization of the transposon in the blood-stage development may be the inability to modify genes that are essential for the parasite in this stage. Based on the high number potential insertion sites within each gene, we can speculate that there may be a spectrum of loss-of-function effects, depending on the location of the insertion site. However, this limitation can be easily overcome by mobilizing piggyBac in the other life-cycle stages of the parasite. Because piggyBac mobilization depends on transposase expression by the helper plasmid, the current helper-plasmid design can be easily modified for transposase expression in sexual-stage or liver-stage development by changing the regulatory sequences. Generally, the transposable element and the helper plasmid can be modified without difficulty for a variety of whole-genome applications, from promoter trapping to epitope tagging. Probably one of the most valuable applications will be stable transgene-expressing parasite clones.
Here, we have demonstrated that the piggyBac transposition system is an important genetic tool for manipulation of the P. falciparum genome. With this report of high-efficiency transposition in this deadly human pathogen and an efficient integration system, many genetic strategies that have eluded Plasmodium researchers will now be feasible. The ability of the piggyBac transposon mutagenesis system to be used for large-scale mutagenesis of P. falciparum will provide new insight into the complex biology of the malaria parasite to greatly accelerate efforts to develop intervention strategies.
Acknowledgments
We thank F. Collins, N. Lobo, and P. Romans for helpful discussions and critical reading of the manuscript and X. Li for valuable technical assistance. This work was supported, in part, by National Institutes of Health Grants R01AI33656 (to J.H.A.) and R01AI48561 (to M.J.F.).
Author contributions: M.J.F., B.B., and J.H.A. designed research; B.B. and D.A.S. performed research; M.J.F. contributed new reagents/analytic tools; B.B. and J.H.A. analyzed data; and B.B. and J.H.A. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: hdhfr, human dihydrofolate reductase; ITR, inverted terminal repeat.
References
- 1.Greenwood, B. & Mutabingwa, T. (2002) Nature 415, 670-672. [DOI] [PubMed] [Google Scholar]
- 2.Maitland, K., Makanga, M. & Williams, T. N. (2004) Curr. Opin. Infect. Dis. 17, 405-412. [DOI] [PubMed] [Google Scholar]
- 3.Winstanley, P., Ward, S., Snow, R. & Breckenridge, A. (2004) Clin. Microbiol. Rev. 17, 612-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Breman, J. G., Alilio, M. S. & Mills, A. (2004) Am. J. Trop. Med. Hyg. 71, 1-15. [PubMed] [Google Scholar]
- 5.Hartl, D. L. (2004) Nat. Rev. Microbiol. 2, 15-22. [DOI] [PubMed] [Google Scholar]
- 6.O'Donnell, R. A., Freitas-Junior, L. H., Preiser, P. R., Williamson, D. H., Duraisingh, M., McElwain, T. F., Scherf, A., Cowman, A. F. & Crabb, B. S. (2002) EMBO J. 21, 1231-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crabb, B. S. & Cowman, A. F. (1996) Proc. Natl. Acad. Sci. USA 93, 7289-7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Dijk, M. R., Waters, A. P. & Janse, C. J. (1995) Science 268, 1358-1362. [DOI] [PubMed] [Google Scholar]
- 9.Waters, A. P., Thomas, A. W., van Dijk, M. R. & Janse, C. J. (1997) Methods 13, 134-147. [DOI] [PubMed] [Google Scholar]
- 10.Wu, Y., Kirkman, L. A. & Wellems, T. E. (1996) Proc. Natl. Acad. Sci. USA 93, 1130-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kadekoppala, M., Cheresh, P., Catron, D., Ji, D. D., Deitsch, K., Wellems, T. E., Seifert, H. S. & Haldar, K. (2001) Mol. Biochem. Parasitol. 112, 211-218. [DOI] [PubMed] [Google Scholar]
- 12.Sarkar, A., Sim, C., Hong, Y. S., Hogan, J. R., Fraser, M. J., Robertson, H. M. & Collins, F. H. (2003) Mol. Genet. Genomics 270, 173-180. [DOI] [PubMed] [Google Scholar]
- 13.Gueiros-Filho, F. J. & Beverley, S. M. (1997) Science 276, 1716-1719. [DOI] [PubMed] [Google Scholar]
- 14.Mamoun, C. B., Gluzman, I. Y., Beverley, S. M. & Goldberg, D. E. (2000) Mol. Biochem. Parasitol. 110, 405-407. [DOI] [PubMed] [Google Scholar]
- 15.Fraser, M. J., Smith, G. E. & Summers, M. D. (1983) J. Virol. 47, 287-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fraser, M. J., Brusca, J. S., Smith, G. E. & Summers, M. D. (1985) Virology 145, 356-361. [DOI] [PubMed] [Google Scholar]
- 17.Cary, L. C., Goebel, M., Corsaro, B. G., Wang, H. G., Rosen, E. & Fraser, M. J. (1989) Virology 172, 156-169. [DOI] [PubMed] [Google Scholar]
- 18.Wang, H. H., Fraser, M. J. & Cary, L. C. (1989) Gene 81, 97-108. [DOI] [PubMed] [Google Scholar]
- 19.Ding, S., Wu, X., Li, G., Han, M., Zhuang, Y. & Xu, T. (2005) Cell 122, 473-483. [DOI] [PubMed] [Google Scholar]
- 20.Thibault, S. T., Singer, M. A., Miyazaki, W. Y., Milash, B., Dompe, N. A., Singh, C. M., Buchholz, R., Demsky, M., Fawcett, R., Francis-Lang, H. L., et al. (2004) Nat. Genet. 36, 283-287. [DOI] [PubMed] [Google Scholar]
- 21.Ryder, E. & Russell, S. (2003) Brief. Funct. Genomics Proteomics 2, 57-71. [DOI] [PubMed] [Google Scholar]
- 22.Lobo, N. F., Hua-Van, A., Li, X., Nolen, B. M. & Fraser, M. J., Jr. (2002) Insect Mol. Biol. 11, 133-139. [DOI] [PubMed] [Google Scholar]
- 23.Tamura, T., Thibert, C., Royer, C., Kanda, T., Abraham, E., Kamba, M., Komoto, N., Thomas, J. L., Mauchamp, B., Chavancy, G., et al. (2000) Nat. Biotechnol. 18, 81-84. [DOI] [PubMed] [Google Scholar]
- 24.Grossman, G. L., Rafferty, C. S., Fraser, M. J. & Benedict, M. Q. (2000) Insect Biochem. Mol. Biol. 30, 909-914. [DOI] [PubMed] [Google Scholar]
- 25.Reed, M. B., Saliba, K. J., Caruana, S. R., Kirk, K. & Cowman, A. F. (2000) Nature 403, 906-909. [DOI] [PubMed] [Google Scholar]
- 26.Li, X., Harrell, R. A., Handler, A. M., Beam, T., Hennessy, K. & Fraser, M. J., Jr. (2005) Insect Mol. Biol. 14, 17-30. [DOI] [PubMed] [Google Scholar]
- 27.Deitsch, K., Driskill, C. & Wellems, T. (2001) Nucleic Acids Res. 29, 850-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Makler, M. T. & Hinrichs, D. J. (1993) Am. J. Trop. Med. Hyg. 48, 205-210. [DOI] [PubMed] [Google Scholar]
- 29.Earp, D. J., Lowe, B. & Baker, B. (1990) Nucleic Acids Res. 18, 3271-3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kissinger, J. C., Brunk, B. P., Crabtree, J., Fraunholz, M. J., Gajria, B., Milgram, A. J., Pearson, D. S., Schug, J., Bahl, A., Diskin, S. J., et al. (2002) Nature 419, 490-492. [DOI] [PubMed] [Google Scholar]
- 31.Trang, D. T., Huy, N. T., Kariu, T., Tajima, K. & Kamei, K. (2004) Malar. J. 3, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosario, V. (1981) Science 212, 1037-1038. [DOI] [PubMed] [Google Scholar]
- 33.Handler, A. M. (2002) Insect Biochem. Mol. Biol. 32, 1211-1220. [DOI] [PubMed] [Google Scholar]
- 34.Lorenzen, M. D., Berghammer, A. J., Brown, S. J., Denell, R. E., Klingler, M. & Beeman, R. W. (2003) Insect Mol. Biol. 12, 433-440. [DOI] [PubMed] [Google Scholar]
- 35.Allen, M. L., Handler, A. M., Berkebile, D. R. & Skoda, S. R. (2004) Med. Vet. Entomol. 18, 1-9. [DOI] [PubMed] [Google Scholar]
- 36.de Koning-Ward, T. F., Fidock, D. A., Thathy, V., Menard, R., van Spaendonk, R. M., Waters, A. P. & Janse, C. J. (2000) Mol. Biochem. Parasitol. 106, 199-212. [DOI] [PubMed] [Google Scholar]
- 37.Crabb, B. S., Rug, M., Gilberger, T. W., Thompson, J. K., Triglia, T., Maier, A. G. & Cowman, A. F. (2004) Methods Mol. Biol. 270, 263-276. [DOI] [PubMed] [Google Scholar]
- 38.Fidock, D. A. & Wellems, T. E. (1997) Proc. Natl. Acad. Sci. USA 94, 10931-10936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li, X., Lobo, N., Bauser, C. A. & Fraser, M. J., Jr. (2001) Mol. Genet. Genomics 266, 190-198. [DOI] [PubMed] [Google Scholar]
- 40.Mi, H., Vandergriff, J., Campbell, M., Narechania, A., Majoros, W., Lewis, S., Thomas, P. D. & Ashburner, M. (2003) Genome Res. 13, 2118-2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spradling, A. C., Stern, D., Beaton, A., Rhem, E. J., Laverty, T., Mozden, N., Misra, S. & Rubin, G. M. (1999) Genetics 153, 135-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gardner, M. J., Hall, N., Fung, E., White, O., Berriman, M., Hyman, R. W., Carlton, J. M., Pain, A., Nelson, K. E., Bowman, S., et al. (2002) Nature 419, 498-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Watanabe, J., Sasaki, M., Suzuki, Y. & Sugano, S. (2001) Nucleic Acids Res. 29, 70-71. [DOI] [PMC free article] [PubMed] [Google Scholar]