Abstract
Léri-Weill dyschondrosteosis (LWD) is a pseudoautosomal dominant disorder characterized by disproportionate short stature and a characteristic curving of the radius, known as the “Madelung deformity.” SHOX mutations resulting in SHOX haploinsufficiency have been found in LWD and in a variable proportion of patients with idiopathic short stature (ISS), whereas homozygous loss of SHOX results in the more severe Langer mesomelic dysplasia (LMD). Defects in SHOX have been identified in ∼60% of LWD cases, whereas, in the remaining ∼40%, the molecular basis is unknown. This suggests either genetic heterogeneity or the presence of mutations in unanalyzed regions of SHOX, such as the upstream, intragenic, or downstream regulatory sequences. Therefore, the pseudoautosomal region 1 (PAR1) of 80 patients with LWD, in whom SHOX deletions and mutations had been excluded, was screened for deletions by use of a new panel of microsatellite markers. We identified 12 patients with LWD who presented with a novel class of PAR1 deletions that did not include SHOX. The deletions were of variable size and mapped at least ∼30–530 kb downstream of SHOX. In our cohort, this type of deletion accounted for 15% of cases. In all cases, the deletions cosegregated with the phenotype. No apparent phenotypic differences were observed between patients with SHOX deletions and those with this new class of PAR1 deletions. Thus, we present here the identification of a second PAR1 region implicated in the etiopathogenesis of LWD. Our findings suggest the presence of distal regulatory elements of SHOX transcription in PAR1 or, alternatively, the existence of an additional locus apparently involved in the control of skeletal development. Deletion analysis of this newly identified region should be included in the mutation screening of patients with LWD, LMD, and ISS.
Introduction
Léri-Weill dyschondrosteosis (LWD [MIM 127300]) is a dominantly inherited skeletal dysplasia characterized by disproportionate short stature, mesomelic limb shortening, and the Madelung deformity of the forearm: the bowing of the radius and the distal dislocation of the ulna. Heterozygous deletions or mutations of the short-stature homeobox-containing gene (SHOX [MIM 312865]) have been identified as the molecular basis of LWD (Belin et al. 1998; Shears et al. 1998), whereas homozygous loss of SHOX results in the more severe Langer mesomelic dysplasia (LMD [MIM 249700]) (Belin et al. 1998). SHOX is located in the pseudoautosomal region 1 (PAR1) of the short arms of the X and Y chromosomes (Ellison et al. 1997; Rao et al. 1997). Thus, two active copies are required for normal function in both males and females. It has also been demonstrated that SHOX haploinsufficiency is responsible in part for the short stature associated with Turner syndrome (Ellison et al. 1997; Rao et al. 1997; Clement-Jones et al. 2000) and also for a variable proportion (2%–22%) of cases of idiopathic short stature (ISS) (Rao et al. 1997; Binder et al. 2000; Rappold et al. 2002; Stuppia et al. 2003; Huber et al. 2004). Those with ISS represent a heterogeneous group of patients in whom the etiology of short stature (height <3rd percentile) is undefined.
SHOX encodes a cell-specific homeodomain protein involved in cell cycle and growth regulation (Marchini et al. 2004) that has been shown to act as a transcription activator in osteogenic cells (Rao et al. 2001), although its targets have yet to be identified.
A defect in SHOX has been identified in ∼60% of LWD cases, whereas, in the remaining ∼40%, the molecular basis is still unknown (Schiller et al. 2000; Falcinelli et al. 2002; Flanagan et al. 2002). This suggests the presence of mutations in previously unanalyzed regions, such as the promoters found in the noncoding exon 1 and part of exon 2 (Blaschke et al. 2003) and/or intragenic, upstream, and downstream regulatory sequences, or, alternatively, the involvement of additional genes. We therefore screened the PAR1 of 80 patients with LWD in whom SHOX deletions and mutations had been excluded, and we identified a novel class of PAR1 deletions that lie ∼30–530 kb downstream of SHOX in 12 patients with LWD. In this cohort, the frequency of such deletions was 15%. A panel of 25 (18 novel) microsatellites and 41 (10 unreported) SNPs spanning 650 kb of PAR1 was used to delimit the deletion boundaries and to define a minimal commonly deleted region. Our results suggest the presence of long-range regulatory elements of SHOX transcription or, alternatively, the existence of an additional locus in PAR1 that, when deleted or mutated, results in a phenotype apparently identical to that observed in patients who have LWD with SHOX mutations or deletions.
Material and Methods
Patients and Controls
Ethical approval was obtained from the respective institutions. Probands with LWD were recruited from three clinics: the genetic clinic at Hopital Necker Enfants Malades, Paris; the Endocrinology Service at Hospital Infantil Universitario Niño Jesús, Madrid; and the Wessex Regional Genetics Laboratory at Salisbury District Hospital, United Kingdom. All participants provided informed consent for the studies performed or were diagnostic referrals for SHOX mutation testing. Peripheral blood was drawn from probands and, when possible, from relevant family members for DNA extraction and, in some cases, for FISH analysis. The control cohorts consisted of 50 normal white control individuals and 70 white families. Genomic DNA was isolated from whole blood by use of the salt-precipitation method (Nicolaides and Stoeckert 1990).
Clinical Evaluation
Clinical details were obtained for all patients recruited into the study (table 1). Whenever possible, these included birth details, actual height and standard deviation of height according to age and sex, physical examination of extremities, and the results of X-radiography of the lower arm. Their family histories were also documented, including parental heights.
Table 1.
Clinical Characteristics of Subjects Who Have LWD with PAR1 Deletions Downstream of SHOX[Note]
| Family | Sex ofProband | ProbandHeight(SD)a | Clinical Features | No. ofFamily MembersAnalyzed | Nationalityb |
| 1 | F | .85 | Short stature, Madelung | 11 | Spanish |
| 2 | M | −2.5 | Short stature, Madelung | 1 | British |
| 3 | M | ND | Short stature, Madelung | 4 | French |
| 4 | F | −2.7 | Short stature, Madelung | 9 | British |
| 5 | M | −2.0 | Short stature, no Madelungc | 2 | French |
| 6 | F | ND | Short stature, Madelung | 2 | French |
| 7 | M | −2.5 | Short stature, Madelung | 3 | French |
| 8 | F | −2.6 | Short stature, Madelung | 5 | French |
| 9 | F | −3.0 | Short stature, Madelung | 2 | French |
| 10 | F | −.3 | Short stature, Madelung | 2 | French |
| 11 | F | ND | Short stature, no Madelungd | 3 | French |
| 12 | F | −4.6 | Short stature, Madelung | 3 | French |
Note.— All probands are from families with a family history of LWD.
Standard deviations were corrected for age, sex, and nationality. ND = no data available.
All probands are of white descent.
Received diagnosis of LWD because the mother presented with the Madelung deformity.
Patient was prepubertal at time of diagnosis.
SHOX Genetic Screening
The presence of SHOX whole-gene deletions was excluded by at least one of the following techniques: (1) multiplex ligation-dependent probe amplification (MLPA), performed using the SHOX Salsa (MRC-Holland); (2) FISH, with two cosmids covering the entire SHOX gene; or (3) microsatellite analysis, performed using markers located intragenically to and 5′ and 3′ flanking the SHOX gene. Analyzed microsatellites included previously reported loci, SHOX CA, DYS290, DXYS233, and DXYS234, and two newly identified microsatellites, DXYS10092 and DXYS10093, which are 5′ flanking and intragenic to SHOX, respectively.
The exclusion of point mutations and small deletions and insertions in SHOX coding sequences and intron/exon boundaries was done either by direct sequencing or by denaturing high-performance liquid chromatography (dHPLC) (with Transgenomic Wave 3500HT), and subsequent sequencing of any identified heteroduplex was peformed using the BigDye Terminator V3.1 kit (ABI Biosystems). Further information on primer sequences and dHPLC conditions is available in tables 2 and 3.
Table 2.
Oligonucleotide Sequences for the Amplification of SHOX Coding Exons and Intron/Exon Boundaries[Note]
|
Oligonucleotide Sequence(5′→3′) |
||||
| SHOX Exon | Sense | Antisense | Annealing Temperature(°C) | Amplicon Size(bp) |
| 2 | AATGGAAAGGCGTAAATAACAGC | GAGACGGGAGCTGCAAATGT | 56 | 507 |
| 3 | GGCTTTTGCGTTATGGACCCC | GCTCCCGAGGACCAGGCGATG | 62 | 416 |
| 4/5 | CTTGGTTCAGCCTCATGGGAAG | CAAATAGGGGAAAGGGGAAGG | 52 | 424 |
| 6a | AGAGGCACGTTGGGGGTTTC | GCGGGGTTGAGTGCAGGACG | 64 | 420 |
Note.— Amplicons were subsequently screened for point mutations and small insertions and deletions, by dHPLC analysis.
Table 3.
dHPLC Conditions for Mutation Screening of SHOX Coding Exons and Intron/Exon Boundaries
| SHOX Exonand dHPLC Gradient of Transgenomic Buffer B (%) | dHPLC Oven Temperature(°C) |
| Exon 2: | |
| 61.7–67.7 | 63.5 |
| 60.7–66.7 | 64.5 |
| 57.7–63.7 | 66.0 |
| Exon 3: | |
| 60.3–66.3 | 64.4 |
| 58.3–64.3 | 65.5 |
| 56.3–62.3 | 67.0 |
| Exon 4/5: | |
| 59.5–65.5 | 60.3 |
| 58.5–64.5 | 61.8 |
| 56.5–62.5 | 63.3 |
| Exon 6a: | |
| 57.4–66.4 | 66.9 |
| 59.4–65.4 | 68.4 |
| 59.4–65.4 | 68.9 |
Design and Genotyping of Microsatellites
The X-chromosome contig (766,173 bp) (National Center for Biotechnology Information [NCBI] accession number NT_028413), including the telomeric part of the PAR1 region, downloaded from the Ensembl Genome Browser and GenBank Web sites, was used as the input for the program Tandem Repeats Finder (Benson 1999; see Tandem Repeats Finder Web site). Sequence positions of microsatellites and SNPs were determined using Ensembl-listed coordinates. A set of microsatellite markers was chosen from the output files by visual examination, on the basis of the following criteria: (1) unit size of 2–4 bp, (2) a match of at least 90%, and (3) an array of six or more copies. Primer pairs were then designed to flank the repeat by use of the program Oligo 6 (Molecular Biology Insights). One primer from each pair was fluorescently labeled. Primer sequences are shown in table 4 and are also available at the GDB Human Genome Database Web site. Initially, new microsatellites were evaluated using 10 control individuals to investigate their polymorphic nature; subsequently, several large families were studied to verify Mendelian inheritance. Once a marker was shown to exhibit both variability and Mendelian inheritance, the heterozygosity indices were determined by analyzing at least 100 chromosomes from healthy white control individuals.
Table 4.
Oligonucleotide Sequences, PCR Conditions, and Amplicon Sizes of the Identified PAR1 Microsatellites
|
Oligonucleotide Sequence(5′→3′) |
|||||
| Microsatellitea | RepeatUnit | Sense | Antisense | AnnealingTemperature(°C) | Amplicon Size Range(bp) |
| DXYS10092 | (GA) | TTCGTGACAAAGGCCTTTGC | CTACAAGTCCTAGTACCTAC | 53 | 317–367 |
| DXYS10093 | (CT) | GCCCGTGATCCCAGTACTG | CAACTTCCTTGGAAATCTTC | 55 | 233–259 |
| DXYS10090 | (TGTT) | TGGCTCTCTGAAGGGGCACC | CCTGGGTGACAAAGTGACTC | 55 | 195–199 |
| DXYS10091 | (AAAC) | GAATTGCTTGAACCTGGC | AGGGAGGTCATACCTTGTTGAA | 55 | 202–222 |
| DXYS10083 | (TG) | GGGGGTGTTTGGAAATGGTATAAT | TTACAATGTATGTCAGCAGAGACC | 51 | 150–172 |
| DXYS10094 | (AT) | GGGGCAGACAGAAGACACACAAAAT | GCCTGGGCGACAGAGCGAGACTCA | 57 | 221–259 |
| DXYS10095 | (CA) | GGGAAAGCCTCATATATCTGAC | CTGGTGTCTCTCCATTGTCCCCAT | 55 | 229–239 |
| DXYS10085 | (TG) | AGCCTGTTAGTGCCTCAACTTC | GAGTGGCAGCACGTTAATGTCA | 54 | 206–228 |
| DXYS10086 | (GAAA) | CTGACCCAGACTGTGTCTGAAG | AAGGAGATTCCTCAGCAGTGTA | 53 | 135–219 |
| DXYS10087 | (CA) | TGTGGCCGGGGCAAGATTC | CCCCAGCACAATCACTGT | 54b | 160–196 |
| DXYS10096 | (TG) | TTTAACAAACCGCATTCTCCAA | GTGGTGGAGCTTGCAGTGA | 57 | 240–252 |
| DXYS10088 | (TTTA) | CAGCCAATCTGTTCAATC | CACGCCTATAGTCCCAG | 53 | 266–278 |
| DXYS10089 | (TTAT) | CGTGCCTGGCTAAGCTGTT | GCTGAGGCAGGAGAATCACTTC | 54 | 183–203 |
| DXYS10080 | (TG) | CCTCTGTGGGACAACATGATA | GTTGTGGACAGGACCTCGTTC | 56 | 145–161 |
| DXYS10081 | (TTTA) | TTCTTGTCGGCTAGTCCTTC | TGCCTGTAATCCCAACTACCC | 55 | 267–283 |
| DXYS10082 | (CA) | TGGTAAGTGCCTGTGG | TGTGAACCATAATTCGTAAGA | 56 | 208–222 |
| DXYS10084 | (CA) | ATTTGAATGTGCACCTCATC | TACAAGAAAAGAAGAATCGTG | 53 | 204–222 |
| DXYS10097 | (TG) | TGAGTGGGATGAGGATGAGA | CAAGAGTGAAACTCCATCTC | 50 | 148–176 |
Microsatellites are listed in order from telomere to centromere.
3.0 mM MgCl2 was added to the PCR reaction.
The PCR conditions for microsatellite amplification were 1 × Qiagen Hotstart Taq buffer and 1 U of Hotstart Taq polymerase, 2.0 mM MgCl2, 200 μM dNTPs (50 μM of each), and 400 nM of each primer. Cycling conditions were an initial denaturation at 94°C for 15 min and 32 cycles at 94°C for 30 s, at 50°C–57°C for 30 s, and at 72°C for 40 s, with a final extension at 72°C for 8 min. Amplification products were diluted and then electrophoresed using an ABI 3100 Sequencer. Allele sizes were determined in relation to internal size standards (Genescan 400HDRox) by use of GeneMapper 3.5 software (Applied Biosystems).
SNP Analysis
To further delimit the deletion boundaries in regions for which no microsatellites were predicted and for which deletion confirmation was required, we identified a series of SNPs within the region of interest by using the dbSNP database at NCBI. Amplicons were designed to incorporate two or more SNPs. We designed eight amplicons to encompass 31 known polymorphisms (table 5). The SNPs were genotyped by direct sequencing of the eight amplicons in each proband and family member. SNP haplotypes were subsequently determined, to finely map the presence of deletions.
Table 5.
Oligonucleotide Sequences and PCR Conditions of the Analyzed PAR1 SNPs
|
Oligonucleotide Sequence(5′→3′) |
|||||
| Amplicon and dbSNP | Variation | Sense | Antisense | Annealing Temperature(°C) | Size(bp) |
| S2: | GCATTCACACGGTAGTCTTG | ATGCCACGCCAGGTATTCTC | 54 | 446 | |
| rs5946526 | C/G | ||||
| rs7053485 | T/G | ||||
| S3: | TGCAGGGTCTTCAGATAGGTG | GCATCTCTTAAAGGGTGGCT | 54 | 680 | |
| rs5946342 | T/C | ||||
| rs5988459 | C/G | ||||
| rs5988460 | A/G | ||||
| S4: | ACAGACACATCAGTTTCCCTTG | GCCAAAGCCCCCTCATTATTAA | 55 | 757 | |
| rs5988291 | T/G | ||||
| rs7061267 | T/C | ||||
| rs4504827 | T/A | ||||
| rs4497142 | G/T | ||||
| S6: | TCAGCTCACGGCAACG | GTTCCTCTTGTCATCTCCC | 57 | 358 | |
| rs5946327 | A/G | ||||
| rs5946503 | C/A/T | ||||
| rs5946504 | C/T | ||||
| rs38346958 | T/A | ||||
| rs5946328 | G/A | ||||
| rs5988279 | C/T | ||||
| rs38346959 | T/C | ||||
| rs38346960 | T/C | ||||
| S8: | GTAGGTTTCCTGCCGTTTTAG | GAGGACGCCAAAGTTCAGGAG | 55 | 530 | |
| rs5946518 | G/A | ||||
| rs5946519 | G/C | ||||
| rs5946520 | T/G | ||||
| S11: | CAAGTGAGCCAAGTGAGTCG | CTTGTCGGTGGGCACAG | 56 | 677 | |
| rs38346961 | G/C | ||||
| rs38346962 | C/T | ||||
| rs38346963 | T/C | ||||
| rs38346964 | A/G | ||||
| rs7067102 | A/G | ||||
| rs5946533 | C/T | ||||
| rs7049502 | G/C | ||||
| S14: | TCACCTCTCAATGCTGCCATGT | GCAGAAGACAGTACCCCTCACC | 54a | 594 | |
| rs6644574 | A/G | ||||
| rs4556269 | C/G | ||||
| rs6644575 | C/T | ||||
| rs1468111 | A/G | ||||
| rs5946759 | A/G | ||||
| rs38346965 | C/T | ||||
| rs5946760 | A/G | ||||
| rs7052082 | C/T | ||||
| S15: | CCATCAGAAATGCCTCTCAGAT | TAAGCACGTGAGCAGCTATTAA | 54 | 301 | |
| rs38346966 | G/A | ||||
| rs5988448 | A/T | ||||
| rs5988449 | C/T | ||||
| rs5988450 | C/G | ||||
| rs5988451 | G/A | ||||
| rs38346967 | G/A | ||||
10% dimethyl sulfoxide (DMSO) was added to the PCR reaction.
To score the SHOXb 657A→G SNP, exon 6b was amplified using the sense primer 5′-TGCACTTGGCCTTTTTTTTT-3′ and the antisense primer 5′-AGTTCCAAGCGATTCTCCTGC-3′. The products were then digested with XmaI (New England Biolabs) and were scored after electrophoresis on a 4% NuSieve agarose gel.
FISH Analysis
FISH analysis was performed on metaphase chromosomes prepared from peripheral blood lymphocytes by standard techniques (Pinkel et al. 1986). FISH was performed with cosmids from the distal PAR1, including two that span the entire SHOX gene, 15D10 (LLNOYCO3′M′15D10) and 34F5 (LLNOYCO3′M′34F5) (Rao et al. 1997), and additional BACs from the Ensembl tiling path. The centromeric probes DXZ2 and DYZ3 were used as controls.
Repeat Analysis
In silico examination of repetitive sequences within the PAR1 contig was performed with the use of the RepeatMasker program.
Results
Clinical Data
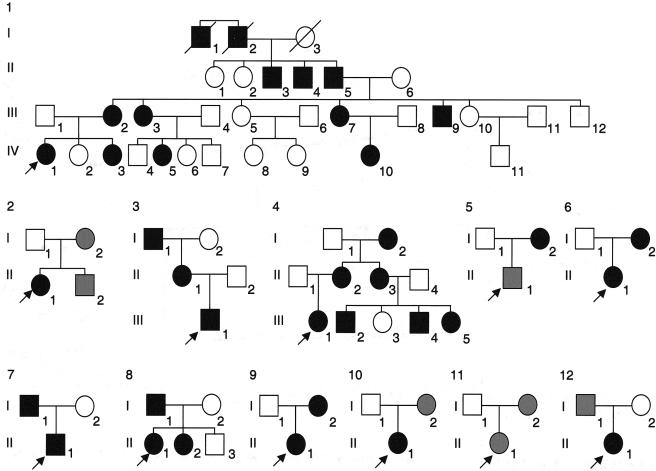
A total of 80 patients with LWD, for whom SHOX coding-sequence changes and deletions had been excluded, were screened for novel mutations in the PAR1 sequence flanking SHOX. Twelve patients with LWD presented with previously undescribed PAR1 deletions located downstream of SHOX. The pedigrees of the 12 families are depicted in figure 1. Clinical details of the probands are summarized in table 1. Of the 12 probands with LWD, 8 were from families in which dominant inheritance of short stature and Madelung deformity was confirmed by assessment of other family members and/or was strongly suspected in the previous generations. Probands 1 and 4 belong to large multigenerational families with a strong family history of short stature and Madelung deformity, although of variable severity.
Figure 1.
Pedigrees of the 12 families with LWD. The blackened symbols indicate individuals affected with LWD (i.e., Madelung deformity is present), whereas the gray symbols indicate those with a classification of ISS, since no Madelung deformity was observed.
Mutation Analysis of SHOX and the PAR1 Region
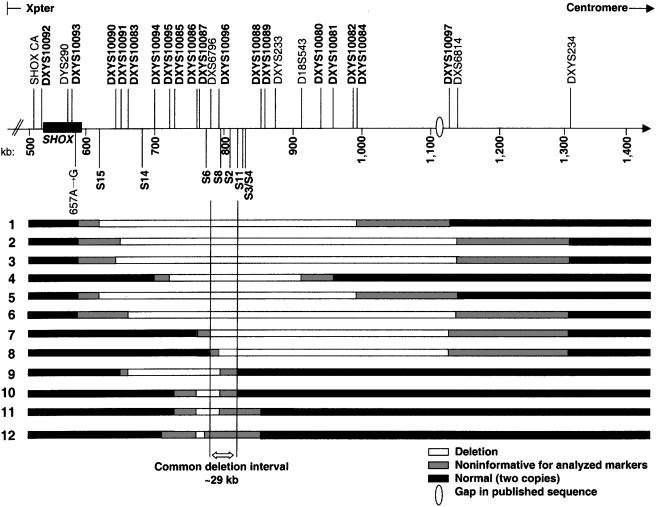
PAR1 deletions downstream of SHOX were characterized in 12 families (fig. 2) by use of a panel of 25 microsatellite markers, 18 of which were novel, and 41 SNPs, 10 of which were previously unreported (tables 4–6). In all cases, the presence of two copies of SHOX was confirmed by microsatellite analysis, MLPA, or FISH (fig. 3). No point mutation or small deletion or insertion was detected in the coding exons of SHOX in any of the probands by dHPLC and DNA sequencing. In all families, PAR1 deletions cosegregated with the phenotype (fig. 2). To analyze the incidence of the newly detected deletions in nonaffected individuals, the PAR1 of 70 control families was also examined by microsatellite analysis. No deletion was observed, which thus confirmed that the newly detected PAR1 deletions were not polymorphisms (i.e., not large copy variation). Likewise, no deletion was detected in any of the nonaffected partners or spouses of the probands included in the study.
Figure 2.
Schematic representation of the PAR1 deletion intervals detected using a panel of known and novel microsatellites and SNPs (not to scale). Microsatellites and amplicons incorporating the SNPs listed in tables 4 and 5 are given above and below the scale bar, respectively. Markers in bold indicate novel microsatellites.
Table 6.
Heterozygosity Values and Allele Ranges for Novel PAR1 Microsatellite Markers in a Minimum of 100 Chromosomes from White Control Individuals
| Microsatellitea | RepeatUnit | HeterozygosityValue | No. ofObservedAllelesb |
| DXYS10092 | (GA) | .96 | 18 |
| DXYS10093 | (CT) | .69 | 14 |
| DXYS10090 | (TGTT) | .35 | 3 |
| DXYS10091 | (AAAC) | .60 | 3 |
| DXYS10083 | (TG) | .82 | 8 |
| DXYS10094 | (AT) | .67 | 20 |
| DXYS10095 | (CA) | .55 | 6 |
| DXYS10085 | (TG) | .87 | 11 |
| DXYS10086 | (GAAA) | .78 | 21 |
| DXYS10087 | (CA) | .68 | 7 |
| DXYS10096 | (TG) | .50 | 6 |
| DXYS10088 | (TTTA) | .46 | 4 |
| DXYS10089 | (TTAT) | .60 | 4 |
| DXYS10080 | (TG) | .62 | 8 |
| DXYS10081 | (TTTA) | .78 | 5 |
| DXYS10082 | (CA) | .73 | 6 |
| DXYS10084 | (CA) | .70 | 9 |
| DXYS10097 | (TG) | .94 | 11 |
Microsatellites are listed in order from telomere to centromere.
The number of observed alleles in a cohort of 50 healthy white individuals. Further information can be found at the GDB Web site.
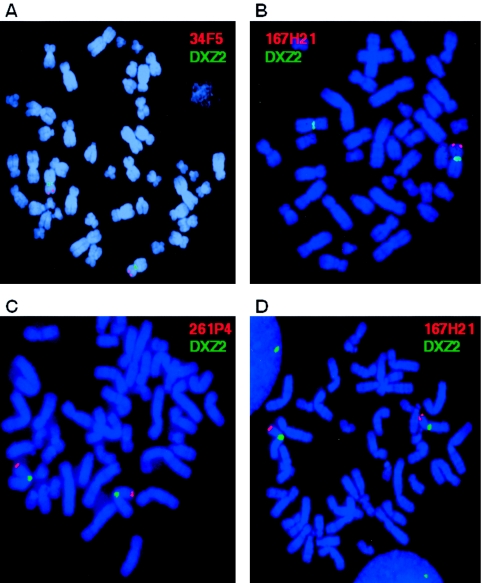
Figure 3.
FISH analyses of proband 1 and her unaffected sister. A, Two copies of SHOX observed as two signals with cosmid 34F5, which hybridizes to exons 2–6a. B, The deletion observed with cosmid 167H21, which hybridizes to sequences downstream of SHOX. C, Cosmid 261P4, which maps proximally to the deletion, present as two copies. D, Cosmid 167H21, present as two copies in the unaffected sister of proband 1. The red signal indicates the pseudoautosomal cosmid/BAC, and the green signal indicates the control probe DXZ2, which maps to the X centromere.
Deletion Mapping
The telomeric ends of the deletions were mapped to ∼30–205 kb downstream of SHOX, between SNP rs38346966 in amplicon S15 and DXS6796, and the centromeric ends were mapped to ∼480–530 kb downstream, between amplicon S11 and DXS6814. Deletions varied in size from the smallest deletion, detected in proband 10 and spanning <81 kb from microsatellite DXYS10086 to possibly amplicon S2, to the largest, detected in proband 3 and spanning ∼501 kb, from rs38346966 in amplicon S15 to DXS6814 (fig. 2).
We identified a null allele at micrrosatellite DXYS233, ∼270 kb downstream of SHOX, in two-thirds of the families with deletions (families 1–8). Subsequent screening of this region with additional microsatellite markers, 18 of which were novel (table 4), revealed the presence of four additional deletions that did not extend to microsatellite DXYS233 (families 9–12). Fine mapping of all 12 deletions enabled us to delimit a minimal commonly deleted region of 29 kb, defined distally by microsatellite DXYS10086 and centromerically by SNP rs7067102 in amplicon S11. The 5′ limit was determined from the deletion found in proband 8, whereas the 3′ limit was determined from probands 9 and 10.
Although most of the deletions encompassed multiple loci, an independent confirmation of the presence of the deletions was necessary in families 11 and 12 because their detected deletions encompassed just one informative microsatellite (fig. 2). Confirmation was obtained by various means: (1) the designing of alternative primer pairs for the microsatellite repeat, (2) the exclusion of polymorphisms below the primers, and (3) SNP analysis in the flanking regions, to replicate the presence of a deletion. In family 11, confirmation was obtained by observing the deletion at SNP rs5946520 in amplicon S8, whereas the deletion in family 12 was verified using SNPs rs5946327, rs5946503, rs5946328, and rs5988279 in amplicon S6 and the previously unreported SNP rs38346960. Therefore, in all cases, we were able to confirm the presence of all identified deletions and to delimit their extensions.
Although the precise breakpoint boundaries could not be determined, our observations suggest the presence of some hotspots for deletion breakpoints. Five of the 12 families (families 1–3, 5, and 6) have deletions that possibly share a 5′ limit between the end of SHOX and amplicon S15, and 3 (families 10–12) have deletions that share a 5′ limit between DXYS10085 and DXYS10086 (fig. 2). Deletions in families 2, 3, and 6 may share the same 3′ boundary, between microsatellites DXS6814 and DXYS234; in families 7 and 8, between DXYS10097 and DX6814; and in families 10 and 11, between amplicons S8 and S11 (fig. 2).
FISH Analysis
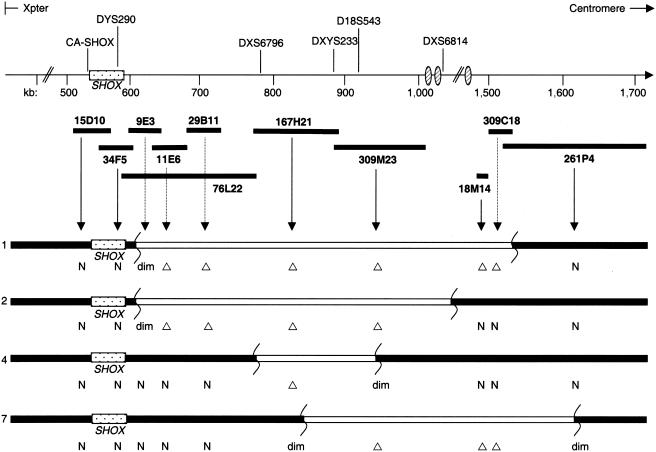
In four probands (of families 1, 2, 4, and 7), FISH analysis was performed to confirm the presence of a large-scale hemizygous deletion downstream of SHOX (fig. 3). A series of cosmids and BACs covering the targeted PAR1 segment were analyzed to determine the extent of the deletions (fig. 4). The FISH data corroborated the results obtained by microsatellite marker analysis (figs. 3 and 4) and confirmed that the deletions extended to different 5′ and 3′ locations (fig. 4). In addition, the presence of two hybridization signals with the overlapping cosmids 15D10 and 34F5, which together contain the entire SHOX coding sequence (Rao et al. 1997), confirmed that all four probands had two copies of SHOX and a heterozygous PAR1 deletion downstream of SHOX (figs. 3A and 4). The deletions were contiguous in all four cases, and the FISH probes that cover the deleted PAR1 region did not hybridize to other regions of the genome. Therefore, we can exclude the occurence of an inversion or translocation of this region to another part of the X or Y chromosome or to an autosome.
Figure 4.
Schematic representation of the FISH analysis in probands 1, 2, 4, and 7, by use of a series of PAR1 cosmids and BACs (not to scale). Microsatellites present in the respective cosmids or BACs are shown above the scale bar. The deletions (Δ) are represented below the corresponding cosmids or BACs. dim = diminished FISH signal, which indicates that the sequence was partly deleted and partly normal.
In family 1, the proband and her unaffected sibling were investigated by FISH. The results confirmed that the deletion was present in the affected sibling and was absent in the unaffected sister, thus validating the segregation pattern observed in the microsatellite analysis (fig. 3).
Discussion
SHOX mutations have been identified elsewhere in ∼60% of LWD cases, whereas the genetic defect in the remaining ∼40% remains unknown (Schiller et al. 2000; Falcinelli et al. 2002; Flanagan et al. 2002). In our study, we have identified the presence of a new class of PAR1 deletions that do not include the SHOX locus, in 12 (15%) of 80 patients with LWD who tested negative for SHOX mutations or deletions. The deletions were shown to cosegregate with the phenotype and were absent in 70 control families. Hence, our results reveal the existence of a second PAR1 region apparently implicated in the etiopathogenesis of LWD.
Affected probands displayed characteristic LWD phenotypic features indistinguishable from those displayed by other patients with LWD in whom SHOX deletions or point mutations were detected. Short stature was observed in all affected individuals, although the presence and severity of the Madelung deformity was variable. Interestingly, the presence of PAR1 deletions downstream of SHOX were also detected in probands with short stature and in affected relatives of probands with LWD who did not display the Madelung deformity—for example, family 12, in which the same PAR1 deletion is associated with a milder degree of affectation in the father (with short stature) than in the proband (with short stature and Madelung deformity). Thus, as described elsewhere for cases in which SHOX deletions were detected (Schiller et al. 2000), there seems to be a variable degree of affectation also in carriers of this new class of PAR1 deletions, probably as a result of sex, age, and other unknown modifying factors.
Short stature in LWD predominantly arises from a shortening of the distal bones of the extremities. The Madelung deformity is the defining feature of LWD and is represented by a short forearm with bowing of the radius and distal hypoplasia of the dorsally dislocated ulna. In general, the severity of the phenotype appears to increase with age, with much milder effects on stature and body proportion in childhood. It is also more severe in females than in males, thus indicating that the presence of female sex hormones plays an important role in determining the expression of the clinical phenotype. However, there are no clear phenotype-genotype correlations, and there is considerable interfamilial variation (Grigelioniene et al. 2001; the present study). Each of these factors may complicate diagnosis.
The characterization of a total of 12 different PAR1 deletions by use of a new panel of PAR1 microsatellites and SNPs revealed that the deleted regions were located within a range of ∼30–530 kb downstream of the SHOX 3′ end. Deletions varied in size, from the smallest at <81 kb to the largest at ∼501 kb. Fine mapping of all deletions allowed us to delimit a minimal commonly deleted region of 29 kb, located between microsatellite DXS6796 and amplicon S11. Both this new class of PAR1 deletions and the deletions described elsewhere, encompassing SHOX display genetic heterogeneity with regard to location and extent (Schiller et al. 2000; Schneider et al. 2005; authors' unpublished data). Nevertheless, there appears to be some clustering of the breakpoint boundaries, probably as a consequence of nonhomologous or homologous recombination events.
When we analyzed the contig of interest, which covers 766,173 bp of the PAR1 including SHOX, the overall percentage of repetitive sequence was found to be 55%, consistent with that of the overall X chromosome (Ross et al. 2005). However, a higher incidence of Alu repeats (28% vs. 8%) and a lower incidence of L1 long interspersed nuclear elements (7% vs. 29%) were found in this region, compared with the entire X chromosome. Low-copy repeats favor homologous recombination, thus predisposing the region to rearrangements (Stankiewicz and Lupski 2002). Indeed, the recombination fraction seems to be higher (26–38 fold) near the pseudoautosomal boundary than at the telomere (Lien et al. 2000; May et al. 2002), for which the recurrent incidence of deletions within this contig, including both SHOX and the new region of interest, is not unexpected. Unfortunately, because none of the deletions were de novo, a detailed investigation of their origin was not possible.
Two hypotheses may be postulated to explain why the deletions in this region may result in the LWD phenotype: the presence of a second, unknown gene implicated in the pathogenesis of LWD and short stature or, alternatively, a position effect on SHOX transcription. With regard to the first hypothesis, we examined, in each case, the sequence in the vicinity of the deletion-spanning region for evidence of transcript(s) that might be interrupted or encompassed by the deletion. No known transcripts were identified, but various bioinformatically predicted genes were present. Recently, the DNA sequence of the X chromosome was published by Ross et al. (2005). On the basis of their predictions, 33% of the X chromosome is transcribed, which is lower than the recent estimates for autosomes 6, 9, 10, and 13 (reviewed by Ross et al. [2005]). Consistent with the low gene density, the frequency of predicted CpG islands on the X chromosome is 50% below the estimated genome average. The PAR1 region to date consists of 34 known and novel protein-coding genes. Further work is therefore required to investigate whether any of these predicted transcripts are expressed and, if so, to analyze their functional relevance with respect to the pathogenesis of LWD and short stature.
For an alternative and more likely hypothesis, we can postulate a positional effect on SHOX transcription. Evidence supporting the position-effect hypothesis was previously obtained in a family with LWD carrying a deletion at microsatellite DXYS233 (Flanagan et al. 2002). Although no further characterization was undertaken to identify the deletion size and breakpoints, RNA expression analysis of bone-marrow fibroblasts obtained during corrective physiolysis procedure for the Madelung deformity revealed the presence of monoallelic expression of the SHOXb transcript (Flanagan et al. 2002). Although the underlying molecular mechanism remains unclear, the observation by Flanagan et al. (2002) seems to support the existence of a positional effect that affects SHOXb expression in association with the deletion at microsatellite DXYS233.
Gene regulatory elements have the ability to modulate gene expression over very long distances and are often found in gene deserts (Nobrega et al. 2003). Interestingly, the majority of loci associated with position effects have been described in genes that encode transcription factors involved in development (reviewed by Kleinjan and van Heyningen [2005]). The mechanism has been identified in some cases (reviewed by Kleinjan and van Heyningen [2005]), but it is often difficult to assess in human disease because access to the affected tissues is often impossible.
Further work is needed to identify the factor affected by these deletions, its role in the etiopathogenesis of LWD, and its significance in the control of skeletal development. Point mutations or small deletions or insertions may be present within this unknown factor, which, along with the identified deletions, may account for the remaining ∼40% of patients with LWD in whom no SHOX mutation or deletion has been identified.
In conclusion, we have identified a novel class of PAR1 deletions that do not include SHOX in 12 families with phenotypes fulfilling the diagnostic criteria for LWD. On the basis of our findings, deletion analysis of this additional PAR1 locus should be included in the routine mutation screening of patients for whom LWD, LMD, or ISS is suspected.
Acknowledgments
This work was supported by grants from the Ministerio de Ciencia y Tecnología (SAF2003-02511 [to K.E.H.]), Fondo de Investigación Sanitaria (PI021663 [to A.C.B.]), Red de Centros de Genética Clínica y Molecular, Fondo de Investigación Sanitaria (C03/07 [to J.A., A.C.B., and K.E.H.]), the Comunidad de Madrid (GR/SAL/0032/2004 [to K.E.H. and A.C.B.]), the Fundación Endocrinología y Nutrición, and the Fundación de Investigación Médica Mutua Madrileña (to K.E.H., A.C.B., and J.A.). This study was also partly supported by Serono España and a research grant from Lilly France. The French patients were included in the “Ed Lilly GeNeSIS SHOX sub-study module.” K.E.H. and A.C.B. were supported by investigator awards from the Program Ramón y Cajal, Ministerio de Ciencia y Tecnología and Fondo de Investigación Sanitaria (003145), respectively. S.B.S. and M.A.C. were recipients of Ph.D. fellowships from the Ministerio Ciencia y Tecnología (SAF2003-02511) and the Comunidad de Madrid, respectively. We thank all the clinicians and patients who participated in the study; Dr. Santiago Rodríguez de Córdoba, for providing DNA from control families; Dr. David Bunyan, who performed the MLPA work in Salisbury; and staff at the Wessex Regional Genetics Laboratory, for growing the FISH probes. FISH cosmids were kindly provided by Dr. Debbie Shears.
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- dbSNP, http://www.ncbi.nlm.nih.gov/SNP/ (for SNPs, including the newly identified rs38346958, rs38346959,rs38346960, rs38346961, rs38346962, rs38346963,rs38346964, rs38346965, rs38346966, and rs38346967)
- Ensembl Genome Browser, http://www.ensembl.org/Homo_sapiens/ (for sequence information of the human X and Y chromosomes)
- MRC-Holland B.V., http://www.mrc-holland.com/ (for details of the SHOX MLPA kit)
- GDB Human Genome Database, http://www.gdb.org/ (for locus information, including details of the new microsatellites DXYS10080, DXYS10081, DXYS10082, DXYS10083, DXYS10084, DXYS10085, DXYS10086, DXYS10087, DXYS10088, DXYS10089, DXYS10090, DXYS10091, DXYS10092, DXYS10093, DXYS10094, DXYS10095, DXYS10096, and DXYS10097)
- NCBI, http://www.ncbi.nlm.nih.gov/entrez/ (for sequence information on the human X and Y chromosomes, including contig NT_028413)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for SHOX, LWD, and LMD)
- RepeatMasker, http://www.repeatmasker.org/ (for screening DNA sequences for interspersed repeats and low-complexity DNA sequences)
- Tandem Repeats Finder, http://tandem.bu.edu/tools.html (for identifying microsatellites in sequence data)
References
- Belin V, Cusin V, Viot G, Girlich D, Toutain A, Moncla A, Vekmans M, Merrer ML, Munnich A, Cormier-Daire V (1998) SHOX mutations in dyschondrosteosis (Léri-Weill syndrome). Nat Genet 19:67–69 [DOI] [PubMed] [Google Scholar]
- Benson G (1999) Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res 27:573–580 10.1093/nar/27.2.573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder G, Schwarze CP, Ranke MB (2000) Identification of short stature caused by SHOX defects and therapeutic effect of recombinant human growth hormone. J Clin Endocrinol Metab 85:245–249 10.1210/jc.85.1.245 [DOI] [PubMed] [Google Scholar]
- Blaschke RJ, Topfer C, Marchini A, Steinbeisser H, Jansen JWG, Rappold GA (2003) Transcriptional and translational regulation of the Léri-Weill and Turner syndrome homeobox gene SHOX. J Biol Chem 278:47820–47826 10.1074/jbc.M306685200 [DOI] [PubMed] [Google Scholar]
- Clement-Jones M, Schiller S, Rao E, Blaschke R, Zuniga A, Zeller R, Robson S, Binder G, Glass I, Strachan T, Rappold G (2000) The short stature homeobox gene SHOX is involved in skeletal abnormalities in Turner syndrome. Hum Mol Genet 9:695–702 10.1093/hmg/9.5.695 [DOI] [PubMed] [Google Scholar]
- Ellison JW, Wardak Z, Young MF, Gehron Robey P, Laig-Webster M, Chiong W (1997) PHOG, a candidate gene for involvement in the short stature of Turner syndrome. Hum Mol Genet 6:1341–1347 10.1093/hmg/6.8.1341 [DOI] [PubMed] [Google Scholar]
- Falcinelli C, Lughetti L, Percescepe A, Calabrese G, Chiarelli F, Cisternino M, De Sanctis L, Pucarelli I, Radetti G, Wasniewska M, Weber G, Stuppia L, Bernasconi S, Forabosco A (2002) SHOX point mutations and deletions in Leri-Weill dyschondrosteosis. J Med Genet 39:e33 10.1136/jmg.39.6.e33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan SF, Munns CFJ, Hayes M, Williams B, Berry M, Vickers D, Rao E, Rappold GA, Batch JA, Hyland CJ, Glass IA (2002) Prevalence of mutations in the short stature homeobox containing gene (SHOX) in Madelung deformity of childhood. J Med Genet 39:758–763 10.1136/jmg.39.10.758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigelioniene G, Schoumans J, Neumeyer L, Ivarsson A, Eklof O, Enkvist O, Tordai P, Fosdal I, Myhre AG, Westphal O, Nilsson NO, Elfving M, Ellis I, Anderlid BM, Fransson I, Tapia-Paez I, Nordenskjold M, Hagenas L, Dumanski JP (2001) Analysis of short stature homeobox-containing gene (SHOX) and auxological phenotype in dyschondrosteosis and isolated Madelung deformity. Hum Genet 109:551–558 10.1007/s00439-001-0609-y [DOI] [PubMed] [Google Scholar]
- Huber C, Ipsas-Jouron S, Rosilio M, Salaun-Martin C, Munnich A, Cormier-Daire V (2004) Molecular analysis of the SHOX gene in a series of 100 patients with short stature [abstract 187]. Paper presented at the 54th Annual Meeting of the American Society of Human Genetics, Toronto, October 26–30 [Google Scholar]
- Kleinjan DA, van Heyningen V (2005) Long-range control of gene expression: emerging mechanisms and disruption in disease. Am J Hum Genet 76:8–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lien S, Szyda J, Schechinger B, Rappold G, Arnheim N (2000) Evidence for heterogeneity in recombination in the human pseudoautosomal region: high resolution analysis by sperm typing and radiation-hybrid mapping. Am J Hum Genet 66:557–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini A, Marttila T, Winter A, Caldeira S, Malanchi I, Blaschke RJ, Hacker B, Rao E, Karperien M, Wit JM, Richter W, Tommasino M, Rappold GA (2004) The short stature homeodomain protein SHOX induces cellular growth arrest and apoptosis and is expressed in human growth plate chondrocytes. J Biol Chem 279:37103–37114 10.1074/jbc.M307006200 [DOI] [PubMed] [Google Scholar]
- May CA, Shone AC, Kalydjieva L, Sajantila A, Jeffreys AJ (2002) Crossover clustering and rapid decay of linkage disequilibrium in the Xp/Yp pseudoautosomal gene SHOX. Nat Genet 31:272–275 10.1038/ng918 [DOI] [PubMed] [Google Scholar]
- Nicolaides NC, Stoeckert CJ Jr (1990) A simple, efficient method for the separate isolation of RNA and DNA from the same cells. Biotechniques 8:154–156 [PubMed] [Google Scholar]
- Nobrega MA, Ocharenko I, Afzal V, Rubin EM (2003) Scanning human gene deserts for long-range enhancers. Science 302:413 10.1126/science.1088328 [DOI] [PubMed] [Google Scholar]
- Pinkel D, Straume T, Gray JW (1986) Cytogenetic analysis using quantitative, high sensitivity, fluorescence hybridization. Proc Natl Acad Sci USA 83:2934–2938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao E, Blaschke RJ, Marchini A, Niesler B, Burnett M, Rappold GA (2001) The Léri-Weill and Turner syndrome homeobox gene SHOX encodes a cell-type specific transcriptional activator. Hum Mol Genet 10:3083–3091 10.1093/hmg/10.26.3083 [DOI] [PubMed] [Google Scholar]
- Rao E, Weiss B, Fukami M, Rump A, Niesler B, Mertz A, Muroya K, Binder G, Kirsch S, Winkelmann M, Bordsiek G, Heinrich U (1997) Pseudoautosomal deletions encompassing a novel homeobox gene cause growth failure in idiopathic short stature and Turner syndrome. Nat Genet 16:54–63 10.1038/ng0597-54 [DOI] [PubMed] [Google Scholar]
- Rappold GA, Fukami M, Niesler B, Schiller S, Zumkeller W, Bettendorf M, Heinrich U, Vlachopapadoupoulou E, Reinehr T, Onigata K, Ogata T (2002) Deletions of the homeobox gene SHOX (short stature homeobox) are an important cause of growth failure in children with short stature. J Clin Endocrinol Metab 87:1402–1406 10.1210/jc.87.3.1402 [DOI] [PubMed] [Google Scholar]
- Ross MT, Grafham DV, Coffey AJ, Scherer S, McLay K, Muzny D, Platzer M, et al (2005) The DNA sequence of the human X chromosome. Nature 434:325–337 10.1038/nature03440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller S, Spranger S, Schechinger B, Fukami M, Merker S, Drop S, Troger J, Knoblauch H, Kunze J, Seidel J, Rappold G (2000) Phenotypic variation and genetic heterogeneity in Léri-Weill syndrome. Eur J Hum Genet 8:54–62 10.1038/sj.ejhg.5200402 [DOI] [PubMed] [Google Scholar]
- Schneider KU, Sabherwal N, Jantz K, Röth R, Muncke N, Blum WF, Cutler GB Jr, Rappold G (2005) Identification of a major recombination hotspot in patients with short stature and SHOX deficiency. Am J Hum Genet 77:89–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shears DJ, Vassal HJ, Goodman FR, Palmer RW, Reardon W, Superti-Furga A, Scambler PJ, Winter RM (1998) Mutation and deletion of the pseudoautosomal gene SHOX cause Léri-Weill dyschondrosteosis. Nat Genet 19:70–73 [DOI] [PubMed] [Google Scholar]
- Stankiewicz P, Lupski JR (2002) Genome architecture, rearrangements and genomic disorders. Trends Genet 18:74–82 10.1016/S0168-9525(02)02592-1 [DOI] [PubMed] [Google Scholar]
- Stuppia L, Calabrese G, Gatta V, Pintor S, Morizio E, Fantasia D, Guanciali Franchi P, Rinaldi MM, Scarano G, Concolino D, Giannotti A, Petreschi F, Anzellotti MT, Pomilio M, Chiarelli D, Tumini DS, Palka G (2003) SHOX mutations detected by FISH and direct sequencing in patients with short stature. J Med Genet 40:e11 10.1136/jmg.40.2.e11 [DOI] [PMC free article] [PubMed] [Google Scholar]