Abstract
Previous evidence suggests that the inheritance of bipolar disorder (BP) may vary depending on the age at onset (AAO). Therefore, we sought to incorporate AAO as a covariate in linkage analyses of BP using two different methods, LODPAL and ordered-subset analysis (OSA), in genomewide scans of 150 multiplex pedigrees with 874 individuals. The LODPAL analysis identified two loci, on chromosomes 21q22.13 (LOD = 3.29; empirical chromosomewide P value = .009) and 18p11.2 (LOD = 2.83; empirical chromosomewide P = .05), with increased linkage among subjects who had early onset (AAO ⩽ 21 years) and later onset (AAO >21 years), respectively. The finding on 21q22.13 was significant at the chromosomewide level, even after correction for multiple testing. Moreover, a similar finding was observed in an independent sample of 65 pedigrees (LOD = 2.88; empirical chromosomewide P = .025). The finding on 18p11.2 was only nominally significant and was not observed in the independent sample. However, 18p11.2 emerged as one of the strongest regions in the OSA (LOD = 2.92; empirical P = .001), in which it was the only finding to meet chromosomewide levels of significance after correction for multiple testing. These results suggest that 21q22.13 and 18p11.2 may harbor genes that increase the risks for early-onset and later-onset forms of BP, respectively. There have been previous reports of linkage on 21q22.13 and 18p11.2, but the findings have not been consistent. This inconsistency may be due to differences in the AAO characteristics of the samples examined. Future studies to fine map susceptibility genes for BP on chromosomes 21q22.13 and 18p11.2 should take AAO into account.
Introduction
Bipolar disorder (BP) is a severe psychiatric disorder that manifests with alternating episodes of mania and depression. Although family, twin, and adoption studies suggest that BP has a strong genetic component, efforts to identify susceptibility genes have thus far yielded inconsistent results. The difficulties may be attributable to genetic (locus) heterogeneity. If a sample consists of subjects harboring different susceptibility genes, then the linkage signal at any one locus may be attenuated. Moreover, even if a linkage signal is identified, it may be difficult to replicate the finding in another sample that contains a different mixture of affected subjects. By using a clinical feature that is related to the underlying genetic heterogeneity, we may be able to identify more etiologically homogeneous subgroups and thereby increase the chances of successfully mapping susceptibility genes for BP.
Age at onset (AAO) is a potential clinical marker of genetic heterogeneity in BP. Previous studies suggest that there may be several subgroups of BP with distinct but overlapping distributions of AAO (Bellivier et al. 2001, 2003; Lin et al., in press). It has been shown that the rates of psychiatric comorbidities and clinical indicators of severity (e.g., a suicide attempt) vary across different AAO subgroups (Carlson et al. 1977, 2000; Carter et al. 2003; Lin et al., in press), and AAO subgroups aggregate in families, such that affected relatives typically have similar AAOs (McElroy et al. 2001; O’Mahony et al. 2002; Lin et al., in press). Additionally, a segregation analysis (Grigoroiu-Serbanescu et al. 2001) found that the pattern of transmission of BP in families differs with AAO. These lines of evidence support the argument that AAO is a heritable characteristic of BP and may be used to facilitate the search for susceptibility genes.
There are several approaches to incorporating information about AAO, as well as other covariates, into linkage analyses of complex disorders (e.g., see Greenwood and Bull 1997; Devlin et al. 2002; Glidden et al. 2003). One approach is to test for linkage directly to AAO as a quantitative trait. Other approaches treat AAO as a covariate rather than the phenotype of interest. These approaches may be classified into two categories. The first considers the family as the unit of analysis. In this case, families are stratified (or are grouped using something conceptually akin to stratification) according to AAO, and linkage is examined in the resulting subgroups. Hauser and colleagues (2004) recently introduced a method called “ordered-subset analysis” (OSA) that allows a systematic search for linkage in defined subgroups of families. The second category considers the relative pairs as the unit of analysis. An example of such an approach has been introduced by Olson (1999) and is implemented in the program LODPAL (Goddard et al. 2001). It builds on the affected sib pair (ASP) LOD score analysis proposed by Risch (1990) and allows for inclusion of covariates defined at the level of the relative pair.
The National Institute of Mental Health (NIMH) Genetics Initiative for Bipolar Disorder was established as a multisite collaborative effort to collect one of the largest samples of families for genetic investigations of BP by use of common methods of ascertainment and assessment. These families were genotyped in two waves consisting of 97 and 56 families. Analyses of the first wave of families (Detera-Wadleigh et al. 1997; Edenberg et al. 1997; Rice et al. 1997; Stine et al. 1997) yielded some evidence of linkage on chromosomes 5q, 6q, 7p, 7q, 9q, 10q, 13q, 15p, 16p, 21q, Xp, and Xq. However, the findings were not sustained in analyses of the second wave of families (Dick et al. 2002; Willour et al. 2003; Zandi et al. 2003), except for the finding on 16p. McInnis et al. (2003a) subsequently combined the data from the two waves of families and performed a genomewide scan of the entire data set. They observed three peak linkage signals with a LOD score >2.0, on 16p13, 20p12, and 10p12, but none of the findings reached conventional levels of significance. The inconsistent findings across samples and the lack of results reaching conventional levels of significance are characteristic of linkage studies of BP and point to the heterogeneity of the condition.
By incorporating information about AAO, we may be able to reveal the underlying heterogeneity in BP and thereby facilitate the mapping of bipolar susceptibility genes. Faraone and colleagues (2004) recently examined the first wave of the NIMH Genetics Initiative families by using AAO as a quantitative trait, and they reported linkage on chromosomes 12, 14, and 15. Here, we report the results of a linkage analysis of the combined data from the first and second waves of families (McInnis et al. 2003a), using both relative-pair and family-level covariate methods.
Subjects and Methods
Ascertainment and Assessment
The ascertainment and assessment of the subjects have been described in detail elsewhere (McInnis et al. 2003a). In brief, the sample was collected as part of the NIMH Genetics Initiative for Bipolar Disorder. Pedigrees were ascertained through probands with bipolar I disorder (BPI) by inclusion criteria consistent across four sites: Indiana University, Washington University in St. Louis, The Johns Hopkins University, and the NIMH. Inclusion criteria for families were as follows: (1) there was at least one other first-degree relative affected with either BPI or schizoaffective disorder, bipolar type (SABP); (2) either the proband or a second affected relative had at least two living siblings aged ⩾18 years; and (3) only one parent was affected with BPI or SABP (i.e., it was a unilineal family). The diagnoses of BPI and SABP were based on Diagnostic and Statistical Manual of Mental Disorders, 3rd edition (DSM-IIIR) criteria (American Psychiatric Association 1987). The diagnoses of bipolar II disorder (BPII) and recurrent major depression (RMD) were based on modified Research Diagnostic Criteria (Gershon et al. 1989). Investigators used the semistructured Diagnostic Interview for Genetic Studies (DIGS) (Nurnberger et al. 1994) and the Family Interview for Genetic Studies to assess key diagnoses, comorbid psychiatric diagnoses, and clinical features. All diagnoses were made by best-estimate procedures based on the consensus of two noninterviewing psychiatrists. We chose two phenotype models for linkage analysis: a narrow-phenotype model that included SABP, BPI, and BPII and a broad-phenotype model that included SABP, BPI, BPII, and RMD. AAO was defined as the self-reported age at the first episode of depression or mania that met diagnostic criteria.
Genotyping
A total of 153 pedigrees were genotyped in two waves. There were 97 pedigrees in the first wave and 56 pedigrees in the second wave. Genotyping was done at one of the four above-mentioned institutions by use of PCR-based methods performed using either fluorescent dye–labeled primers resolved on a DNA sequencer or radiolabeled primers with PAGE and autoradiography (Detera-Wadleigh et al. 1997; Edenberg et al. 1997; Rice et al. 1997; Stine et al. 1997; Dick et al. 2002; Willour et al. 2003; Zandi et al. 2003). A total of 425 markers were genotyped in the first 97 pedigrees, and 378 markers were genotyped in the remaining 56 pedigrees (further information about the genotyped markers is available at the Center for Collaborative Genetic Studies on Mental Disorders Web site). Of these, 298 markers were genotyped in common across both sets. To align the allele bins of the common markers, we used data from two CEPH control subjects who were genotyped across pedigree sets and/or allele frequencies. For quality control, we used the programs Genetic Analysis System (Young 1995) and UNKNOWN (Lathrop et al. 1984) to check for inheritance inconsistencies. We then used the chrompic routine of CRIMAP (Lander et al. 1987) to check for unlikely double recombinations. Finally, we used the pedigree relationship statistical test (Sun et al. 2002) to check for family-relationship inconsistencies by use of genomewide data. Identified inconsistencies were resolved by deleting the problematic genotypes. The inconsistencies could not be resolved in three pedigrees; these pedigrees were therefore removed from the sample. This left 150 pedigrees containing 874 individuals for the linkage analyses.
Replication Sample
We used an independent sample consisting of 65 pedigrees, to further explore our positive linkage findings. This sample has been described in detail elsewhere (McInnis et al. 2003b). The pedigrees were ascertained through probands with BPI who had to have at least two affected first-degree relatives. All subjects were assessed by trained psychiatrists using the Schedule for Affective Disorders and Schizophrenia–Lifetime Version (Endicott and Spitzer 1978), and diagnoses were assigned using best-estimate procedures with Research Diagnostic Criteria (Endicott and Spitzer 1979). The genotyping for these 65 families was performed at The Johns Hopkins University, Stanford University, and the Center for Inherited Disease Research (for further details of the markers studied, see the work by McInnis et al. [2003b]). Quality control for the genotyping was performed using the same procedures described above.
Statistical Analyses
We used the following procedures to construct genetic maps of the markers genotyped in both the initial and replication samples. First, we ordered the markers using the physical sequence databases. We then obtained genetic map distances between markers by analyzing our data with CRIMAP. Finally, we compared our maps for consistency with the published maps from Marshfield and deCODE.
We then analyzed the genotype data using two different covariate-based linkage methods. First, we used an affected relative pair approach implemented in the program LODPAL of the S.A.G.E. package (version 4.4). The approach was first introduced by Olson (1999) and was later modified by Goddard et al. (2001). It is based on a conditional logistic parameterization of the recurrence risk ratios for offspring and MZ twins, conditional on any number of covariates. The model estimates a parameter for the “average” linkage in the sample, as well as parameters for the change in linkage as a function of the included covariates. These parameters can be used to estimate the relative recurrence risk ratios as a function of the covariates.
We examined the effect of AAO on linkage by comparing models with and without AAO entered as a covariate. We treated AAO as either a continuous or a dichotomous variable. We dichotomized AAO at age 21 years (⩽21 vs. >21) on the basis of previous evidence that this cutoff empirically distinguished early- and later-onset forms of the disease (Lin et al., in press). The AAO covariates were entered into the linkage model as the sum of the values for the two individuals in each affected relative pair. The summed covariates were then centered on their sample mean before parameter estimation.
Results are reported as LOD scores, where the LOD score equals the likelihood-ratio statistic (LRS) from the fitted model divided by 4.605 (i.e., 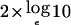 ). Significance values were derived on the basis of the asymptotic distribution of the LRS. The distribution of the LRS for the baseline model with no covariates is a 50:50 mixture of a point mass at 0 and a χ2 distribution with 1 df. The distribution of the LRS for the one-covariate model is a 50:50 mixture of χ2 distributions with 1 df and 2 df. To test the significance of the covariate effect, the LRS difference between models with and without the covariate can be compared with a χ2 distribution with 1 df.
). Significance values were derived on the basis of the asymptotic distribution of the LRS. The distribution of the LRS for the baseline model with no covariates is a 50:50 mixture of a point mass at 0 and a χ2 distribution with 1 df. The distribution of the LRS for the one-covariate model is a 50:50 mixture of χ2 distributions with 1 df and 2 df. To test the significance of the covariate effect, the LRS difference between models with and without the covariate can be compared with a χ2 distribution with 1 df.
Because asymptotic P values estimated on the basis of a mixture of χ2 distributions may be liberal, we further evaluated the significance of our best findings using permutation methods. To test the covariate effect of AAO on linkage at a locus, we randomly permuted the AAO covariate values among the affected relative pairs 1,000 times. We then analyzed each replicate with LODPAL to determine the number of times that, across the chromosome, there was an increase in the LOD score from that in the observed data set. This number, divided by 1,000, was taken as the empirical chromosomewide P value for the covariate effect. Similarly, to test the overall linkage at a locus with AAO entered as a covariate, we kept the AAO covariate fixed but randomly permuted the genotypes based on our families and the pattern of missing data 1,000 times, using the simulation package in Merlin. We then analyzed each replicate with LODPAL to determine the number of times that, across the chromosome, there was a LOD score as high as that in the original data. This number, divided by 1,000, was taken as the empirical chromosomewide P value for overall linkage. Because we evaluated two phenotype models and incorporated AAO as both a continuous and a dichotomous covariate, we corrected the chromosomewide significance level by dividing 0.05 by 4 (=0.0125). To account for all 22 autosomal chromosomes tested, we corrected the genomewide significance level by dividing 0.0125 by 22 (=0.0006).
The second covariate-based linkage method we used was OSA, as implemented in OSA software, version 2.1 (Hauser et al. 2004). OSA tests for linkage in subsets of families defined by different covariate values, without having to choose arbitrary cutoff points for the covariate. The first step in performing an OSA is to assign a covariate value to each family. We used two definitions for AAO: the mean and minimum values of AAO for all affected subjects in a family. OSA then uses the ASM module implemented in GENEHUNTER-PLUS (Kong and Cox 1997) to calculate allele-sharing LOD scores with the Spair statistic and a linear model in successive subsets of the families by the following procedures. First, families are ranked according to their covariate values in both low-to-high and high-to-low directions. Families with the same covariate values are assigned the same rank. Second, allele-sharing LOD scores are estimated across a chromosome in the first-ranked family. Third, the second-ranked family is added to the subset of families, and the allele-sharing LOD scores are recalculated. Fourth, the procedures are repeated, as higher-ranked families are added one by one, until all families are analyzed as a final subset. The maximum allele-sharing LOD scores for each subset of families and the positions where the maximum scores occurred are recorded.
To assess the significance of any increment between LOD scores obtained in a subset, we used the permutation tests available in the OSA software package to calculate empirical P values. These permutation tests were performed by randomly ordering the families with the same number of ranks as in the original analysis. A total of 10,000 randomized ordered samples were generated, and maximum LOD scores were obtained through the OSA procedures described above. We counted events for which the maximum allele-sharing LOD score detected at any position along the chromosome was greater than or equal to the original subset maximum LOD score. The number of such events per 10,000 was considered to be the empirical chromosomewide P value. Because we examined two phenotype models, two covariates (i.e., mean and minimum AAO values), and two directional orders (low-to-high and high-to-low), we corrected the chromosomewide significance level by dividing 0.05 by 8 (=0.00625). To adjust for 22 autosomal chromosomes in the genome scan, the genomewide significance level was calculated as 0.00625 divided by 22 (=0.0003).
Results
The NIMH Genetics Initiative sample consisted of 874 genotyped subjects in 150 pedigrees. A total of 638 subjects were diagnosed with a major mood disorder, including 363 (56.9%) with BPI, 37 (5.8%) with SABP, 110 (17.2%) with BPII, and 128 (20.1%) with RMD. The replication sample consisted of 561 genotyped subjects in 65 pedigrees. A total of 298 subjects were diagnosed with a major mood disorder, including 7 (2.4%) with SABP, 127 (42.6%) with BPI, 97 (32.5%) with BPII, and 67 (22.5%) with RMD. The subjects and the AAO characteristics for the two samples are further described in table 1. These two samples did not differ significantly in terms of sex composition or AAO characteristics.
Table 1.
Characteristics of the Families in the NIMH Genetics Initiative and Replication Samples[Note]
|
NIMH Sample |
Replication Sample |
|||
| Characteristic | Narrow Phenotype | Broad Phenotype | Narrow Phenotype | Broad Phenotype |
| No. of affected subjects | 510 | 638 | 231 | 298 |
| No. (%) of affected femalesa | 299 (58.6) | 387 (60.7) | 134 (58.0) | 183 (61.4) |
| Mean no. of affected subjects per family | 3.7 | 4.2 | 3.5 | 4.6 |
| Range of no. of affected subjects per family | 1–8 | 2–10 | 1–9 | 2–10 |
| No. of all possible affected relative pairs | 730 | 1,038 | 260 | 506 |
| Mean ± SD AAOa (years) | 20.8 ± 9.5 | 22.0 ± 10.7 | 22.1 ± 9.9 | 23.1 ± 10.5 |
| Range of AAO (years) | 5–60 | 5–76 | 6–80 | 4–80 |
| No. (%) of subjects with early onset (AAO⩽21 years)a | 327 (64.6) | 385 (61.0) | 137 (59.6) | 163 (54.9) |
| No. (%) of subjects with late onset (AAO>21 years)a | 179 (35.4) | 246 (39.0) | 93 (40.4) | 134 (45.1) |
| Range of mean AAO per family (years) | 7.7–35 | 9.8–39.3 | 13–38 | 14–34 |
| Range of minimum AAO per family (years) | 5–31 | 5–31 | 6–38 | 4–26 |
Note.— The NIMH sample comprised 150 families, including 874 genotyped individuals. The replication sample comprised 65 families, including 561 genotyped individuals.
T tests were used to compare continuous variables, and χ2 tests were used to compare categorical variables across samples; none were found to be significantly different (P<.05).
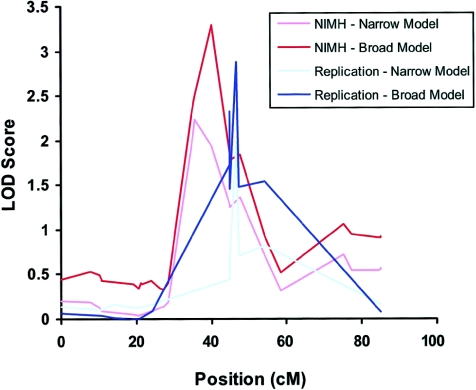
The LODPAL analyses revealed two chromosomal regions of interest where the LOD score increased by >2 after AAO was included as a covariate in analyses under either the narrow-phenotype or the broad-phenotype model. The most significant region was on chromosome 21 (see fig. 1). Under the broad-phenotype model, the LOD score increased from 0.02 to 3.29 (for the increase in LOD, P=.0001; for the overall LOD, P=.0003) on chromosome 21q22.13 near marker D21S1252 (chromosome 21: 36,647,255–36,847,595 bp) with AAO treated as a dichotomous variable. Permutation tests at this locus yielded an empirical chromosomewide P value of .006 for the increase in LOD and .009 for the overall LOD. The 1-LOD support interval was ∼6 cM, from marker D21S1254 to D21S1252. The estimated parameter for AAO at this locus was negative, which suggests that the evidence for linkage was greatest among subjects with early-onset BP. We estimated that the sibling recurrence risk at this locus decreased from 1.60 for subjects with an AAO of 6 years to 0.75 for subjects with an AAO of 34 years.
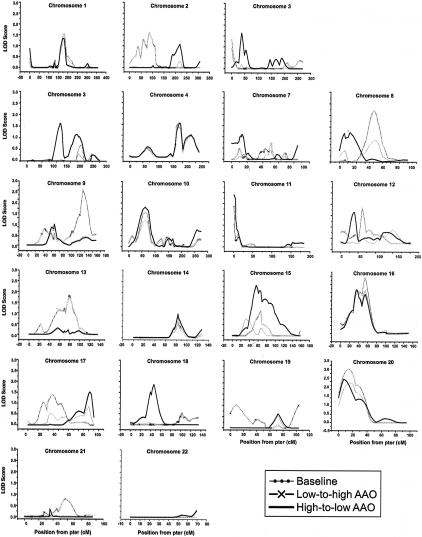
Figure 1.
LOD scores along chromosome 21, from the LODPAL analyses of the NIMH sample (n=150 families) and replication sample (n=65 families). Results from the one-covariate model with AAO as a dichotomous variable under the broad- and narrow-phenotype models are shown. For the sake of clarity, baseline models without AAO, which generally yielded LOD scores <0.05, are not shown. The peak finding on chromosome 22q22.13 in the NIMH sample was ∼2.5 Mb away from the peak finding in the replication sample. In both samples, the increased evidence of linkage was observed among affected subjects with early-onset BP (AAO⩽21 years).
Under the narrow-phenotype model, the LOD score increased from 0.12 to 2.24 (for the increase in LOD, P=.0018; for the overall LOD, P=.0035) on chromosome 21q22.11 near marker D21S65 (chromosome 21: 34,909,475–35,109,672 bp) when AAO was treated as a dichotomous variable. However, the LOD score at this locus increased even more, to 3.20 (for the increase in LOD, P=.0001; for the overall LOD, P=.0004) when AAO was treated as a continuous variable (see figs. 2–5). Additionally, a neighboring peak emerged at marker pfkl (chromosome 21: 44,576,175–44,603,450 bp) in the gene for phosphofructokinase, liver type (PFKL [MIM 171860]). Here, the LOD score increased from 0.00 to 2.98 (for the increase in LOD, P=.0002; for the overall LOD, P=.0006). It is difficult to determine whether the two neighboring peaks represented the same or distinct loci. Consistent with the findings for the broad-phenotype model, the evidence of linkage at these loci was greatest among subjects with early-onset disease.
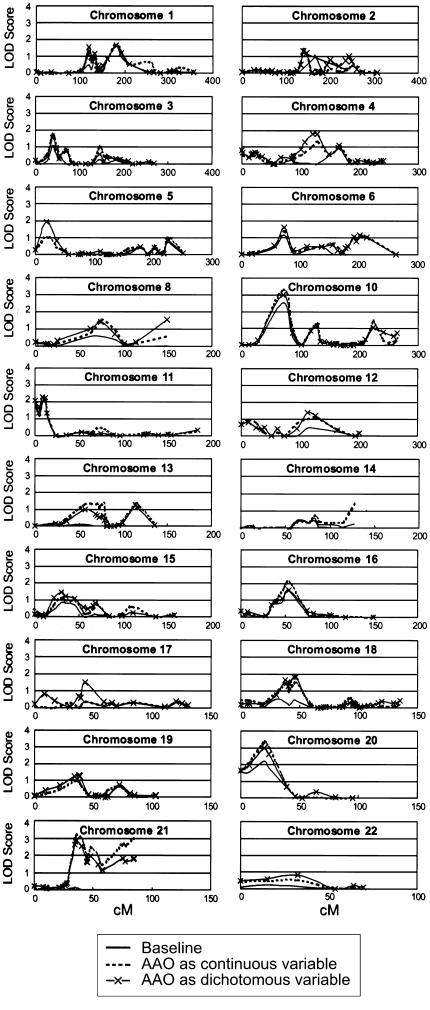
Figure 2.
Genomewide scan results from the LODPAL analysis under the narrow-phenotype model.
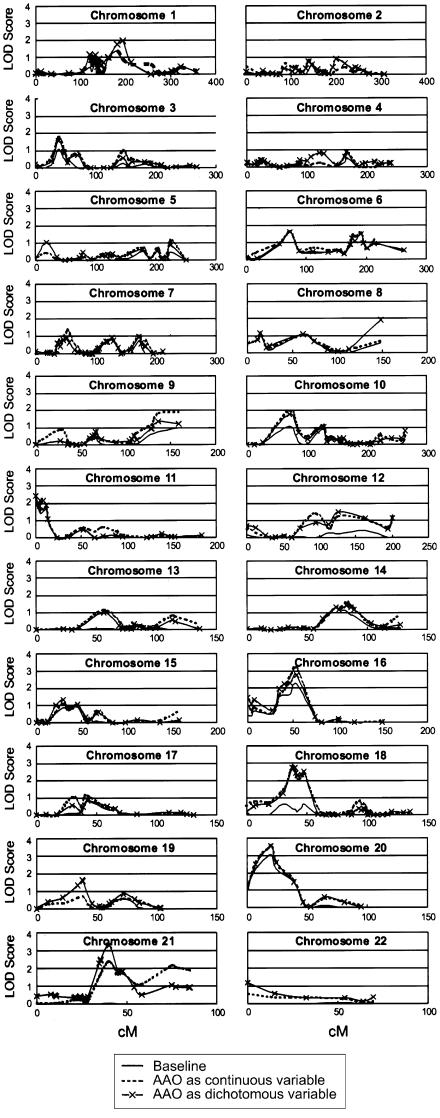
Figure 3.
Genomewide scan results from the LODPAL analysis under the broad-phenotype model.
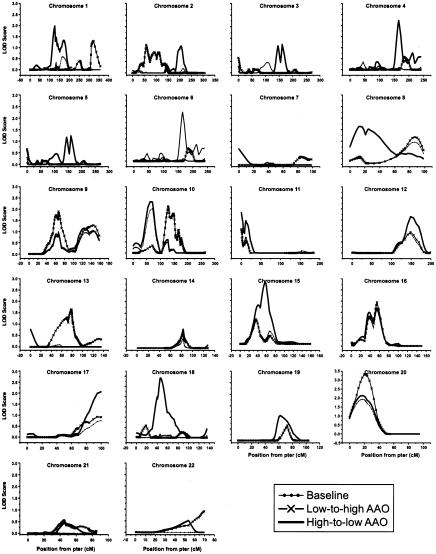
Figure 4.
Genomewide scan results from the OSA under the narrow-phenotype model. “Baseline” denotes the LOD score in the overall sample. “Low-to-high AAO” denotes the LOD score obtained in the subset ranked by family-specific AAO from low to high values, whereas “High-to-low AAO” denotes the LOD score obtained in the subset ranked by family-specific AAO from high to low values.
Figure 5.
Genomewide scan results from the OSA under the broad-phenotype model. “Baseline” denotes the LOD score in the overall sample. “Low-to-high AAO” denotes the LOD score obtained in the subset ranked by family-specific AAO from low to high values, whereas “High-to-low AAO” denotes the LOD score obtained in the subset ranked by family-specific AAO from high to low values.
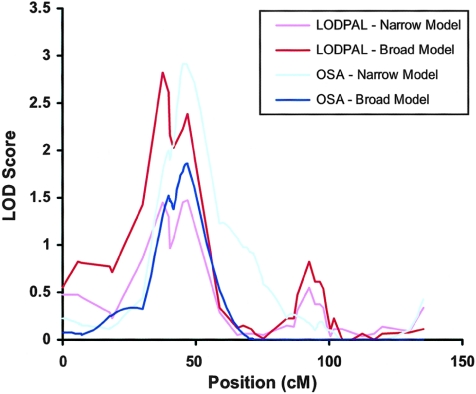
The other finding of interest was on chromosome 18 (see fig. 6). Under the broad-phenotype model, the LOD score increased from 0.35 to 2.83 (for the increase in LOD, P=.0007; for the overall LOD, P=.0009) on chromosome 18p11.2 near marker D18S1150 (chromosome 18: 10,136,591–10,336,929 bp), when AAO was treated as a continuous variable. Permutation tests in this region yielded an empirical chromosomewide P value of .018 for the increase in LOD and .05 for the overall LOD. The 1-LOD support interval was ∼10 cM, from marker D18S1150 to D18S40. The regression coefficient for the AAO covariate was positive, suggesting that the evidence of linkage was greatest among those with later AAO. We estimate that the sibling recurrence risk at this locus ranged from 0.64 for subjects with an AAO of 6 years to 1.58 for subjects with an AAO of 34 years. The findings were not as compelling under the narrow-phenotype model. Here, the LOD score increased from 0.42 to 1.47 (for the increase in LOD, P=.028; for the overall LOD, P=.022) on chromosome 18p11.2 near marker D18S40 (chromosome 18: 13,643,793–13,843,883 bp).
Figure 6.
LOD scores along chromosome 18 from the LODPAL and OSA analyses of the NIMH sample (n=150 families) under the broad- and narrow-phenotype models are presented. For the LODPAL analyses, results from the one-covariate model with AAO as a continuous variable are shown. For the OSA analyses, results from the high-to-low slicing of the data set are shown. Baseline models without AAO, which generally yielded LOD scores <0.05, are not shown, for the sake of clarity. The results from both sets of analyses converged at chromosome 18p11.2 and showed increased evidence of linkage among affected subjects with later-onset BP (AAO>21 years).
We sought to further explore the findings with AAO on chromosomes 21q22.13 and 18p11.2 in an independent sample of 65 pedigrees. The results were similar on chromosome 21q22.13 (see fig. 1) but not 18p11.2. At marker D21S1444 (chromosome 21: 38,179,973–38,380,289 bp) the LOD score increased from 0.28 to 2.32 (for the increase in LOD, P=.0056; for the overall LOD, P=.0022) when examined under the broad-phenotype model and with AAO treated as a dichotomous variable. At a neighboring marker, D21S156 (chromosome 21: 39,156,176–39,356,252 bp), the LOD score similarly increased from 0.65 to 2.88 (for the increase in LOD, P=.0036; for the overall LOD, P=.0014). Permutation tests at this marker yielded an empirical chromosomewide P value of .012 for the increase in LOD and .025 for the overall LOD. Markers D21S144 and D21S156 are ∼1.5 and 2.5 Mb away, respectively, from the peak signal observed in the initial sample. The regression coefficient for the AAO covariate was negative for these two markers, again suggesting that linkage at this locus was greatest among those with younger AAO.
OSA of the NIMH Genetics Initiative sample revealed a total of five loci that were nominally significant (P<.05) under the narrow-phenotype model and five loci under the broad-phenotype model (table 2). Only one locus, on chromosome 18p11.2, had chromosomewide significance in either phenotype model after correction for multiple tests. Under the narrow-phenotype model, the maximum LOD score occurred near marker D18S37 (chromosome 18: 13,080,902–13,280,999 bp) in the subset of families with the oldest mean AAO range: 22–35 years. Here, the maximum LOD score increased from 0.13 to 2.92 (P=.001). This subset comprised 54 families, or 36% of the overall sample. Chromosome 18p11.2 was also one of the top findings under the broad-phenotype model. Under this model, the LOD score increased from 0.04 to 1.86 (P=.039) in a subset of families with a mean AAO age range of 24–35 years. This subset comprised 46 families, or 31% of the sample. The OSA LOD scores along chromosome 18 are shown in figure 6.
Table 2.
Nominally Significant OSA Findings, by Chromosome, with AAO as the Key Covariate[Note]
| Chromosome andPhenotype Model | NearestMarker | Distancefrom pter(cM) | BaselineLOD | PeakLOD | δ LODa | EmpiricalP | No. (%) ofFamilies | AAO Range(years) |
| 3: | ||||||||
| Narrow | D3S1763 | 209.9 | .00 | 2.04 | 2.04 | .044 | 31 (21) | 5–9b |
| 4: | ||||||||
| Narrow | D4S1625 | 165.5 | .10 | 2.23 | 2.13 | .036 | 29 (19) | 20.3–35c |
| 5: | ||||||||
| Narrow | D5S1354 | 250.9 | .19 | 2.22 | 2.03 | .045 | 39 (26) | 18–31b |
| Broad | D5S1354 | 209.9 | .07 | 1.87 | 1.80 | .040 | 32 (21) | 18–31b |
| 7: | ||||||||
| Broad | D7S1799 | 127.5 | .37 | 1.75 | 1.38 | .045 | 90 (60) | 13–31b |
| 9: | ||||||||
| Broad | D9S915 | 136.5 | .48 | 3.03 | 2.55 | .029 | 53 (36) | 9.8–20c |
| 15: | ||||||||
| Narrow | gata153f11 | 56.2 | .20 | 2.83 | 2.63 | .021 | 41 (27) | 24–35c |
| Broad | gata153f11 | 55.1 | .06 | 2.31 | 2.25 | .050 | 24 (16) | 30.3–39.3c |
| 18: | ||||||||
| Narrow | D18S37 | 45.2 | .13 | 2.92 | 2.79 | .001 | 54 (36) | 22–35c |
| Broad | D18S40 | 47.0 | .04 | 1.86 | 1.82 | .039 | 60 (40) | 24.2–39.3c |
Note.— Analysis was performed on the entire sample of 150 families.
δ LOD score denotes the difference between the peak LOD score and the baseline LOD score.
Family-specific minimum values of AAO.
Family-specific mean values of AAO.
To further explore these findings, we examined the clinical features in the subset of 54 families with the oldest mean AAO who appeared to be most linked on chromosome 18 under the narrow-phenotype model. Compared with the rest of the sample, the subjects in those families tended to have lower risks of alcohol abuse/dependence (26.2% vs. 35.7%; P=.004), rapid cycling (36.3% vs. 46.3%; P=.04), and suicide attempts (14.9% vs. 25.2%; P<.001), as well as a lower overall number of comorbidities (1.2 vs. 1.6; P<.001). These findings were consistent with our report elsewhere that AAO in BP is inversely associated with the risks of a number of comorbid psychiatric disturbances (Lin et al., in press). Consideration of these other clinical features, however, did not improve our findings on chromosome 18.
Discussion
We used two different approaches for incorporating AAO as a covariate in linkage analyses of BP and identified two regions of interest. On chromosome 21q22.13, there was evidence of linkage to earlier-onset BP, whereas, on chromosome 18q11.2, there was evidence of linkage to later-onset BP. Linkage has been reported to both loci in previous studies. Interestingly, there was little evidence of linkage in our sample until AAO information was included in the analysis. These findings suggest that AAO may be a clinical marker of the underlying heterogeneity in BP, demarcating subtypes of the disorder with distinct genetic susceptibilities.
The strongest finding was on chromosome 21q22.13 in the relative-pair analysis of LODPAL. Empirical tests with permutations suggested that the finding was significant at the chromosomewide level (threshold P<.0125) after correction for multiple tests. The finding did not quite reach the genomewide level of significance (threshold P<.0006), but the corrections we employed were probably overly conservative, since the multiple tests performed were not independent. Moreover, we have found that the linkage signal on chromosome 21q22.13 was also strongest among individuals with early onset in another independent sample, lending additional support to the finding in this 21q region. There were two neighboring peaks identified under the narrow-phenotype model, but this finding resolved more clearly to become one peak spanning ∼6 cM under the broad-phenotype model. Genotyping a denser map of markers across the region should help clarify the localization of the finding.
The other finding of interest was on chromosome 18p11.2. In the relative-pair analysis of LODPAL, the finding was only nominally significant with permutation tests, and we were unable to replicate the finding in an independent sample. However, the locus emerged as one of the top findings also in the family-level analysis of OSA. The consistent findings with two different covariate-based methods merit attention. Thus, further investigation of the region in this and other samples is needed to confirm the findings related to AAO.
Several previous studies have reported linkage of BP to 21q and 18p. As mentioned above, a linkage signal was detected on chromosome 21q, near markers D21S1254, D21S65, D21S1440, and D21S1255, in the 97 families of the first wave of the NIMH Genetics Initiative (Detera-Wadleigh et al. 1997). However, this linkage signal was not sustained in 53 families of the second wave (Willour et al. 2003). When the two samples were combined, the linkage signal at this locus disappeared altogether (McInnis et al. 2003a). To assess whether the mixed evidence of linkage across samples may be attributable to differences in AAO characteristics, we compared the AAO distribution of the two samples and found that the proportion of affected individuals from families with early-onset disease (proband’s AAO ⩽21 years) was significantly greater in the first wave than in the second (73.4% vs. 63.0%; P<.001).
There have also been several reports of linkage to the PFKL gene, which encodes phosphofructokinase, a key regulatory enzyme in glycolysis. Straub and colleagues (1994) found evidence for linkage to PFKL using a parametric dominant model (LOD=3.41) and an affected-pedigree-member (APM) method (P<.0003). Other studies have reported similar findings (Detera-Wadleigh et al. 1996; Liu et al. 2001). In one follow-up study of 40 U.S. and Israeli pedigrees, Aita and colleagues (1999) obtained a two-point heterogeneity (α=0.5) LOD score of 3.35 (P<.000156) at D21S1260, which is 5 cM from PFKL.
Linkage to chromosome 18p11.2 has been reported in a number of previous studies as well. Berrettini and colleagues (1994) first reported linkage evidence at chromosome region 18p11 in 22 white families. Using ASP and APM methods, they obtained a P value of .0004 at marker D18S21 (∼3 Mb away from our peak finding on chromosome 18 at D18S1150) under a broad-phenotype model similar to the one we tested. A number of studies followed-up this finding and also detected significant linkage signals in the region (e.g., see Stine et al. 1995; De Bruyn et al. 1996; Lin and Bale 1997; Bennett et al. 2002). This region contains the IMPA2 gene, which encodes the enzyme myoinositol monophosphatase (IMPase) in the phospholipase C signaling pathway, which can be inhibited by the mood-stabilizer lithium at therapeutic levels (Sjoholt et al. 2000). It has been suggested that IMPA2 promoter polymorphisms are associated with BP (Sjoholt et al. 2004). In addition, this region may harbor a gene contributing to the etiology of both BP and schizophrenia (Berrettini 2000).
Despite these promising findings, not all studies have observed linkage on chromosomes 21q22.13 and 18p11.2. The conflicting findings may be due to genetic heterogeneity in BP. Some samples may be enriched for affected subjects with susceptibility genes at either of these two loci. AAO may reflect the underlying heterogeneity at these two loci and therefore may be used to uncover previously obscured linkage in samples that are less enriched. The strategy of using clinical features to identify more-homogeneous subsets of affected subjects to facilitate genetic mapping has been used successfully in the past with other complex disorders (e.g., see Hall et al. 1990; Pericak-Vance et al. 1991). The results of the current study encourage wider application of this strategy.
As described above, there are several different approaches to incorporating covariates into linkage analyses. The approaches that we and others (Faraone et al. 2004) have used to examine AAO in BP have yielded some divergent results. The divergent results may simply be due to chance. On the other hand, they may provide some insights into the nature of the underlying heterogeneity. For example, in our analyses, linkage to chromosome 21q22.13 was identified by a relative-pair approach but not a family-level one. In the family-level approach, the covariate of interest must be defined at the level of the family, such as by taking the mean, minimum, or maximum value across all affected subjects in the family. As a result, the family-level approach may not be sensitive to linkage if there is intra- and interfamilial heterogeneity at a locus. The fact that both the relative-pair and family-level approaches identified linkage on chromosome 18p11.2 may suggest that there is little intrafamilial heterogeneity at this locus. Interestingly, none of the loci identified by Faraone and colleagues (2004) were detected in our analyses. Perhaps this is not surprising, given that they examined a slightly different sample and used a quantitative-trait approach that may be better suited to searching for modifier genes that do not increase the susceptibility for a disorder per se, but, rather, modify the AAO. Clearly, more methodological work is needed to help interpret the divergent results from the different covariate-based linkage analyses and to clarify which are most appropriate to use under various circumstances.
A notable limitation of the current study is that AAO was defined retrospectively on the basis of self-reported information. The validity of such information may be degraded by poor recall (i.e., recall bias). Subjects may not be able to accurately report on their history of mood symptoms. As a result, there may have been some error in the assigned AAOs used in the current linkage analyses. However, there is no reason to believe that the direction of this error would be related in any way to the genotype data. Random error of this sort would tend to introduce noise into the linkage analyses and would likely bias the findings toward the null. Despite this, we still observed robust linkage signals on chromosomes 21q22.13 and 18p11.2. In addition, we reasoned that subjects who might not be able to accurately recall the exact age at disease onset would be able to distinguish whether they had an early onset versus a late one. Thus, we decided to examine AAO as both a continuous and a dichotomous covariate. We used findings from a previous investigation of AAO in BP to empirically determine the most reasonable cutoff for the dichotomization of AAO (Lin et al., in press).
The study also has several important strengths. First, the study included one of the largest pedigree samples of BP collected using common ascertainment and assessment criteria. The large sample size provided improved power for identification of linkage with AAO as a covariate. Second, all subjects were assessed with the DIGS, a widely used and well-validated instrument, and experienced clinicians used best-estimate procedures to assign diagnoses of mood disorders. Thus, concerns about misclassification of affection status were minimized. Finally, the replication sample was ascertained and assessed by the same team of researchers who played a leading role in collecting the primary sample, which ensured consistency across samples.
We have shown that AAO is potentially a clinical marker of genetic heterogeneity in BP and that affected subjects with early-onset versus late-onset disease may carry variants of different susceptibility genes. Specifically, we identified a locus on chromosome 21q22.13 that may harbor a susceptibility gene for earlier-onset BP and another locus on chromosome 18p11.2 that may harbor a susceptibility gene for later-onset BP. The findings encourage additional investigations in these regions. Future investigations should take into account information on AAO to facilitate identification of the relevant susceptibility variants.
Acknowledgments
Data and biomaterial were partly collected in four projects as part of the National Institute of Mental Health (NIMH) Bipolar Disorder Genetics Initiative. From 1991 to 1998, the four project institutions and the respective principal investigators and coinvestigators were as follows. Indiana University, Indianapolis: John Nurnberger, Marvin Miller, and Elizabeth Bowman (supported by grant U01 H46282); Washington University, St. Louis: Theodore Reich, Allison Goate, and John Rice (supported by grant U01 MH46280); The Johns Hopkins University, Baltimore: J. Raymond DePaulo, Jr., Sylvia Simpson, and Colin Stine (supported by grant U01 H46274); NIMH Intramural Research Program, Clinical Neurogenetics Branch, Bethesda: Elliot Gershon, Diane Kazuba, and Elizabeth Maxwell. The work of P.P.Z. was supported in part by a grant from the National Alliance for Research on Schizophrenia and Depression.
Web Resources
The URLs for data presented herein are as follows:
- Center for Collaborative Genetic Studies on Mental Disorders, http://zork.wustl.edu/nimh/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for PFKL) [PubMed]
References
- Aita VM, Liu J, Knowles JA, Terwilliger JD, Baltazar R, Grunn A, Loth JE, Kanyas K, Lerer B, Endicott J, Wang Z, Penchaszadeh G, Gilliam TC, Baron M (1999) A comprehensive linkage analysis of chromosome 21q22 supports prior evidence for a putative bipolar affective disorder locus. Am J Hum Genet 64:210–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Psychiatric Association (1987) Diagnostic statistical manual III-R. American Psychiatric Association, Washington, DC [Google Scholar]
- Bellivier F, Golmard JL, Henry C, Leboyer M, Schurhoff F (2001) Admixture analysis of age at onset in bipolar I affective disorder. Arch Gen Psychiatry 58:510–512 10.1001/archpsyc.58.5.510 [DOI] [PubMed] [Google Scholar]
- Bellivier F, Golmard JL, Rietschel M, Schulze TG, Malafosse A, Preisig M, McKeon P, Mynett-Johnson L, Henry C, Leboyer M (2003) Age at onset in bipolar I affective disorder: further evidence for three subgroups. Am J Psychiatry 160:999–1001 10.1176/appi.ajp.160.5.999 [DOI] [PubMed] [Google Scholar]
- Bennett P, Segurado R, Jones I, Bort S, McCandless F, Lambert D, Heron J, Comerford C, Middle F, Corvin A, Pelios G, Kirov G, Larsen B, Mulcahy T, Williams N, O’Connell R, O’Mahony E, Payne A, Owen M, Holmans P, Craddock N, Gill M (2002) The Wellcome Trust UK-Irish bipolar affective disorder sibling-pair genome screen: first stage report. Mol Psychiatry 7:189–200 10.1038/sj.mp.4000957 [DOI] [PubMed] [Google Scholar]
- Berrettini WH (2000) Susceptibility loci for bipolar disorder: overlap with inherited vulnerability to schizophrenia. Biol Psychiatry 47:245–251 10.1016/S0006-3223(99)00226-7 [DOI] [PubMed] [Google Scholar]
- Berrettini WH, Ferraro TN, Goldin LR, Weeks DE, Detera-Wadleigh S, Nurnberger JI Jr, Gershon ES (1994) Chromosome 18 DNA markers and manic-depressive illness: evidence for a susceptibility gene. Proc Natl Acad Sci USA 91:5918–5921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson GA, Bromet EJ, Sievers S (2000) Phenomenology and outcome of subjects with early- and adult-onset psychotic mania. Am J Psychiatry 157:213–219 10.1176/appi.ajp.157.2.213 [DOI] [PubMed] [Google Scholar]
- Carlson GA, Davenport YB, Jamison K (1977) A comparison of outcome in adolescent- and later-onset bipolar manic-depressive illness. Am J Psychiatry 134:919–922 [DOI] [PubMed] [Google Scholar]
- Carter TD, Mundo E, Parikh SV, Kennedy JL (2003) Early age at onset as a risk factor for poor outcome of bipolar disorder. J Psychiatr Res 37:297–303 10.1016/S0022-3956(03)00052-9 [DOI] [PubMed] [Google Scholar]
- De Bruyn A, Souery D, Mendelbaum K, Mendlewicz J, Van Broeckhoven C (1996) Linkage analysis of families with bipolar illness and chromosome 18 markers. Biol Psychiatry 39:679–688 10.1016/0006-3223(95)00293-6 [DOI] [PubMed] [Google Scholar]
- Detera-Wadleigh SD, Badner JA, Goldin LR, Berrettini WH, Sanders AR, Rollins DY, Turner G, Moses T, Haerian H, Muniec D, Nurnberger JI Jr, Gershon ES (1996) Affected-sib-pair analyses reveal support of prior evidence for a susceptibility locus for bipolar disorder, on 21q. Am J Hum Genet 58:1279–1285 [PMC free article] [PubMed] [Google Scholar]
- Detera-Wadleigh SD, Badner JA, Yoshikawa T, Sanders AR, Goldin LR, Turner G, Rollins DY, et al (1997) Initial genome scan of the NIMH genetics initiative bipolar pedigrees: chromosomes 4, 7, 9, 18, 19, 20, and 21q. Am J Med Genet 74:254–262 [DOI] [PubMed] [Google Scholar]
- Devlin B, Jones BL, Bacanu SA, Roeder K (2002) Mixture models for linkage analysis of affected sibling pairs and covariates. Genet Epidemiol 22:52–65 10.1002/gepi.1043 [DOI] [PubMed] [Google Scholar]
- Dick DM, Foroud T, Edenberg HJ, Miller M, Bowman E, Rau NL, DePaulo JR, McInnis M, Gershon E, McMahon F, Rice JP, Bierut LJ, Reich T, Nurnberger J Jr (2002) Apparent replication of suggestive linkage on chromosome 16 in the NIMH genetics initiative bipolar pedigrees. Am J Med Genet 114:407–412 10.1002/ajmg.10380 [DOI] [PubMed] [Google Scholar]
- Edenberg HJ, Foroud T, Conneally PM, Sorbel JJ, Carr K, Crose C, Willig C, Zhao J, Miller M, Bowman E, Mayeda A, Rau NL, Smiley C, Rice JP, Goate A, Reich T, Stine OC, McMahon F, DePaulo JR, Meyers D, Detera-Wadleigh SD, Goldin LR, Gershon ES, Blehar MC, Nurnberger JI Jr (1997) Initial genomic scan of the NIMH genetics initiative bipolar pedigrees: chromosomes 3, 5, 15, 16, 17, and 22. Am J Med Genet 74:238–246 [DOI] [PubMed] [Google Scholar]
- Endicott J, Spitzer RL (1978) A diagnostic interview: the schedule for affective disorders and schizophrenia. Arch Gen Psychiatry 35:837–844 [DOI] [PubMed] [Google Scholar]
- ——— (1979) Use of the Research Diagnostic Criteria and the Schedule for Affective Disorders and Schizophrenia to study affective disorders. Am J Psychiatry 136:52–56 [DOI] [PubMed] [Google Scholar]
- Faraone SV, Glatt SJ, Su J, Tsuang MT (2004) Three potential susceptibility loci shown by a genome-wide scan for regions influencing the age at onset of mania. Am J Psychiatry 161:625–630 10.1176/appi.ajp.161.4.625 [DOI] [PubMed] [Google Scholar]
- Gershon ES, Goldin LR, Guroff JJ, Hamovit JR (1989) Description of the National Institute of Mental Health family study of affective disorders. Genet Epidemiol 6:183–185 10.1002/gepi.1370060132 [DOI] [PubMed] [Google Scholar]
- Glidden DV, Liang KY, Chiu YF, Pulver AE (2003) Multipoint affected sibpair linkage methods for localizing susceptibility genes of complex diseases. Genet Epidemiol 24:107–117 10.1002/gepi.10215 [DOI] [PubMed] [Google Scholar]
- Goddard KA, Witte JS, Suarez BK, Catalona WJ, Olson JM (2001) Model-free linkage analysis with covariates confirms linkage of prostate cancer to chromosomes 1 and 4. Am J Hum Genet 68:1197–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood CM, Bull SB (1997) Incorporation of covariates into genome scanning using sib-pair analysis in bipolar affective disorder. Genet Epidemiol 14:635–640 [DOI] [PubMed] [Google Scholar]
- Grigoroiu-Serbanescu M, Martinez M, Nothen MM, Grinberg M, Sima D, Propping P, Marinescu E, Hrestic M (2001) Different familial transmission patterns in bipolar I disorder with onset before and after age 25. Am J Med Genet 105:765–773 10.1002/ajmg.10047 [DOI] [PubMed] [Google Scholar]
- Hall JM, Lee MK, Newman B, Morrow JE, Anderson LA, Huey B, King MC (1990) Linkage of early-onset familial breast cancer to chromosome 17q21. Science 250:1684–1689 [DOI] [PubMed] [Google Scholar]
- Hauser ER, Watanabe RM, Duren WL, Bass MP, Langefeld CD, Boehnke M (2004) Ordered subset analysis in genetic linkage mapping of complex traits. Genet Epidemiol 27:53–63 10.1002/gepi.20000 [DOI] [PubMed] [Google Scholar]
- Kong A, Cox NJ (1997) Allele-sharing models: LOD scores and accurate linkage tests. Am J Hum Genet 61:1179–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newburg L (1987) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1:174–181 10.1016/0888-7543(87)90010-3 [DOI] [PubMed] [Google Scholar]
- Lathrop GM, Lalouel JM, Julier C, Ott J (1984) Strategies for multilocus linkage analysis in humans. Proc Natl Acad Sci USA 81:3443–3446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JP, Bale SJ (1997) Parental transmission and D18S37 allele sharing in bipolar affective disorder. Genet Epidemiol 14:665–668 [DOI] [PubMed] [Google Scholar]
- Lin PI, McInnis MG, Potash JB, MacKinnon DF, Willour VL, DePaulo JR, Zandi PP. Clinical correlates and familial aggregation of age at onset in bipolar disorder. Am J Psychiatry (in press) [DOI] [PubMed] [Google Scholar]
- Liu J, Juo SH, Terwilliger JD, Grunn A, Tong X, Brito M, Loth JE, Kanyas K, Lerer B, Endicott J, Penchaszadeh G, Gilliam TC, Baron M (2001) A follow-up linkage study supports evidence for a bipolar affective disorder locus on chromosome 21q22. Am J Med Genet 105:189–194 10.1002/ajmg.1195 [DOI] [PubMed] [Google Scholar]
- McElroy SL, Altshuler LL, Suppes T, Keck PE Jr, Frye MA, Denicoff KD, Nolen WA, Kupka RW, Leverich GS, Rochussen JR, Rush AJ, Post RM (2001) Axis I psychiatric comorbidity and its relationship to historical illness variables in 288 patients with bipolar disorder. Am J Psychiatry 158:420–426 10.1176/appi.ajp.158.3.420 [DOI] [PubMed] [Google Scholar]
- McInnis MG, Dick DM, Willour VL, Avramopoulos D, MacKinnon DF, Simpson SG, Potash JB, et al (2003a) Genome-wide scan and conditional analysis in bipolar disorder: evidence for genomic interaction in the National Institute of Mental Health Genetics Initiative bipolar pedigrees. Biol Psychiatry 54:1265–1273 10.1016/j.biopsych.2003.08.001 [DOI] [PubMed] [Google Scholar]
- McInnis MG, Lan TH, Willour VL, McMahon FJ, Simpson SG, Addington AM, MacKinnon DF, Potash JB, Mahoney AT, Chellis J, Huo Y, Swift-Scanlan T, Chen H, Koskela R, Stine OC, Jamison KR, Holmans P, Folstein SE, Ranade K, Friddle C, Botstein D, Marr T, Beaty TH, Zandi P, DePaulo JR (2003b) Genome-wide scan of bipolar disorder in 65 pedigrees: supportive evidence for linkage at 8q24, 18q22, 4q32, 2p12, and 13q12. Mol Psychiatry 8:288–298 10.1038/sj.mp.4001277 [DOI] [PubMed] [Google Scholar]
- Nurnberger JI Jr, Blehar MC, Kaufmann CA, York-Cooler C, Simpson SG, Harkavy-Friedman J, Severe JB, Malaspina D, Reich T (1994) Diagnostic interview for genetic studies: rationale, unique features, and training. NIMH Genetics Initiative. Arch Gen Psychiatry 51:849–859 [DOI] [PubMed] [Google Scholar]
- Olson JM (1999) A general conditional-logistic model for affected-relative-pair linkage studies. Am J Hum Genet 65:1760–1769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Mahony E, Corvin A, O’Connell R, Comerford C, Larsen B, Jones R, McCandless F, Kirov G, Cardno AG, Craddock N, Gill M (2002) Sibling pairs with affective disorders: resemblance of demographic and clinical features. Psychol Med 32:55–61 [DOI] [PubMed] [Google Scholar]
- Pericak-Vance MA, Bebout JL, Gaskell PC Jr, Yamaoka LH, Hung WY, Alberts MJ, Walker AP, Bartlett RJ, Haynes CA, Welsh KA (1991) Linkage studies in familial Alzheimer disease: evidence for chromosome 19 linkage. Am J Hum Genet 48:1034–1050 [PMC free article] [PubMed] [Google Scholar]
- Rice JP, Goate A, Williams JT, Bierut L, Dorr D, Wu W, Shears S, Gopalakrishnan G, Edenberg HJ, Foroud T, Nurnberger J Jr, Gershon ES, Detera-Wadleigh SD, Goldin LR, Guroff JJ, McMahon FJ, Simpson S, MacKinnon D, McInnis M, Stine OC, DePaulo JR, Blehar MC, Reich T (1997) Initial genome scan of the NIMH Genetics Initiative bipolar pedigrees: chromosomes 1, 6, 8, 10, and 12. Am J Med Genet 74:247–253 [DOI] [PubMed] [Google Scholar]
- Risch N (1990) Linkage strategies for genetically complex traits. II. The power of affected relative pairs. Am J Hum Genet 46:229–241 [PMC free article] [PubMed] [Google Scholar]
- Sjoholt G, Ebstein RP, Lie RT, Berle JO, Mallet J, Deleuze JF, Levinson DF, Laurent C, Mujahed M, Bannoura I, Murad I, Molven A, Steen VM (2004) Examination of IMPA1 and IMPA2 genes in manic-depressive patients: association between IMPA2 promoter polymorphisms and bipolar disorder. Mol Psychiatry 9:621–629 10.1038/sj.mp.4001460 [DOI] [PubMed] [Google Scholar]
- Sjoholt G, Gulbrandsen AK, Lovlie R, Berle JO, Molven A, Steen VM (2000) A human myo-inositol monophosphatase gene (IMPA2) localized in a putative susceptibility region for bipolar disorder on chromosome 18p11.2: genomic structure and polymorphism screening in manic-depressive patients. Mol Psychiatry 5:172–180 10.1038/sj.mp.4000681 [DOI] [PubMed] [Google Scholar]
- Stine OC, McMahon FJ, Chen L, Xu J, Meyers DA, MacKinnon DF, Simpson S, McInnis MG, Rice JP, Goate A, Reich T, Edenberg HJ, Foroud T, Nurnberger JI Jr, Detera-Wadleigh SD, Goldin LR, Guroff J, Gershon ES, Blehar MC, DePaulo JR (1997) Initial genome screen for bipolar disorder in the NIMH genetics initiative pedigrees: chromosomes 2, 11, 13, 14, and X. Am J Med Genet 74:263–269 [DOI] [PubMed] [Google Scholar]
- Stine OC, Xu J, Koskela R, McMahon FJ, Gschwend M, Friddle C, Clark CD, McInnis MG, Simpson SG, Breschel TS (1995) Evidence for linkage of bipolar disorder to chromosome 18 with a parent-of-origin effect. Am J Hum Genet 57:1384–1394 [PMC free article] [PubMed] [Google Scholar]
- Straub RE, Lehner T, Luo Y, Loth JE, Shao W, Sharpe L, Alexander JR, Das K, Simon R, Fieve RR, Lerer B, Endicott J, Ott J, Gilliam TC, Baron M (1994) A possible vulnerability locus for bipolar affective disorder on chromosome 21q22.3. Nat Genet 8:291–296 10.1038/ng1194-291 [DOI] [PubMed] [Google Scholar]
- Sun L, Wilder K, McPeek MS (2002) Enhanced pedigree error detection. Hum Hered 54:99–110 10.1159/000067666 [DOI] [PubMed] [Google Scholar]
- Willour VL, Zandi PP, Huo Y, Diggs TL, Chellis JL, MacKinnon DF, Simpson SG, McMahon FJ, Potash JB, Gershon ES, Reich T, Foroud T, Nurnberger JI Jr, DePaulo JR Jr, McInnis MG (2003) Genome scan of the fifty-six bipolar pedigrees from the NIMH Genetics Initiative replication sample: chromosomes 4, 7, 9, 18, 19, 20, and 21. Am J Med Genet B Neuropsychiatr Genet 121:21–27 10.1002/ajmg.b.20051 [DOI] [PubMed] [Google Scholar]
- Young A (1995) Genetic Analysis System, version 2.0 (http://users.ox.ac.uk/~ayoung/gas.html; accessed August 9, 2005)
- Zandi PP, Willour VL, Huo Y, Chellis J, Potash JB, MacKinnon DF, Simpson SG, McMahon FJ, Gershon E, Reich T, Foroud T, Nurnberger J Jr, DePaulo JR Jr, McInnis MG (2003) Genome scan of a second wave of NIMH genetics initiative bipolar pedigrees: chromosomes 2, 11, 13, 14, and X. Am J Med Genet B Neuropsychiatr Genet 119:69–76 10.1002/ajmg.b.10063 [DOI] [PubMed] [Google Scholar]