Abstract
The minor allele of the R620W missense single-nucleotide polymorphism (SNP) (rs2476601) in the hematopoietic-specific protein tyrosine phosphatase gene, PTPN22, has been associated with multiple autoimmune diseases, including rheumatoid arthritis (RA). These genetic data, combined with biochemical evidence that this SNP affects PTPN22 function, suggest that this phosphatase is a key regulator of autoimmunity. To determine whether other genetic variants in PTPN22 contribute to the development of RA, we sequenced the coding regions of this gene in 48 white North American patients with RA and identified 15 previously unreported SNPs, including 2 coding SNPs in the catalytic domain. We then genotyped 37 SNPs in or near PTPN22 in 475 patients with RA and 475 individually matched controls (sample set 1) and selected a subset of markers for replication in an additional 661 patients with RA and 1,322 individually matched controls (sample set 2). Analyses of these results predict 10 common (frequency >1%) PTPN22 haplotypes in white North Americans. The sole haplotype found to carry the previously identified W620 risk allele was strongly associated with disease in both sample sets, whereas another haplotype, identical at all other SNPs but carrying the R620 allele, showed no association. R620W, however, does not fully explain the association between PTPN22 and RA, since significant differences between cases and controls persisted in both sample sets after the haplotype data were stratified by R620W. Additional analyses identified two SNPs on a single common haplotype that are associated with RA independent of R620W, suggesting that R620W and at least one additional variant in the PTPN22 gene region influence RA susceptibility.
Introduction
Autoimmune diseases (MIM 109100) afflict up to 5% of the population and are characterized by an aberrant immune response to self-antigens (Marrack et al. 2001; Wandstrat and Wakeland 2001). The mechanisms of disease initiation and persistence are poorly understood, but genetic factors appear to play a major role. In humans, disease-specific familial clustering has been demonstrated for most common autoimmune diseases, and concordance is generally higher for MZ twins than for DZ twins. Furthermore, it appears that different autoimmune diseases share susceptibility loci. These diseases can overlap in single individuals and in families (Lin et al. 1998; Prahalad et al. 2002; Alkhateeb et al. 2003). For example, parents of children with type 1 diabetes (T1D [MIM 222100]) have increased rates of T1D and other autoimmune diseases (Tait et al. 2004), and a recent study of relatives of 1,214 patients with systemic lupus erythematosus (SLE [MIM 152700]) showed familial aggregation of not only SLE but also RA and other autoimmune diseases (Alarcon-Segovia et al. 2005). Additionally, loci identified by linkage studies in different human autoimmune diseases and in mouse models of autoimmunity cluster together (Becker et al. 1998; Wandstrat and Wakeland 2001).
With the exclusion of the human leukocyte antigen (HLA) region, attempts to identify genetic variants that confer risk of multiple autoimmune diseases have proven difficult, and, although some genes in the HLA region affect risk of multiple autoimmune diseases, different alleles are typically risk factors for different diseases. Ueda et al. (2003) have reported that a common variant of CTLA4 is associated with both T1D and Graves disease (MIM 275000), and several other reports suggest that a variant of another member of the CTLA4 family of T-cell regulatory receptors, PDCD1, may also predispose individuals to several autoimmune diseases (Prokunina et al. 2002, 2004; Nielsen et al. 2003; Lin et al. 2004). In addition, variants in the CARD15 and SLC22A4/SLC22A5 genes appear to play a role in both Crohn disease (MIM 266600) and psoriatic arthritis (Hugot et al. 2001; Rahman et al. 2003; Ho et al. 2004; Peltekova et al. 2004), and a recent study suggests a functional variant of FCRL3 may be associated with RA, SLE, Graves disease, and Hashimoto thyroiditis in Japanese (Kochi et al. 2005).
Perhaps one of the best examples of a non–major histocompatibility complex (MHC) common susceptibility allele for autoimmunity is the R620W SNP (rs2476601) in the cytoplasmic protein tyrosine phosphatase gene, PTPN22 (reviewed by Siminovitch [2004] and Gregersen [2005]). The W620 allele has been consistently associated with increased risk for T1D (Bottini et al. 2004; Onengut-Gumuscu et al. 2004; Smyth et al. 2004; Criswell et al. 2005; Ladner et al. 2005; Qu et al. 2005; Zheng and She 2005; Zhernakova et al. 2005), rheumatoid arthritis (RA [MIM 180300]) (Begovich et al. 2004; Criswell et al. 2005; Hinks et al. 2005; Lee et al. 2005; Orozco et al. 2005; Simkins et al. 2005; Steer et al. 2005; Van Oene et al. 2005; Viken et al. 2005; Zhernakova et al. 2005), SLE (Kyogoku et al. 2004; Criswell et al. 2005; Orozco et al. 2005), Graves disease (Smyth et al. 2004; Velaga et al. 2004; Skorka et al. 2005), and juvenile idiopathic arthritis (Hinks et al. 2005; Viken et al. 2005), and there are single reports of association with autoimmune Addison disease (Velaga et al. 2004) and Hashimoto thyroiditis (Criswell et al. 2005). However, no association has been seen with multiple sclerosis (MS [MIM 126200]) (Begovich et al. 2005; Criswell et al. 2005; Hinks et al. 2005; Matesanz et al. 2005), primary sclerosing cholangitis (Viken et al. 2005), Crohn disease (Van Oene et al. 2005), or psoriasis vulgaris (MIM 177900) (Hinks et al. 2005; M. Cargill, unpublished data), which indicates that this SNP is a risk allele for some but not all autoimmune diseases.
These genetic data suggest that autoimmune diseases associated with the W620 risk allele of PTPN22 may share a common etiology. Although phosphatases are known to be crucial for maintaining immune-cell homeostasis, the specific function of PTPN22, otherwise known as “Lyp” (lymphocyte phosphatase) (Cohen et al. 1999), is poorly understood. More information is available for the mouse ortholog, PEP, which serves as a negative regulator of T-cell activation via interaction with the c-Src tyrosine kinase, Csk (Cloutier and Veillette 1996). PEP also appears to modify the phosphorylation state of regulatory tyrosines on other Src family kinases, such as Lck, Fyn, and ZAP-70 (Cloutier and Veillette 1999; Gjorloff-Wingren et al. 1999). Correspondingly, knockout mice deficient in PEP show selective disregulation of the effector/memory T-cell compartment with enhanced activation of Lck, hyperproliferation, and exaggerated early-signaling responses in restimulated T cells. These mice also spontaneously develop germinal centers and increased serum levels of certain immunoglobulin isotypes; however, they do not display overt signs of autoimmunity (Hasegawa et al. 2004).
The R620W SNP lies in the protein’s N-terminal SH3-binding domain, which is necessary for interaction with Csk (Gregorieff et al. 1998). In vitro experiments show that the W620 variant of PTPN22 binds less efficiently to Csk than the R620 variant does (Begovich et al. 2004; Bottini et al. 2004), suggesting that T cells expressing the W620 allele may be hyperresponsive; consequently, individuals carrying this allele may be more prone to autoimmunity.
Given the fundamental role of PTPN22 in autoimmunity, we wished to characterize the extent of linkage disequilibrium (LD) across this gene, define the common PTPN22 haplotypes, and determine whether variants other than W620 predispose individuals to the development of RA. Accordingly, we sequenced the coding regions of this gene in 48 white North Americans with RA to identify novel SNPs, developed assays for a subset of these SNPs as well as others from public databases, and genotyped two large RA case-control sample sets. The results not only support the notion that R620W is a major RA risk site in this region but also suggest that at least one other genetic variant in this region, independent of R620W, predisposes individuals to RA.
Material and Methods
Samples
A detailed description of the case and control samples is provided elsewhere (Begovich et al. 2004). In brief, sample set 1, which consists of 475 individuals with RA and 475 individually matched controls, was obtained by Genomics Collaborative. All case samples were from white North Americans who were rheumatoid factor (RF) positive and whose condition met the 1987 American College of Rheumatology diagnostic criteria for RA. Control samples were taken from a pool of healthy white individuals with no medical history of RA. A single control was matched to each case on the basis of sex, age (±5 years), and ethnicity (grandparental country/region of origin). All protocols and recruitment sites were approved by national and/or local institutional review boards, and informed written consent was obtained from all subjects.
Cases in sample set 2 were obtained by the North American Rheumatoid Arthritis Consortium (NARAC) (see NARAC Web site) and consisted of members from 661 white North American multiplex families (Jawaheer et al. 2001, 2004). Both RF-positive and RF-negative patients are included in this sample set. Controls were selected from 20,000 healthy individuals who are part of the New York Cancer Project, a population-based prospective study of the genetic and environmental factors that cause disease (see AMDeC Web site). Two control individuals were matched to a single, randomly chosen, affected sib from each NARAC family on the basis of sex, age (decade of birth), and ethnicity (grandparental country/region of origin). Informed written consent was obtained from every subject.
PTPN22 Sequencing
To identify novel variants in PTPN22, 48 patients from sample set 1 who represented all three R620W genotypes (CC, CT, and TT) were selected for resequencing. Sequence data from all 21 annotated exons of PTPN22, which spans close to 58 kb, were extracted from the R27 draft of the Celera human genome sequence (see Celera Web site). Primers were designed using the Primer3 program and included the M13 forward (5′ primer) or reverse (3′ primer) universal sequencing primer-binding sequence at their 5′ ends. (Primer sequences are available on request.) Standard PCR reactions were performed in a 96-well format by use of the Applied Biosystems 9700 thermocycler (denaturation at 96° for 5 min, followed by 40 amplification cycles at 94° for 30 s, 60° for 45 s, and 70° for 45 s, and then an extension at 72° for 10 min). Unincorporated dNTPs and primers were removed with shrimp alkaline phosphatase/exonuclease, and the product was used as the template in a 7-μl Big Dye Terminator (Applied Biosystems) sequencing reaction on the AB9700 (denaturation at 96° for 2 min, followed by 35 amplification cycles at 96° for 10 s, 50° for 30 s, and 60° for 1 min). Samples were precipitated using the EtOH/NaOAc protocol recommended by Applied Biosystems, were resuspended in deionized distilled water, and were loaded on the Applied Biosystems Prism 3730 DNA Sequencer. Sequence traces were assembled and analyzed using a modified version of the PolyPhred software package. Skilled sequence annotators reviewed all predicted variants.
SNP Selection
In addition to R620W, assays for 36 SNPs in and around PTPN22 were successfully built and genotyped in sample set 1. To minimize the amount of genotyping yet retain as much statistical power as possible to detect disease-associated SNPs, we applied the program Redigo (Hu et al. 2004), which identified seven tagging SNPs for replication (SNPs 1, 18, 20, 22, 27, 35, and 36 in table 1). We included three additional SNPs (SNPs 2, 32, and 37) on the basis of functional categorization (putative transcription factor binding sites [TFBSs] and UTR) and one SNP (SNP 21) with the most significant allelic disease-association P value in sample set 1. Three other SNPs with a minor-allele frequency <5% (SNPs 23, 28, and 34), which were not in the Redigo selected set, were included to increase the power of detecting association for rare SNPs. In all, 14 SNPs, including the R620W SNP, were genotyped in sample set 2 (see table 1 for a complete list of SNPs). Elsewhere, we have reported the results for R620W in sample set 1 and the first 463 families and 926 controls of sample set 2 (Begovich et al. 2004).
Table 1.
PTPN22 SNP Allele Frequencies and Allele-Based Association with RA
|
Sample Set 1d |
Sample Set 2d |
||||||||||||
| Frequency in |
Frequency in |
||||||||||||
| SNPa | dbSNP ID | Typeb | Positionc | Cases(n=475) | Controls(n=475) | OR | 95% CI | P | Cases (n=661) | Controls(n=1,322) | OR | 95% CI | P |
| 1 | rs1217414 | Intron | T10498782C | .261 | .269 | .96 | .78–1.18 | .71 | .241 | .275 | .84 | .72–.98 | .02 |
| 2 | rs2488458 | TFBS | A10492566G | .270 | .224 | 1.28 | 1.04–1.58 | .024 | .292 | .245 | 1.27 | 1.10–1.48 | .0016 |
| 3 | rs2476604 | Intron | A10492559T | .485 | .388 | 1.48 | 1.24–1.78 | 2.7E−05 | … | … | … | … | … |
| 4 | rs1775754 | Intron | G10491016A | .273 | .224 | 1.30 | 1.06–1.61 | .014 | … | … | … | … | … |
| 5 | rs1217421 | Intron | C10488960G | .274 | .225 | 1.30 | 1.05–1.60 | .016 | … | … | … | … | … |
| 6 | rs1217420 | Intron | T10488865C | .484 | .387 | 1.48 | 1.24–1.78 | 2.7E−05 | … | … | … | … | … |
| 7 | rs1217417 | Intron | C10487019G | .245 | .248 | .99 | .80–1.21 | .91 | … | … | … | … | … |
| 8 | rs3789609 | Intron | A10483914G | .260 | .351 | .65 | .53–.79 | 2.4E−05 | … | … | … | … | … |
| 9 | ss38346947 | R183X | T10483780C | .001 | .000 | NC | … | 1.00 | … | … | … | … | … |
| 10 | ss38346946 | Intron | A10483096T | .006 | .005 | 1.21 | .37–3.94 | .77 | … | … | … | … | … |
| 11 | rs2476602 | Intron | T10483070C | .251 | .258 | .96 | .78–1.18 | .75 | … | … | … | … | … |
| 12 | rs1217410 | Intron | G10482931A | .274 | .223 | 1.31 | 1.06–1.62 | .014 | … | … | … | … | … |
| 13 | rs1217408 | Intron | G10482395A | .256 | .261 | .97 | .79–1.19 | .83 | … | … | … | … | … |
| 14 | ss38346945 | R263Q | A10480804G | .024 | .017 | 1.39 | .73–2.66 | .33 | … | … | … | … | … |
| 15 | rs3765598 | Intron | A10480578G | .205 | .156 | 1.40 | 1.11–1.77 | .0056 | … | … | … | … | … |
| 16 | rs1217407 | Intron | T10479863C | .273 | .225 | 1.29 | 1.05–1.59 | .019 | … | … | … | … | … |
| 17 | rs1217406 | Intron | T10479268G | .483 | .388 | 1.47 | 1.22–1.76 | 4.6E−05 | … | … | … | … | … |
| 18 | rs12760457 | Intron | A10475862G | .262 | .354 | .65 | .53–.79 | 2.0E−05 | .258 | .297 | .82 | .71–.96 | .01 |
| 19 | rs974404 | Intron | C10468140A | .488 | .390 | 1.49 | 1.24–1.79 | 2.4E−05 | … | … | … | … | … |
| 20 | rs11102685 | Intron | G10467748A | .084 | .085 | .98 | .71–1.35 | .93 | .099 | .079 | 1.27 | 1.01–1.60 | .04 |
| 21 | rs12730735 | Intron | G10467572A | .261 | .353 | .65 | .53–.79 | 2.0E−05 | .258 | .300 | .81 | .70–.94 | .0064 |
| 22 | rs2476601 | R620W | T10463683C | .138 | .089 | 1.65 | 1.23–2.20 | 6.6E−04 | .153 | .085 | 1.93 | 1.58–2.37 | 3.2E−10 |
| 23 | ss38346944 | Intron | T10463342G | .041 | .032 | 1.29 | .79–2.09 | .33 | .031 | .024 | 1.29 | .87–1.93 | .21 |
| 24 | rs1970559 | Intron | G10463263A | .253 | .260 | .97 | .79–1.19 | .79 | … | … | … | … | … |
| 25 | rs2797415 | Intron | T10463208C | .273 | .224 | 1.30 | 1.06–1.61 | .014 | … | … | … | … | … |
| 26 | rs1217395 | Intron | G10460550A | .272 | .224 | 1.29 | 1.05–1.59 | .018 | … | … | … | … | … |
| 27 | rs1310182 | TFBS | T10459618C | .482 | .386 | 1.48 | 1.23–1.77 | 3.2E−05 | .502 | .428 | 1.35 | 1.18–1.54 | 1.0E−05 |
| 28 | ss38346943 | Intron | C10458639T | .026 | .017 | 1.52 | .81–2.87 | .21 | .018 | .028 | .63 | .40–1.01 | .05 |
| 29 | rs2476600 | Intron | T10455849C | .483 | .385 | 1.49 | 1.24–1.79 | 2.5E−05 | … | … | … | … | … |
| 30 | rs2797416 | Intron | A10455374G | .484 | .387 | 1.49 | 1.24–1.79 | 2.2E−05 | … | … | … | … | … |
| 31 | rs1217389 | Intron | G10451867A | .274 | .225 | 1.30 | 1.05–1.60 | .016 | … | … | … | … | … |
| 32 | rs1217388 | TFBS | C10450591T | .274 | .222 | 1.32 | 1.07–1.63 | .010 | .294 | .246 | 1.28 | 1.11–1.49 | .0011 |
| 33 | rs2476599 | Intron | T10449574C | .255 | .262 | .96 | .78–1.18 | .75 | … | … | … | … | … |
| 34 | ss38346942 | Intron | A10448233T | .010 | .013 | .75 | .32–1.78 | .66 | .014 | .016 | 1.03 | .59–1.79 | 1 |
| 35 | rs1217413 | Intron | C10443865T | .245 | .189 | 1.39 | 1.12–1.73 | .0035 | .269 | .212 | 1.37 | 1.18–1.60 | 6.2E−05 |
| 36 | rs3811021 | UTR3 | C10442778T | .207 | .160 | 1.37 | 1.08–1.73 | .010 | .205 | .183 | 1.15 | .97–1.36 | .10 |
| 37 | rs3789604 | TFBS | C10441057A | .210 | .160 | 1.39 | 1.10–1.76 | .0071 | .206 | .185 | 1.15 | .97–1.35 | .11 |
Alleles are listed in order from the 5′ end of the gene to the 3′ end.
All TFBSs are putative and have not been experimentally confirmed.
Positions are according to genomic contig NT_019273. The minor allele is listed first, followed by the position and then the major allele. The alleles are oriented according to transcript NM_012411, which is the reverse complement of the genomic contig sequence.
ORs and 95% CIs are estimated for the minor allele of each SNP. P values for allelic association with disease were calculated using Fisher's exact test. NC = not calculated.
Genotyping Methods
Genotypes were generated by kinetic, allele-specific PCR (Germer et al. 2000). In brief, 0.3 ng of DNA was amplified in a 15-μl reaction containing allele-specific primers. (Primer sequences are available on request.) Genotyping calls were made automatically using custom software and were subsequently hand-curated before statistical analysis. Genotyping accuracy has been estimated to be >99.8% by comparison with an independent method.
LD Analysis
To better understand the genetic structure of this region, we calculated the LD measures D′ and r2 between every marker pair in each sample set, using the LDMAX program in the GOLD package (Abecasis and Cookson 2000) (the nonsense SNP, SNP 9, which was found on only one chromosome, was excluded). We then employed Spotfire software to generate a graphical representation of the r2 LD matrix for the sample set 1 control data.
Haplotype Estimation
The Haplo.stats package was used to predict haplotypes and to directly estimate their frequencies for the 14-marker data in both sample sets as well as to test for haplotype association with RA via a permutation procedure (Schaid et al. 2002). Because of the computational limitations of the haplotype-estimation algorithm within Haplo.stats when looking at large numbers of markers, SNPHAP was used to predict haplotypes for the 36-marker data (SNP 9, the rare nonsense SNP, was excluded). To ensure that these two algorithms yielded similar results, we also used SNPHAP to analyze the 14-marker data from both sample sets and compared the results with those generated by Haplo.stats; the results from the two algorithms were similar.
Haplotype-Method Analysis
To determine whether other PTPN22 variants predispose individuals to RA independent of R620W, the haplotype method was applied to the 14-marker data in both sample sets (Thomson et al. 1988, Valdes and Thomson 1997). Haplotype frequencies were directly estimated using an expectation-maximization algorithm implemented in SNPHAP. To determine whether R620W by itself could explain all the association with RA in this region, we removed all chromosomes carrying the risk T allele (W620), which uniquely defines haplotype 2, from the data and then examined the remaining 13 SNPs on haplotypes carrying the C allele (620R) for disease association, using a permutation procedure to assess significance (Li 2001). We also conducted haplotype-method pairwise analyses between R620W and every other SNP to identify those SNPs that showed evidence of 620W-independent association with disease. To address whether SNP 27 is a risk factor independent of R620W and SNP 37, we removed all chromosomes carrying either W620 or the SNP 37 risk allele (C) and examined the remaining chromosomes for association of SNP 27 with RA.
CLR Analysis
To independently confirm the haplotype-method analysis results, and at the same time to address possible inaccuracies or bias that could have arisen from the estimation of haplotype frequencies in the haplotype method, we used conditional logistic regression (CLR) on the unphased 14-marker genotype data, to examine the genotypic associations of the individual PTPN22 markers in each of the study sample sets after adjusting for the previously identified R620W risk marker (with or without the other known genetic risk factor, HLA-DRB1) and accounting for the matching between cases and controls (Breslow and Day 1980). HLA genotypes were binned as described elsewhere (Begovich et al. 2004). Tests for trend in increasing odds ratios (ORs) were performed under the assumption of an additive model by entering the number of risk alleles into the regression model.
RPE and Haplotype Regression Analyses
To assess whether haplotypes 2 and 4 were sufficient to explain the association with PTPN22, we first employed a test similar to the relative predispositional effects (RPE) method (Payami et al. 1989) in which haplotypes 2 and 4 were removed from the 14-marker haplotype data and a χ2 test of heterogeneity was conducted for the remaining haplotypes. To assess the relative effects of the individual haplotypes on RA status, we used the haplotype regression method (Lake et al. 2003) from the Haplo.stats package to model the disease status as a function of the common haplotypes with or without the other known genetic risk factor, HLA-DRB1. We modeled the haplotype effects under an additive model and only included haplotypes with frequencies >1% in the regression analysis.
Diplotype Analysis
To better understand the disease-predisposing effects at selected SNPs, diplotypes were estimated from the unphased genotype data by use of a pseudo–Gibbs Sampler algorithm (Stephens et al. 2001) implemented by SNPAnalyzer (Yoo et al. 2005). The diplotype estimation procedure was performed separately on cases and controls for individuals with complete genotype data at both sites, and the most likely pair of haplotypes for each individual was selected by the program. CLR was used to assess the association of the inferred diplotypes with RA risk, relative to the most common diplotype.
Results
PTPN22 Sequencing in Patients with RA
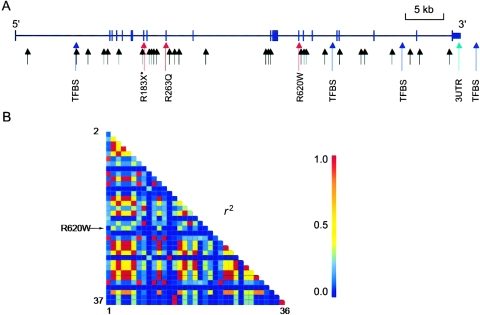
We sequenced 960 bases of 5′ sequence, all exons and intron/exon boundaries, and 1,460 bases of 3′ sequence of PTPN22 in 48 RF-positive individuals from sample set 1. On average, we successfully sequenced 37 individuals for each amplicon (coverage ranged from 21 to 48 individuals) and identified 32 SNPs, 15 of which were not found in public databases (table 2). Of these 15, 2 were coding region SNPs (cSNPs): one cSNP was a missense SNP in a nonconserved residue of the catalytic domain (R263Q), and the other was a nonsense SNP (R183X) also in the catalytic domain (fig. 1A). Minor alleles of both cSNPs were observed on only one chromosome in these 48 individuals.
Table 2.
SNPs Identified through Resequencing in 48 Patients with RA
| dbSNP ID | SNPa | Positionb | Type |
| rs1217419 | C10488019A | Intron | |
| ss38346956 | A10487866T | Intron | |
| ss38346953 | A10487850T | Intron | |
| ss38346954 | A10487465G | Intron | |
| rs1217418 | T10487346C | Intron | |
| rs1217417 | 7 | C10487019G | Intron |
| rs1217416 | G10486919A | TFBS | |
| rs11582409 | A10485548G | TFBS | |
| ss38346952 | G10484936C | Intron | |
| ss38346955 | A10484227G | Intron | |
| rs3789609 | 8 | A10483914G | Intron |
| rs3789608 | A10483903G | Intron | |
| ss38346947 | 9 | T10483780C | Nonsense |
| ss38346946 | 10 | A10483096T | Intron |
| rs2476602 | 11 | T10483070C | Intron |
| rs1217410 | 12 | G10482931A | Intron |
| ss38346945 | 14 | A10480804G | Missense |
| rs3765598 | 15 | A10480578G | Intron |
| ss38346951 | T10477441A | Intron | |
| ss38346957 | C10477397G | Intron | |
| ss38346950 | A10466290T | Intron | |
| rs17509844 | T10463884C | Intron | |
| rs2476601 | 22 | T10463683C | Missense |
| ss38346944 | 23 | T10463342G | Intron |
| rs1970559 | 24 | G10463263A | Intron |
| rs2797415 | 25 | T10463208C | Intron |
| rs3761935 | C10458643A | Intron | |
| ss38346943 | 28 | C10458639T | Intron |
| ss38346949 | A10448217G | Intron | |
| rs1217413 | 35 | C10443865T | Intron |
| ss38346948 | A10443204T | UTR 3 | |
| rs1217411 | C10442240T | TFBS |
Figure 1.
Gene structure and LD of PTPN22. A, Location and functional domain types of the 37 SNPs genotyped in sample set 1. Missense variants are colored in red; the nonsense variant is colored in red and marked by an asterisk (*); putative TFBSs are colored in blue; and sites within the 5′ and 3′ UTRs are colored in green. B, Pairwise LD between 36 SNPs (excluding the rare nonsense SNP 9), as measured by r2 in 475 controls from sample set 1. The indexes of the SNPs (table 1) were arranged vertically from SNP 2 to SNP 37 and horizontally from SNP 1 to SNP 36.
PTPN22 SNP Genotyping and Single-Marker Association
We genotyped 37 SNPs in the region of the PTPN22 gene (fig. 1A) in sample set 1 (475 independent cases and 475 individually matched controls). As we reported elsewhere for this sample set (Begovich et al. 2004), the W620 allele (SNP 22 [rs2476601]) was significantly enriched in cases compared with controls (allele frequency was 13.8% in cases and 8.9% in controls) (P=6.6×10-4; allelic OR=1.65); however, alleles at many of the other 36 markers also showed significant association with RA (table 1). The minor allele of the novel missense SNP (R263Q) was present at a frequency of 2.4% in cases compared with 1.7% in controls and was not significantly associated with disease (P=.33). The minor allele of the nonsense SNP (R183X) appeared on only one chromosome in this sample (in the case patient in whom it was originally identified) and was excluded from all subsequent analyses. Interestingly, this sample was from a woman with RA onset at age 64 who was negative for the HLA-DRB1 shared epitope and who carried two copies of the R620 PTPN22 allele.
Given that the genotypes at many of these 37 SNPs are correlated due to strong LD across this gene (see below), we sought to minimize the amount of genotyping in sample set 2 (661 independent cases and 1,322 individually matched controls) by selecting 14 markers for replication (see the “Material and Methods” section for the selection procedure). Of the 14 markers tested, 7 showed significant association with RA in both sample sets (SNPs 2, 18, 21, 22 [R620W], 27, 32, and 35), but R620W was the most significant SNP in sample set 2 (P=3.2×10-10; OR=1.93) (table 1). For five SNPs (SNPs 2, 22 [R620W], 27, 32, and 35), the minor-allele frequency was increased in the cases, whereas, for SNPs 18 and 21, the major-allele frequency was increased in the cases.
LD and Haplotype Association
To assess the extent of LD across PTPN22, we calculated D′ and r2 values for all SNP pairs, using the 36-marker data from the sample set 1 controls (SNP 9, the rare nonsense SNP, was excluded). The pairwise D′ values in the PTPN22 gene were near 1 among almost all SNP pairs in our white North American samples (data not shown), and a number of these 36 markers were highly correlated with each other (r2 values >0.90) (see fig. 1B). It is interesting to note that the R620W SNP was not completely correlated with any other SNP genotyped in this study (all r2 values were <0.5). Similar results were seen for the cases from sample set 1 as well as both the cases and controls from sample set 2 (14-marker data) (data not shown).
Ten common (frequency ⩾1%) haplotypes were predicted for sample set 1 by use of data from both the 14- and 36-marker sets; the same 10 haplotypes were observed in sample set 2 (table 3). Of the 10 haplotypes, 6 were at a frequency of 5% or more in the controls. The most common haplotype, haplotype 5, was present at a frequency of ∼35% in sample set 1 controls and ∼30% in sample set 2 controls. The W620 risk allele was found on a single haplotype, haplotype 2, which differs from haplotype 1 at only this SNP.
Table 3.
PTPN22 Predicted Haplotypes and Association with RA[Note]
| SNP |
Sample Set 1 |
Sample Set 2 |
||||||||||||||||||||||||||||||||||||||||
| Frequency in |
Frequency in |
|||||||||||||||||||||||||||||||||||||||||
| Haplotype | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | 29 | 30 | 31 | 32 | 33 | 34 | 35 | 36 | 37 | Cases | Controls | P | Cases | Controls | P |
| 1 |
C |
A |
A | G | C | T | G | G | T | C | G | A | G | G | T | T |
G |
C |
A |
A |
C |
G |
A | T | G |
T |
T |
T | A | G |
C |
C |
T |
C |
T |
A |
.105 | .098 | .590 | .116 | .127 | .32 |
| 2 |
C |
A |
A | G | C | T | G | G | T | C | G | A | G | G | T | T |
G |
C |
A |
A |
T |
G |
A | T | G |
T |
T |
T | A | G |
C |
C |
T |
C |
T |
A |
.136 | .089 | .001 | .151 | .081 | <5E−05 |
| 3 |
C |
A |
A | G | C | T | G | G | T | C | G | A | G | G | T | T |
G |
C |
A |
A |
C |
G |
A | T | G |
T |
T |
T | A | G |
C |
C |
T |
T |
T |
A |
.027 | .034 | .416 | .026 | .032 | .26 |
| 4 |
C |
G |
A | A | G | T | G | G | T | C | A | A | G | A | C | T |
G |
C |
A |
A |
C |
G |
A | C | A |
T |
T |
T | A | A |
T |
C |
T |
T |
C |
C |
.203 | .152 | .003 | .201 | .177 | .07 |
| 5 |
C |
G |
T | A | G | C | G | A | T | C | A | A | G | G | C | G |
A |
A |
A |
G |
C |
G |
A | C | A |
C |
T |
C | G | A |
T |
C |
T |
T |
T |
A |
.260 | .350 | 5E−05 | .259 | .298 | .01 |
| 6 |
T |
G |
T | A | G | C | C | G | T | T | A | G | G | G | C | G |
G |
A |
A |
A |
C |
G |
G | C | A |
C |
T |
C | G | A |
T |
T |
T |
T |
T |
A |
.094 | .116 | .15 | .076 | .126 | <5E−05 |
| 7 |
T |
G |
T | A | G | C | C | G | T | T | A | G | G | G | C | G |
G |
A |
A |
A |
C |
T |
G | C | A |
C |
T |
C | G | A |
T |
T |
T |
T |
T |
A |
.041 | .031 | .23 | .031 | .025 | .30 |
| 8 |
T |
G |
T | A | G | C | C | G | T | T | A | G | G | G | C | G |
G |
A |
G |
A |
C |
G |
G | C | A |
C |
T |
C | G | A |
T |
T |
T |
T |
T |
A |
.082 | .086 | .77 | .095 | .077 | .07 |
| 9 |
T |
G |
T | A | G | C | C | G | T | T | A | G | A | G | C | G |
G |
A |
A |
A |
C |
G |
G | C | A |
C |
C |
C | G | A |
T |
T |
T |
T |
T |
A |
.026 | .016 | .18 | .018 | .028 | .06 |
| 10 |
T |
G |
T | A | G | C | G | G | T | T | A | G | G | G | C | G |
G |
A |
A |
A |
C |
G |
G | C | A |
C |
T |
C | G | A |
T |
T |
A |
T |
T |
A |
.010 | .013 | .52 | .013 | .014 | .70 |
Note.— The program SNPHAP was used to estimate common (frequency > 0.01) haplotypes for 36 of the 37 SNPs genotyped in sample set 1. SNP 9, the nonsense SNP found in one individual, was excluded from this analysis. The global P value was .0001 for sample set 1 and <5E−05 for sample set 2. Global P values were estimated from 20,000 simulations following a permutation procedure. Haplotype frequencies in cases and controls and P values were estimated from 14 tagging SNPs (underlined and in bold italics) by use of Haplo.stats. The haplotype frequencies do not sum to 1 because of rare, unlisted haplotypes.
Next, we assessed the association of these predicted haplotypes with RA by using the frequency data generated for the 14 SNPs (table 3). The overall association between the haplotypes and disease status is highly significant in both sample sets (sample set 1, global P=.0001; sample set 2, global P<5×10-5). As expected from the single-marker data, haplotype 2 (which carries the W620 allele) was significantly increased among cases compared with controls, in both samples (sample set 1, P=.001; sample set 2, P<5×10-5). Haplotype 1, which differs from haplotype 2 only at R620W, showed no association with disease in either sample set (sample set 1, P=.59; sample set 2, P=.32). Haplotype 4, which is uniquely marked by the minor alleles of SNPs 15, 36, and 37, appeared to be increased among cases in both sample sets, although the statistical significance in sample set 2 was marginal (P=.07). On the other hand, haplotype 5, uniquely marked by the minor alleles of SNPs 8, 18, and 21, was significantly decreased among cases in both sample sets (sample set 1, P=5×10-5; sample set 2, P=.01). Haplotype 6 was also decreased among cases, compared with controls, in both sample sets, but this association was only significant in sample set 2 (P<5×10-5).
The differences in haplotype frequencies between the two sample sets, combined with the lack of replication of some of the haplotype associations, led us to examine first how consistent case haplotype frequencies were between the two studies and then how consistent control haplotype frequencies were. A χ2 test showed no significant differences between cases (P=.51) but did reveal a significant difference between controls in the two studies (P=.015), which appears to be largely driven by differences in the frequencies of haplotypes 1, 4, and 5.
Does R620W Explain All the Association Seen with PTPN22?
The known association of the W620 allele with RA, in addition to the strong LD across PTPN22 (fig. 1B), may confound our ability to determine which, if any, of the other SNPs in this region are independently associated with susceptibility to RA. Therefore, we used the haplotype method to assess whether the observed association of other PTPN22 markers (table 1) could be explained entirely by LD with R620W. Using the 14-marker data, we applied the haplotype method to both sample sets. Statistical significance, assessed using a permutation procedure, allowed us to reject the null hypothesis that R620W by itself accounts for all the predisposing effects in this region in both data sets (sample set 1, P=.002; sample set 2, P=.0086).
Association of Other PTPN22 SNPs after Adjustment for R620W: The Haplotype Method
Having established that R620W by itself does not account for all the association observed between RA and PTPN22 in both sample sets, we used the haplotype method to identify which SNPs, independent of R620W, were associated with RA. Analysis of the 14-marker data in both samples sets identified three SNPs (SNPs 27, 36, and 37) that were consistently associated with RA (table 4) independent of R620W. These three SNPs represent, at most, two independent associations, since SNPs 36 and 37 are in nearly complete LD (r2=0.993 in sample set 1 cases and 0.984 in sample set 1 controls), and the minor allele of each is found on a single haplotype, haplotype 4 (table 3); consequently, all subsequent analyses used only one of these two SNPs, SNP 37.
Table 4.
Association of PTPN22 SNPs with RA after Conditioning on R620W: The Haplotype Method
|
P Valuea for |
||
| SNP | Sample Set 1 | Sample Set 2 |
| 1 | 1 | .29 |
| 2 | 1 | 1.00 |
| 18 | .0012 | .21 |
| 20 | 1 | .01 |
| 21 | .0006 | .20 |
| 23 | .22 | .24 |
| 27 | .001 | .03 |
| 28 | .15 | .004 |
| 32 | 1 | 1.00 |
| 34 | 1 | 1.00 |
| 35 | 1 | 1.00 |
| 36 | .0009 | .01 |
| 37 | .0003 | .01 |
Association of PTPN22 SNPs after Adjustment for R620W: CLR
To further investigate the association of SNPs 27 and 37 with risk of RA independent of R620W, we analyzed the genotype data by using CLR. ORs and 95% CIs for each genotype, relative to the major-allele homozygote genotype after adjustment for the R620W SNP, are shown in table 5 along with a trend test to assess overall significance. SNP 37 showed significant association with RA after adjustment for R620W in both data sets (sample set 1, Ptrend=.002; sample set 2, Ptrend=.014). Evidence of R620W-adjusted RA association for SNP 27 was significant in sample set 1 (Ptrend=.002) but was marginally significant in sample set 2 (Ptrend=.052). Further adjustment for HLA-DRB1 genotype, the strongest known genetic risk factor for RA (Seldin et al. 1999; Newton et al. 2004), had little impact on the risk estimates for these two SNPs (data not shown). The substantial risk estimated for heterozygotes suggests that a recessive mode of inheritance is an unlikely explanation for these data.
Table 5.
CLR Analysis, Adjusted for R620W Genotypes[Note]
|
Sample Set 1 |
Sample Set 2 |
|||||
| Model,SNP Tested,and Genotype | ORa | 95% CI | Ptrend | ORa | 95% CI | Ptrend |
| SNP 27 and R620W: | ||||||
| SNP 27: | ||||||
| TT | 1.85 | 1.23–2.77 | .0022 | 1.32 | .98–1.77 | .0523 |
| TC | 1.42 | 1.04–1.93 | … | 1.22 | .97–1.54 | … |
| CC | 1.00 | … | … | 1.00 | … | … |
| R620W: | ||||||
| TT | 1.63 | .39–6.76 | .0823 | 2.17 | 1.02–4.61 | <.0001 |
| TC | 1.34 | .95–1.88 | … | 1.89 | 1.47–2.43 | … |
| CC | 1.00 | … | … | 1.00 | … | … |
| SNP 37 and R620W: | ||||||
| SNP 37: | ||||||
| CC | 1.73 | .78–3.82 | .0022 | 1.78 | 1.13–2.79 | .0143 |
| CA | 1.55 | 1.16–2.06 | … | 1.15 | .93–1.42 | … |
| AA | 1.00 | … | … | 1.00 | … | … |
| R620W: | ||||||
| TT | 2.53 | .62–10.26 | .0004 | 2.90 | 1.37–6.18 | <.0001 |
| TC | 1.77 | 1.27–2.46 | … | 2.19 | 1.73–2.78 | … |
| CC | 1.00 | … | … | 1.00 | … | … |
Note.— Case-control status was modeled within a CLR framework with both R620W and the second SNP (either SNP 27 or 37) genotypes coded as indicator variables and included in the model.
ORs were calculated relative to the major-allele homozygote, mutually adjusted for the other marker in the model.
We also examined the association of R620W with RA after adjusting for these new risk alleles by using CLR (table 5). Adjustment of R620W by SNP 37 did not markedly alter the R620W OR estimates, which remained significant in both sample sets (Ptrend<.001). Adjustment for SNP 27 did not have an appreciable effect on the R620W association in sample set 2 (Ptrend<.0001), but R620W was no longer significantly associated with RA risk in sample set 1 (Ptrend=.08).
When SNPs 27, 37, and R620W were jointly modeled using CLR, no independent effects were obtained in sample set 1, and only R620W remained significant in sample set 2 (data not shown). These results indicate an inability of CLR to resolve evidence for independent effects from SNPs 27 and 37 in the presence of R620W, reflecting the existence of LD among these markers.
Are SNPs 27 and 37 Independent Risk Factors?
The above results suggest that SNPs 27 and 37 are potential disease-predisposing sites independent of R620W. Whereas the SNP 37 risk allele is found on a single common haplotype, haplotype 4, the risk allele of SNP 27 is present on 4 of the 10 common haplotypes (haplotypes 1–4) (table 3), 2 of which (haplotypes 2 and 4) are associated with RA. It is therefore possible that the R620W-independent association observed between SNP 27 and RA is the result of an effect from haplotype 4, which is defined by the SNP 37 risk allele. To test this hypothesis, we again used the haplotype method, and, this time, from the estimation of haplotype frequencies and their analysis via the haplotype method, we were unable to reject the null hypothesis that just two SNPs, R620W and SNP 37, explain the association observed for all three markers (620W, SNP 37, and SNP 27) (P=1 in sample sets 1 and 2). These results suggest that SNP 27 and SNP 37 are not mutually independent risk factors and that just W620 and the SNP 37 risk allele together can explain the association observed between these three SNPs and RA.
Do R620W and SNP 37 Explain All the Significant Association between PTPN22 and RA?
To assess whether these two SNPs could explain all the observed association between PTPN22 and RA, we conducted a test of heterogeneity between case and control haplotypes after removing the two haplotypes (haplotypes 2 and 4) uniquely marked by the risk alleles at R620W and SNP 37. This analysis is similar to the RPE method of Payami et al. (1989). The test of heterogeneity for the remaining eight haplotypes was significant in sample set 2 (χ2=25.28; P=6.77×10-4) and was marginally significant in sample set 1(χ2=13.19; P=.07); however, none of the remaining haplotypes that demonstrated significant differences between cases and controls in sample set 2—haplotypes 6 (P=.001) and 8 (P=.002)—was significant in sample set 1 (haplotype 6, P=.64; haplotype 8, P=.52). These results suggest that haplotypes 2 and 4 capture the major effects of association between PTPN22 and RA in sample set 1 but not in sample set 2.
To explore the relative effects of each haplotype, we performed a haplotype regression analysis in which the most common haplotype, haplotype 5, was set as the reference group. The results show that haplotype 2 (sample set 1, P=7.7×10-6, OR=2.17; sample set 2, P=3.9×10-10, OR=2.11) and haplotype 4 (sample set 1, P=2.5×10-5, OR=1.86; sample set 2, P=.02, OR=1.26) are the most significant and also the two major haplotypes that conferred significantly increased risk in both sample sets, relative to haplotype 5. Further adjustment for HLA status had little impact on the risk estimates for haplotype 2 (sample set 1, P=5.2×10-4, OR=1.93; sample set 2, P=1.2×10-6, OR=1.94) and haplotype 4 (sample set 1, P=5.9 × 10-4, OR=1.74; sample set 2, P=3.7×10-3, OR=1.40). These results provide support for the notion that haplotypes 2 and 4 are the only RA-risk haplotypes that are significantly associated with RA relative to the most common haplotype (haplotype 5) in both sample sets.
The Combined Effects of R620W and SNP 37 in RA: Diplotype Analysis
To further assess the combined effects of R620W and SNP 37 and their potential predisposition to RA risk, we conducted a diplotype analysis. First, we estimated phase for the alleles at the two sites for each individual, using a Bayesian method (Stephens et al. 2001) implemented in SNPAnalyzer (table 6). Consistent with the observation that the D′ value between R620W and SNP 37 is close to 1, only three of the four possible haplotypes were observed; chromosomes with a risk allele at both sites were not seen.
Table 6.
Diplotype Analysis for R620W and SNP 37[Note]
|
Sample Set 1 |
Sample Set 2 |
||||||||||
| Diplotype | Haplotypesa | No. ofCases | No. ofControls | ORb | 95% CI | Pc | No. ofCases | No. ofControls | ORb | 95% CI | Pc |
| T-A/T-A | H2/H2 | 6 | 3 | 2.54 | .62–10.32 | .1934 | 13 | 14 | 2.54 | 1.16–5.54 | .0196 |
| T-A/C-C | H2/H4 | 32 | 14 | 2.67 | 1.36–5.23 | .0043 | 41 | 43 | 2.4 | 1.52–3.79 | .0002 |
| C-C/C-C | H4/H4 | 15 | 11 | 1.73 | .78–3.84 | .1757 | 36 | 52 | 1.75 | 1.12–2.74 | .0148 |
| T-A/C-A | */H2 | 86 | 64 | 1.78 | 1.22–2.61 | .0027 | 135 | 153 | 2.26 | 1.72–2.98 | <.0001 |
| C-A/C-C | */H4 | 133 | 113 | 1.56 | 1.14–2.14 | .0059 | 158 | 336 | 1.19 | .94–1.51 | .1529 |
| C-A/C-A | */* | 194 | 261 | … | … | … | 275 | 706 | … | … | … |
Note.— Case-control status was modeled within a CLR framework with the common diplotypes (frequency >1%) for two loci (R620W and SNP 37) coded as indicator variables and included in the model.
Corresponding haplotypes for each diplotype. H2 = haplotype 2; H4 = haplotype 4 (see table 3). An asterisk (*) indicates all other haplotypes (1, 3, and 5–10).
ORs were calculated relative to the most common diplotype, C-A/C-A.
The Wald test P value was calculated for each diplotype coefficient.
Next, CLR was used to assess association of the inferred diplotypes with RA risk by use of the most common diplotype (C-A/C-A: R620W–SNP 37/R620W–SNP 37), corresponding to the major homozygous (nonrisk) genotypes at both sites, as the reference group (table 6). When R620W heterozygous genotypes (CT) were subdivided by the presence or absence of the SNP 37 risk allele on the other chromosome, we found, in both sample sets, that the diplotype positive for the SNP 37 risk allele (T-A/C-C) had a higher OR point estimate than that of the diplotype without the SNP 37 risk allele (T-A/C-A) (2.67 vs. 1.78 in sample set 1; 2.40 vs. 2.26 in sample set 2). Diplotypes carrying two copies of the SNP 37 risk allele and no copies of W620 (C-C/C-C) also appear to have increased risk (OR=1.73 and 1.75 in sample sets 1 and 2, respectively) relative to the most common diplotype; however, this observation was significant only in sample set 2 (sample set 1, P=.1757; sample set 2, P=.0148). Diplotypes carrying a single copy of the SNP 37 risk allele and no copies of W620 (C-A/C-C) are at increased risk for RA in sample set 1 (P=.0059; OR=1.56); however, this was not confirmed in sample set 2 (P=.1529; OR=1.19). We observed similar results after further adjustment for HLA-DRB1 genotypes.
Discussion
Given the genetic and other biological data implicating the minor allele of the PTPN22 R620W SNP as a risk factor for RA and other autoimmune diseases, we wanted to determine whether additional PTPN22 variants were associated with RA, independent of R620W. Accordingly, we sequenced all exons and intron/exon boundaries of PTPN22 as well as 5′ and 3′ sequences in 48 RF-positive patients with RA and identified 15 previously unreported SNPs, including two cSNPs in the catalytic domain. Using this information and that in public databases, we generated assays for 36 SNPs in or near PTPN22, in addition to R620W, and genotyped them in an RA sample set.
Neither of the two new cSNPs showed significant association with disease in this sample set. The minor allele of R263Q was present on ∼2% of both case and control chromosomes, and, although arginine (R263) is the major allele in humans, glutamine (Q263) appears to be the ancestral allele. Q263 is found in chimpanzees, dogs, rodents, and chickens. The predicted stop codon (X183) lies in the middle of the catalytic domain and likely leads to loss of function; however, it was seen only once in our study, in an individual with RA (existence of this allele was independently confirmed by both sequencing and genotyping). Although this nonsense SNP was not genotyped in our second sample set, we did screen 1,104 individuals from a Dutch early-arthritis cohort (van Aken et al. 2003) and failed to detect it (authors' unpublished results), which suggests that this nonsense SNP is very rare in whites.
Many of the other 34 markers spanning this 58-kb region were strongly associated with RA in this sample set (table 1), and several markers were also replicated in a second sample set. However, the strong LD across this region, revealed by pairwise D′ values near 1 (data not shown) and r2 values >0.8 between some of these SNPs (fig. 1B), made it difficult to determine whether these associated SNPs are independent risk factors for RA or are in LD with the known W620 risk allele. To help resolve this issue, we conducted haplotype analyses. The same 10 common haplotypes (frequency >1%) were predicted in both data sets. Comparison of these results with the HapMap data, which predict 6 haplotypes from the 30 CEPH family trios, shows that the minor-allele and haplotype frequencies are noticeably different between the HapMap data and our control samples. For example, haplotype 2, which contains the W620 risk allele and has a frequency of 8%–9% in our control populations, has a frequency of 13.3% in the CEPH samples. Haplotype 5, the most common haplotype in our sample sets, is also the most common haplotype in the HapMap data; however, it is found on 27.5% of the CEPH chromosomes, as opposed to 35% of the control chromosomes in sample set 1 and 29.9% of the control chromosomes in sample set 2. These differences may be due to the smaller sample size used to generate the HapMap data and/or population-based differences. Since the CEPH samples represent a relatively narrow slice of the European gene pool, they may not be representative of all the genetic diversity found in white North Americans. Indeed, evidence is emerging that the frequency of the PTPN22 W620 variant may vary between different European populations, in which it appears to be more frequent in northern than in southern populations (Gregersen 2005; Seldin et al., in press). Since our controls are individually matched to the cases on the basis of grandparental country of origin, it is unlikely that different ethnicities influenced the disease-association results reported here.
Analysis of the haplotype data (table 3) shows that the lone haplotype carrying the W620 risk allele (haplotype 2) was significantly associated with RA in both sample sets. Haplotype 1, identical at all other SNPs but carrying the R620 allele, was not associated with RA. These results suggest that the increased risk associated with haplotype 2 is unlikely to be explained by the alleles at the other 35 SNPs on this haplotype. Although it is possible that another unknown polymorphism on this haplotype could explain the association with RA, the data presented here, together with the finding that the W620 variant of PTPN22 shows impaired function (Begovich et al. 2004; Bottini et al. 2004), provide evidence that W620 is a disease-predisposing allele on this haplotype.
We also observed that some variants in this region are associated with RA independent of R620W. Further analyses identified three SNPs (SNPs 27, 36, and 37) that were consistently associated with RA independent of R620W in both sample sets. SNP 36 and 37 are in nearly complete LD (r2 of ∼1), and the risk alleles of each are found on a single common haplotype (haplotype 4 in table 3) present on ∼15%–18% of control chromosomes, which suggests that they represent a single association. Although we cannot exclude the possibility that SNP 27 is a disease-predisposing locus, its association with RA is not independent of both R620W and SNP 37. Furthermore, our results suggest that R620W and SNP 37 uniquely define two major risk haplotypes in both sample sets that may be sufficient to explain the significant associations of PTPN22 with RA in sample set 1 but not in sample set 2. The residual significant association in sample set 2 after removal of haplotypes 2 and 4 indicates that other haplotypes are required to account for the remaining genetic heterogeneity observed in this sample set.
The diplotype analysis suggests that individuals who are homozygous for the W620-positive haplotype 2 might have an increased risk for RA compared with individuals who carry one copy of haplotype 2, except when the other chromosome carries the risk allele at SNP 37 (i.e., haplotype 4) (table 6). On the basis of the data from sample set 1, we estimated the population-attributable risk percentage (Rothman 2002) for the H2/H2, H2/H4, H4/H4, */H2, and */H4 diplotypes to be 1.7%, 7.8%, 2.9%, 13.3%, and 14.5% respectively, relative to the */* diplotype (where an asterisk [*] indicates haplotypes 1, 3, and 5–10). However, these results should be viewed with caution. Because the 95% CIs for the OR estimates overlapped in our case-control sample sets (table 6), they may not be significantly different. In addition, because these results were obtained from a case-control study design, they should be replicated in large cohort studies to accurately estimate absolute risks associated with these markers and their diplotypes in the general population.
SNPs 36 and 37 are also in near absolute LD with a third SNP (SNP 15) that was not genotyped in the second sample set, and minor alleles of all three SNPs are carried by a single common haplotype, haplotype 4 (table 3). Biologically, SNPs 36 and 37 may be particularly interesting. SNP 36 lies in the 3′ UTR of the major PTPN22 transcript and may influence transcript stability and/or balance of the two known alternatively spliced PTPN22 transcripts (Cohen et al. 1999).
SNP 37 lies 1,496 bases downstream of PTPN22 at the 5′ end of the round spermatid basic protein 1 gene (RSBN1), where it encodes either a silent mutation or a putative TFBS, depending on the transcript. RSBN1 is a somewhat unlikely RA-susceptibility candidate gene. Although its function is unknown, its closest homologue in Caenorhabditis elegans, dpy-21, is involved in X-chromosome dosage compensation. This SNP also lies in a putative TFBS for Pax-5, Pax-4, Nrf-1, and c-Myb (TRANSFAC positional weight matrices) (Wingender et al. 2000). The minor allele at SNP 37 is predicted to change the binding activity for Pax-4 and Pax-5, and, although it lies 3′ of PTPN22, it may still regulate expression of that gene. Pax-5 (B-cell–specific activator protein) is required for commitment to the B-lymphoid lineage. In the absence of Pax-5, pro-B cells are capable of differentiating into T cells, macrophages, osteoclasts, dendritic cells, granulocytes, and natural killer cells (Nutt et al. 1999).
Although these observations are provocative, we must keep in mind the possibility that these SNPs are in LD with a true causal SNP(s), which could reside in PTPN22 or in other genes in the same haplotype block. Although the focus of this work was on variation in the PTPN22 gene, the HapMap data indicate that this gene is found in a region of high LD that extends ∼300 kb and includes six other genes: the 3′ end of MAGI3 (encoding a membrane-associated guanylate kinase-related protein), PHTF1 (encoding a putative homeodomain transcription factor), RSBN1 (encoding round spermatid basic protein 1), FLJ22588 (encoding hypothetical protein LOC440603 of unknown function), AP4B1 (encoding adaptor-related protein complex 4, β1 subunit), and the 5′ end of DCLRE1B (the DNA cross-link repair 1B gene) (International HapMap Project, 16th release, March 2005). However, only RSBN1 lies in the same haplotype block predicted by the method of Gabriel et al. (2002). Given the strong LD between these various SNPs, there is the definite possibility that alternative sites in these other genes may play a role in disease susceptibility, although given what is currently known about the function of these genes, none is as biologically compelling as PTPN22.
In conclusion, we have found that 620W defines one major risk haplotype that is strongly and consistently associated with RA in our two sample sets. These genetic data, along with other biological data (Begovich et al. 2004; Bottini et al. 2004), point to W620 as a disease-predisposing allele. Furthermore, we find that alleles of other SNPs in this region that are on a relatively common haplotype are positively associated with RA, independent of R620W. These SNPs and the haplotype on which they are found should be investigated in greater detail in other populations with RA, as well as in populations with other autoimmune diseases that show association with the R620W SNP, to fully elucidate their role in autoimmunity. Finally, it will be of interest to determine whether R620W and these newly defined disease-susceptibility alleles also play a role in the response to specific therapies.
Acknowledgments
We are grateful to the patients with RA, the control individuals, and the collaborating clinicians, for participation in this study; members of the Celera Diagnostics (CDx) High Throughput and Computational Biology groups, for invaluable help; J. Law and V. Garcia, for development and implementation of the automated CDx statistical programs; R. Lundsten, M. Kern, H. Khalili, A. Rodrigues-Brown, and A. T. Lee, for database and sample management of the NARAC collection; J. Lemaire and S. Mahan, for database and sample management at Genomics Collaborative; and S. Broder, M. Cargill, W. V. Chen, A. Clark, C. Rowland, J. Sninsky, T. White, and X. Zhou, for discussions and insightful comments on the manuscript. Collection of the NARAC cohort has been funded by a National Arthritis Foundation grant and by the National Institutes of Health, acting through the National Institute of Arthritis and Musculoskeletal and Skin Diseases and the National Institute of Allergy and Infectious Diseases (contracts N01-AR-7-2232 and R01-AR44222). These studies were performed in part at the General Clinical Research Center, Moffitt Hospital, University of California–San Francisco, with funds provided by the National Center for Research Resources (U.S. Public Health Service grant 5 M01 RR-00079). Support was also provided by National Institutes of Health grants HG02275 and GM35326.
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- AMDeC, http://www.amdec.org (for New York Cancer Project)
- Celera, http://www.celera.com/
- dbSNP, http://www.ncbi.nlm.nih.gov/SNP/ (for novel SNPs discovered by resequencing: ss38346943, ss38346944, ss38346945, ss38346946, ss38346947, ss38346948, ss38346949, ss38346950, ss38346951, ss38346952,ss38346953, ss38346954, ss38356955, ss38346956, and ss38346957)
- HapMap, http://www.hapmap.org/
- NARAC, http://www.naracdata.org/index.asp
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for autoimmune diseases, Crohn disease, Graves disease, MS, psoriasis vulgaris, RA, SLE, and T1D)
- Polyphred, http://droog.mbt.washington.edu/PolyPhred.html
- Primer3, http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi
- SNPAnalyzer, http://www.istech21.com/download/download_a01.html
- SNPHAP, http://www-gene.cimr.cam.ac.uk/clayton/software/
References
- Abecasis GR, Cookson WO (2000) GOLD—graphical overview of linkage disequilibrium. Bioinformatics 16:182–183 [DOI] [PubMed] [Google Scholar]
- Alarcón-Segovia D, Alarcón-Riquelme ME, Cardiel MH, Caeiro F, Massardo L, Villa AR, Pons-Estel BA (2005) Familial aggregation of systemic lupus erythematosus, rheumatoid arthritis and other autoimmune diseases in 1,177 lupus patients from the GLADEL cohort. Arthritis Rheum 52:1138–1147 [DOI] [PubMed] [Google Scholar]
- Alkhateeb A, Fain PR, Thody A, Bennett DC, Spritz RA (2003) Epidemiology of vitiligo and associated autoimmune diseases in Caucasian probands and their families. Pigment Cell Res 16:208–214 [DOI] [PubMed] [Google Scholar]
- Becker KG, Simon RM, Bailey-Wilson JE, Freidlin B, Biddison WE, McFarland HF, Trent JM (1998) Clustering of non-major histocompatibility complex susceptibility candidate loci in human autoimmune diseases. Proc Natl Acad Sci USA 95:9979–9984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begovich AB, Caillier SJ, Alexander HC, Penko JM, Hauser SL, Barcellos LF, Oksenberg JR (2005) The R620W polymorphism in the protein tyrosine phosphatase PTPN22 is not associated with multiple sclerosis. Am J Hum Genet 76:184–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begovich AB, Carlton VEH, Honigberg LA, Schrodi SJ, Chokkalingam AP, Alexander HC, Ardlie KG, et al (2004) A missense single-nucleotide polymorphism in a gene encoding a protein tyrosine phosphatase (PTPN22) is associated with rheumatoid arthritis. Am J Hum Genet 75:330–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottini N, Musumeci L, Alonso A, Rahmouni S, Nika K, Rostamkhani M, MacMurray J, Meloni GF, Lucarelli P, Pellecchia M, Eisenbarth GS, Comings D, Mustelin T (2004) A functional variant of lymphoid tyrosine phosphatase is associated with type I diabetes. Nat Genet 36:337–338 [DOI] [PubMed] [Google Scholar]
- Breslow NE, Day NE (1980) Statistical methods in cancer research. Volume I. The analysis of case-control studies. IARC Sci Publ 32:5–338 [PubMed] [Google Scholar]
- Cloutier JF, Veillette A (1996) Association of inhibitory tyrosine protein kinase p50csk with protein tyrosine phosphatase PEP in T cells and other hemopoietic cells. EMBO J 15:4909–4918 [PMC free article] [PubMed] [Google Scholar]
- ——— (1999) Cooperative inhibition of T-cell antigen receptor signaling by a complex between a kinase and a phosphatase. J Exp Med 189:111–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S, Dadi H, Shaoul E, Sharfe N, Roifman CM (1999) Cloning and characterization of a lymphoid-specific, inducible human protein tyrosine phosphatase, Lyp. Blood 93:2013–2024 [PubMed] [Google Scholar]
- Criswell LA, Pfeiffer KA, Lum RF, Gonzales B, Novitzke J, Kern M, Moser KL, Begovich AB, Carlton VEH, Li W, Lee AT, Ortmann W, Behrens TW, Gregersen PK (2005) Analysis of families in the Multiple Autoimmune Disease Genetics Consortium (MADGC) collection: the PTPN22 620W allele associates with multiple autoimmune phenotypes. Am J Hum Genet 76:561–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D (2002) The structure of haplotype blocks in the human genome. Science 296:2225–2229 [DOI] [PubMed] [Google Scholar]
- Germer S, Holland MJ, Higuchi R (2000) High-throughput SNP allele-frequency determination in pooled DNA samples by kinetic PCR. Genome Res 10:258–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gjorloff-Wingren A, Saxena M, Williams S, Hammi D, Mustelin T (1999) Characterization of TCR-induced receptor-proximal signaling events negatively regulated by the protein tyrosine phosphatase PEP. Eur J Immunol 29:3845–3854 [DOI] [PubMed] [Google Scholar]
- Gregersen PK (2005) Pathways to gene identification in rheumatoid arthritis: PTPN22 and beyond. Immunol Rev 204:74–86 [DOI] [PubMed] [Google Scholar]
- Gregorieff A, Cloutier JF, Veillette A (1998) Sequence requirements for association of protein-tyrosine phosphatase PEP with the Src homology 3 domain of inhibitory tyrosine protein kinase p50(csk). J Biol Chem 273:13217–13222 [DOI] [PubMed] [Google Scholar]
- Hasegawa K, Martin F, Huang G, Tumas D, Diehl L, Chan AC (2004) PEST domain-enriched tyrosine phosphatase (PEP) regulation of effector/memory T cells. Science 303:685–689 [DOI] [PubMed] [Google Scholar]
- Hinks A, Barton A, John S, Bruce I, Hawkins C, Griffiths CE, Donn R, Thomson W, Silman A, Worthington J (2005) Association between the PTPN22 gene and rheumatoid arthritis and juvenile idiopathic arthritis in a UK population: further support that PTPN22 is an autoimmunity gene. Arthritis Rheum 52:1694–1699 [DOI] [PubMed] [Google Scholar]
- Ho P, Bowes J, Eyre S, Bradburn P, Newman B, Bruce I, Silman A, Worthington J, Barton A (2004) Association of the organic cation transporter (OCTN) SLC22A4 and SLC22A5 genes with psoriatic arthritis. Arthritis Rheum 50:S258 [Google Scholar]
- Hu X, Schrodi SJ, Ross DA, Cargill M (2004) Selecting tagging SNPs for association studies using power calculations from genotype data. Hum Hered 57:156–170 [DOI] [PubMed] [Google Scholar]
- Hugot JP, Chamaillard M, Zouali H, Lesage S, Cezard JP, Belaiche J, Almer S, Tysk C, O’Morain CA, Gassull M, Binder V, Finkel Y, Cortot A, Modigliani R, Laurent-Puig P, Gower-Rousseau C, Macry J, Colombel JF, Sahbatou M, Thomas G (2001) Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature 411:599–603 [DOI] [PubMed] [Google Scholar]
- Jawaheer D, Lum RF, Amos CI, Gregersen PK, Criswell LA (2004) Clustering of disease features within 512 multicase rheumatoid arthritis families. Arthritis Rheum 50:736–741 [DOI] [PubMed] [Google Scholar]
- Jawaheer D, Seldin MF, Amos CI, Chen WV, Shigeta R, Monteiro J, Kern M, Criswell LA, Albani S, Nelson JL, Clegg DO, Pope R, Schroeder HW Jr, Bridges SL Jr, Pisetsky DS, Ward R, Kastner DL, Wilder RL, Pincus T, Callahan LF, Flemming D, Wener MH, Gregersen PK (2001) A genomewide screen in multiplex rheumatoid arthritis families suggests genetic overlap with other autoimmune diseases. Am J Hum Genet 68:927–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochi Y, Yamada R, Suzuki A, Harley JB, Shirasawa S, Sawada T, Bae S-C, Tokuhiro S, Chang X, Sekine A, Takahasi A, Tsunoda T, Ohnishi Y, Kaufman KM, Kang CP, Kang C, Otsubo S, Yumura W, Mimori A, Kioke T, Nakamura Y, Sasazuki T, Yamamoto K (2005) A functional variant in FCRL3, encoding Fc receptor-like 3, is associated with rheumatoid arthritis and several autoimmunities. Nat Genet 37:478–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyogoku C, Langefeld CD, Ortmann WA, Lee A, Selby S, Carlton VEH, Chang M, Ramos P, Baechler EC, Batliwalla FM, Novitzke J, Williams AH, Gillett C, Rodine P, Graham RR, Ardlie KG, Gaffney PM, Moser KL, Petri M, Begovich AB, Gregersen PK, Behrens TW (2004) Genetic association of the R620W polymorphism of protein tyrosine phosphatase PTPN22 with human SLE. Am J Hum Genet 75:504–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladner MB, Bottini N, Valdes AM, Noble JA (2005) Association of the single nucleotide polymorphism C1858T of the PTPN22 gene with type 1 diabetes. Hum Immunol 66:60–64 [DOI] [PubMed] [Google Scholar]
- Lake SL, Lyon H, Tantisira K, Silverman EK, Weiss ST, Laird NM, Schaid DJ (2003) Estimation and tests of haplotype-environment interaction when linkage phase is ambiguous. Hum Hered 55:56–65 [DOI] [PubMed] [Google Scholar]
- Lee AT, Li W, Liew A, Bombardier C, Weisman M, Massarotti EM, Kent J, Wolfe F, Begovich AB, Gregersen PK (2005) The PTPN22 R620W polymorphism associates with RF positive rheumatoid arthritis in a dose-dependent manner but not with HLA-SE status. Genes Immun 6:129–133 [DOI] [PubMed] [Google Scholar]
- Li H (2001) A permutation procedure for the haplotype method for identification of disease-predisposing variants. Ann Hum Genet 65:189–196 [DOI] [PubMed] [Google Scholar]
- Lin JP, Cash JM, Doyle SZ, Peden S, Kanik K, Amos CI, Bale SJ, Wilder RL (1998) Familial clustering of rheumatoid arthritis with other autoimmune diseases. Hum Genet 103:475–482 [DOI] [PubMed] [Google Scholar]
- Lin SC, Yen JH, Tsai JJ, Tsai WC, Ou TT, Liu HW, Chen CJ (2004) Association of a programmed death 1 gene polymorphism with the development of rheumatoid arthritis, but not systemic lupus erythematosus. Arthritis Rheum 50:770–775 [DOI] [PubMed] [Google Scholar]
- Marrack P, Kappler J, Kotzin BL (2001) Autoimmune disease: why and where it occurs. Nat Med 7:899–905 [DOI] [PubMed] [Google Scholar]
- Matesanz F, Rueda B, Orozco G, Fernandez O, Leyva L, Alcina A, Martin J (2005) Protein tyrosine phosphatase gene (PTPN22) polymorphism in multiple sclerosis. J Neurol (http://www.blackwell-synergy.com/doi/abs/10.1111/j.0022-202X.2005.23802.x) (electronically published March 17, 2005; accessed August 10, 2005) [DOI] [PubMed] [Google Scholar]
- Newton JL, Harney SMJ, Wordsworth BP, Brown MA (2004) A review of the MHC genetics of rheumatoid arthritis. Genes Immun 5:151–157 [DOI] [PubMed] [Google Scholar]
- Nielsen C, Hansen D, Husby S, Jacobsen BB, Lillevang ST (2003) Association of a putative regulatory polymorphism in the PD-1 gene with susceptibility to type 1 diabetes. Tissue Antigens 62:492–497 [DOI] [PubMed] [Google Scholar]
- Nutt SL, Heavey B, Rolink AG, Busslinger M (1999) Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature 401:556–562 [DOI] [PubMed] [Google Scholar]
- Onengut-Gumuscu S, Ewens KG, Spielman RS, Concannon P (2004) A functional polymorphism (1858C/T) in the PTPN22 gene is linked and associated with type I diabetes in multiplex families. Genes Immun 5:678–680 [DOI] [PubMed] [Google Scholar]
- Orozco G, Sanchez E, Gonzalez-Gay MA, Lopez-Nevot MA, Torres B, Caliz R, Ortego-Centeno N, Jimenez-Alonso J, Pascual-Salcedo D, Balsa A, de Pablo R, Nunez-Roldan A, Gonzalez-Escribano MF, Martin J (2005) Association of a functional single-nucleotide polymorphism of PTPN22, encoding lymphoid protein phosphatase, with rheumatoid arthritis and systemic lupus erythematosus. Arthritis Rheum 52:219–224 [DOI] [PubMed] [Google Scholar]
- Payami H, Joe S, Farid NR, Stenszky V, Chan SH, Yeo PPB, Cheah JS, Thomson G (1989) Relative predispositional effects (RPEs) of marker alleles with disease: HLA-DR alleles and Graves disease. Am J Hum Genet 45:541–546 [PMC free article] [PubMed] [Google Scholar]
- Peltekova VD, Wintle RF, Rubin LA, Amos CI, Huang Q, Gu X, Newman B, Van Oene M, Cescon D, Greenberg G, Griffiths AM, St George-Hyslop PH, Siminovitch KA (2004) Functional variants of OCTN cation transporter genes are associated with Crohn disease. Nat Genet 36:471–475 [DOI] [PubMed] [Google Scholar]
- Prahalad S, Shear ES, Thompson SD, Giannini EH, Glass DN (2002) Increased prevalence of familial autoimmunity in simplex and multiplex families with juvenile rheumatoid arthritis. Arthritis Rheum 46:1851–1856 [DOI] [PubMed] [Google Scholar]
- Prokunina L, Castillejo-López C, Öberg F, Gunnarsson I, Berg L, Magnusson V, Brookes AJ, Tentler D, Kristjansdóttir H, Gröndal G, Bolstad AI, Svenungsson E, Lundberg I, Sturfelt G, Jönssen A, Truedsson L, Lima G, Alcocer-Varela J, Jonsson R, Gyllensten UB, Harley JB, Alarcón-Segovia D, Steinsson K, Alarcón-Riquelme ME (2002) A regulatory polymorphism in PDCD1 is associated with susceptibility to systemic lupus erythematosus in humans. Nat Genet 32:666–669 [DOI] [PubMed] [Google Scholar]
- Prokunina L, Padyukov L, Bennet A, de Faire U, Wiman B, Prince J, Alfredsson L, Klareskog L, Alarcon-Riquelme M (2004) Association of the PD-1.3A allele of the PDCD1 gene in patients with rheumatoid arthritis negative for rheumatoid factor and the shared epitope. Arthritis Rheum 50:1770–1773 [DOI] [PubMed] [Google Scholar]
- Qu H, Tessier MC, Hudson TJ, Polychronakos C (2005) Confirmation of the association of the R620W polymorphism in the protein tyrosine phosphatase PTPN22 with type 1 diabetes in a family based study. J Med Genet 42:266–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman P, Bartlett S, Siannia S, Pellett FJ, Farewell VT, Peddle L, Schentag CT, Alderdice CA, Hamilton S, Khraishi M, Tobin Y, Hefferton D, Gladman DD (2003) CARD15: a pleiotropic autoimmune gene that confers susceptibility to psoriatic arthritis. Am J Hum Genet 73:677–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman KJ (2002) Epidemiology: an introduction. Oxford University Press, New York [Google Scholar]
- Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA (2002) Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet 70:425–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seldin MF, Amos CI, Ward R, Gregersen PK (1999) The genetics revolution and the assault on rheumatoid arthritis. Arthritis Rheum 42:1071–1079 [DOI] [PubMed] [Google Scholar]
- Seldin MF, Shigeta R, Laiho K, Li H, Saila H, Savolainen A, Leirisalo-Repo M, Aho K, Tuomilehto-Wolf E, Kaarela K, Kauppi M, Alexander HC, Begovich AB, Tuomilehto J. Finnish case control and family studies support PTPN22 R620W polymorphism as a risk factor in rheumatoid arthritis but suggest only minimal or no effect in juvenile idiopathic arthritis. Genes Immun (in press) [DOI] [PubMed] [Google Scholar]
- Siminovitch KA (2004) PTPN22 and autoimmune disease. Nat Genet 36:1248–1249 [DOI] [PubMed] [Google Scholar]
- Simkins HM, Merriman ME, Highton J, Chapman PT, O’Donnell JL, Jones PB, Gow PJ, McLean L, Pokorny V, Harrison AA, Merriman TR (2005) Association of the PTPN22 locus with rheumatoid arthritis in a New Zealand Caucasian cohort. Arthritis Rheum 52:2222–2225 [DOI] [PubMed] [Google Scholar]
- Skorka A, Bednarczuk T, Bar-Andziak E, Nauman J, Ploski R (2005) Lymphoid tyrosine phosphatase (PTPN22/LYP) variant and Graves’ disease in a Polish population: association and gene dose-dependent correlation with age of onset. Clin Endocrinol (Oxf) 62:679–682 [DOI] [PubMed] [Google Scholar]
- Smyth D, Cooper JD, Collins JE, Heward JM, Franklyn JA, Howson JM, Vella A, Nutland S, Rance HE, Maier L, Barratt BJ, Guja C, Ionescu-Tirgoviste C, Savage DA, Dunger DB, Widmer B, Strachan DP, Ring SM, Walker N, Clayton DG, Twells RC, Gough SC, Todd JA (2004) Replication of an association between the lymphoid tyrosine phosphatase locus (LYP/PTPN22) with type 1 diabetes, and evidence for its role as a general autoimmunity locus. Diabetes 53:3020–3023 [DOI] [PubMed] [Google Scholar]
- Steer S, Lad B, Grumley JA, Kingsley GH, Fisher SA (2005) Association of R620W in a protein tyrosine phosphatase gene with a high risk of rheumatoid arthritis in a British population: evidence for an early onset/disease severity effect. Arthritis Rheum 52:358–360 [DOI] [PubMed] [Google Scholar]
- Stephens M, Smith NJ, Donnelly P (2001) A new statistical method for haplotype reconstruction from population data. Am J Hum Genet 68:978–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tait KF, Marshall T, Berman J, Carr-Smith J, Rowe B, Todd JA, Bain SC, Barnett AH, Gough SC (2004) Clustering of autoimmune disease in parents of siblings from the type 1 diabetes Warren repository. Diabet Med 21:358–362 [DOI] [PubMed] [Google Scholar]
- Thomson G, Robinson WP, Kuhner MK, Joe S, MacDonald MJ, Gottschall JL, Barbosa J, Rich SS, Bertrams J, Baur MP, Partanen J, Tait B, Schober E, Mayr WR, Ludvigsson J, Lindblom B, Farid NR, Thompson C, Deschamps I (1988) Genetic heterogeneity, modes of inheritance, and risk estimates for a joint study of Caucasians with insulin dependent diabetes mellitus. Am J Hum Genet 43:799–816 [PMC free article] [PubMed] [Google Scholar]
- Ueda H, Howson JM, Esposito L, Heward J, Snook H, Chamberlain G, Rainbow DB, et al (2003) Association of the T-cell regulatory gene CTLA4 with susceptibility to autoimmune disease. Nature 423:506–511 [DOI] [PubMed] [Google Scholar]
- Valdes AM, Thomson G (1997) Detecting disease-predisposing variants: the haplotype method. Am J Hum Genet 60:703–716 [PMC free article] [PubMed] [Google Scholar]
- van Aken J, van Bilsen JH, Allaart CF, Huizinga TW, Breedveld FC (2003) The Leiden Early Arthritis Clinic. Clin Exp Rheumatol 21:S100–S105 [PubMed] [Google Scholar]
- Van Oene M, Wintle RF, Liu X, Yazdanpanah M, Gu X, Newman B, Kwan A, Johnson B, Owen J, Greer W, Mosher D, Maksymowych W, Keystone E, Rubin LA, Amos CI, Siminovitch KA (2005) Association of the lymphoid tyrosine phosphatase R620W variant with rheumatoid arthritis, but not Crohn’s disease, in Canadian populations. Arthritis Rheum 52:1993–1998 [DOI] [PubMed] [Google Scholar]
- Velaga MR, Wilson V, Jennings CE, Owen CJ, Herington S, Donaldson PT, Ball SG, James RA, Quinton R, Perros P, Pearce SH (2004) The codon 620 tyrptophan allele of the lymphoid tyrosine phosphatase (LYP) gene is a major determinant of Graves’ disease. J Clin Endocrinol Metab 89:5862–5865 [DOI] [PubMed] [Google Scholar]
- Viken MK, Amundsen SS, Kvien TK, Boberg KM, Gilboe IM, Lilleby V, Sollid LM, Forre OT, Thorsby E, Smerdel A, Lie BA (2005) Association analysis of the 1858C>T polymorphism in the PTPN22 gene in juvenile idiopathic arthritis and other autoimmune diseases. Genes Immun 6:271–273 [DOI] [PubMed] [Google Scholar]
- Wandstrat A, Wakeland E (2001) The genetics of complex autoimmune diseases: non-MHC susceptibility genes. Nat Immunol 2:802–809 [DOI] [PubMed] [Google Scholar]
- Wingender E, Chen X, Hehl R, Karas H, Liebich I, Matys V, Meinhardt T, Pruss M, Reuter I, Schacherer F (2000) TRANSFAC: an integrated system for gene expression regulation. Nucleic Acids Res 28:316–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo J, Seo B, Kim Y (2005) SNPAnalyzer: a web-based integrated workbench for single-nucleotide polymorphism analysis. Nucleic Acids Res (Web Server issue) 33:W483–W488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W, She JX (2005) Genetic association between a lymphoid tyrosine phosphatase (PTPN22) and type 1 diabetes. Diabetes 54:906–908 [DOI] [PubMed] [Google Scholar]
- Zhernakova A, Eerligh P, Wijmenga C, Barrera P, Roep BO, Koeleman BP (2005) Differential association of the PTPN22 variant with autoimmune diseases in a Dutch population. Genes Immun (http://www.nature.com/gene/journal/vaop/ncurrent/full/6364220a.html) (published electronically May 5) [DOI] [PubMed] [Google Scholar]