Abstract
Several independent studies and meta-analyses aimed at identifying genomic regions linked to bipolar disorder (BP) have failed to find clear and consistent evidence of linkage regions. Our hypothesis is that combining the original genotype data provides benefits of increased power and control over sources of heterogeneity that outweigh the difficulty and potential pitfalls of the implementation. We conducted a combined analysis using the original genotype data from 11 BP genomewide linkage scans comprising 5,179 individuals from 1,067 families. Heterogeneity among studies was minimized in our analyses by using uniform methods of analysis and a common, standardized marker map and was assessed using novel methods developed for meta-analysis of genome scans. To date, this collaboration is the largest and most comprehensive analysis of linkage samples involving a psychiatric disorder. We demonstrate that combining original genome-scan data is a powerful approach for the elucidation of linkage regions underlying complex disease. Our results establish genomewide significant linkage to BP on chromosomes 6q and 8q, which provides solid information to guide future gene-finding efforts that rely on fine-mapping and association approaches.
Introduction
Bipolar disorder (BP) is a common and often disabling mood disorder, from which individuals suffer from episodes of mania and depression. The symptoms of mania include an expansive, elevated, or irritable mood; inflated self-esteem; grandiosity; decreased need for sleep; increased talkativeness; racing thoughts; distractibility; increased goal-directed activity; and excessive involvement in pleasurable activities with a high potential for painful consequences (National Institute of Mental Health [NIMH] Genetics Workgroup 1999). The symptoms of depression include depressed mood, diminished interest or pleasure in activities, change in sleeping patterns, psychomotor agitation or retardation, fatigue or loss of energy, feelings of worthlessness or excessive guilt, inability to concentrate or act decisively, and recurrent thoughts of death or suicide (NIMH Genetics Workgroup 1999). Bipolar disorder I (BPI) is defined by the occurrence of one or more manic or mixed (manic/depressive) episodes and is often accompanied by at least one major depressive episode (American Psychiatric Association [APA] 1994). Bipolar disorder II (BPII) is characterized by milder manic episodes (hypomania) and recurrent major depressive episodes (APA 1994).
It is estimated that BPI has a lifetime prevalence of ∼0.5%–1.5%, whereas BPII has a lifetime prevalence of ∼0.5% (Kessler et al. 1994; Weissman et al. 1996). The burden of illness for BP is considerable. It is estimated that completed suicide occurs in 10%–15% of individuals who received a diagnosis of BP (APA 1994), as well as a higher rate of unemployment and marital dysfunction and an increased use of health services (Weissman et al. 1991). Further, treatment of BP is not curative and is not completely effective in mitigating symptoms. Extensive research efforts have been directed at uncovering the etiology of BP, with the hope that more-effective treatment and prevention strategies can be developed.
The importance of genetic factors in BP has been repeatedly confirmed using family, twin, and adoption studies. The recurrence risk ratio is ∼7 for first-degree relatives and as high as 60 for MZ twins (NIMH Genetics Workgroup 1999). Twin studies have suggested heritability estimates of ∼80%, providing evidence that genes contribute strongly to the familial aggregation of BP (Tsuang and Faraone 2000; Smoller and Finn 2003). The mode of inheritance for BP is complex and likely involves multiple genes. The specific number of susceptibility loci, the recurrence risk ratio attributable to each locus, and the degree of the interaction between loci are unknown. Nevertheless, it is clear that a single major locus does not explain any substantial proportion of the familial aggregation of BP (NIMH Genetics Workgroup 1999).
Genetic complexity underlying BP provides at least one plausible explanation for the many insignificant and/or inconsistent findings among gene-mapping efforts over the past 15 years. There have been numerous genomewide scans of BP, and linkage signals from individual studies have been reported throughout the genome, including chromosomes 1, 4, 6, 10, 12, 13, 18, 21, 22, and X (reviewed by Baron [2002]). Consistent with other disorders of complex etiology, independent replication of linkage signals for BP has not been convincing.
The lack of reproducibility of linkage findings among different studies may reflect a variety of study-specific issues. For example, different studies have variable power to detect linkage, depending on the sample size and the number of affected individuals who meet ascertainment criteria for the study. There are often differences in the diagnosis and ascertainment of affected individuals, differences in the phenotypic model chosen for analysis (i.e., inclusion of recurrent unipolar depression [RUD]), or differences in the modeling parameters (i.e., penetrance, disease-allele frequency, etc.) specified for parametric linkage analysis. The genetic markers, marker densities, and the genetic maps used for each study can also vary considerably. Finally, in addition to false-positive results, there may be a considerable amount of genetic heterogeneity of BP among the populations studied.
Meta-analysis is one strategy that offers a systematic and quantitative approach to summarizing evidence from multiple genome scans. Two relevant meta-analyses of BP linkage studies have been published recently. Badner and Gershon (2002) developed the multiple scan probability (MSP) method to conduct a meta-analysis of reported P values in published linkage-analysis studies. Chromosomal regions that had a P value <.01 from 11 independently published BP linkage studies were included in their meta-analysis. The meta-analysis provided evidence of two susceptibility loci residing on chromosomes 13q and 22q. In a similar effort, Segurado et al. (2003) conducted a meta-analysis that included up to 18 independent linkage studies that used the genome scan meta-analysis (GSMA) method (Wise et al. 1999). In contrast to the meta-analysis by Badner and Gershon (2002), Segurado et al. (2003) reported no significant chromosomal regions linked to BP across the genome.
An advantage of the MSP and GSMA approaches is that they require only linkage statistics and/or P values from each study. However, many of the study-specific issues discussed above are also likely to affect these meta-analyses. One method to circumvent many of the issues encountered with the meta-analysis of linkage studies is through the use of the original genotype data rather than summary statistics (Levinson et al. 2003). Here, we explore whether combining the original genotype data provides benefits of increased power and control over sources of heterogeneity that outweigh the difficulty and potential pitfalls of the implementation. We report that, in this case, combining the original genotype data was clearly worth the effort, since we were able to demonstrate significant linkage to loci on chromosomes 6q and 8q while controlling for potential sources of heterogeneity.
Material and Methods
Source and Structure of the Original Data Sets
Our analysis included original genotype data from 11 independent linkage studies. Studies excluded from the present meta-analysis include genome scans that had (1) a unique population origin (i.e., founder populations) or (2) a unique ascertainment scheme (i.e., lithium-dependent BP); (3) studies containing largely multiplex complex pedigrees (these tended to be smaller studies with <20 pedigrees); or (4) no original data available. Table 1 summarizes the populations represented, as well as the numbers of families and individuals per data set. In sum, the 11 data sets include 1,067 families of 5,179 individuals from North America, Italy, Germany, Portugal, the United Kingdom, Ireland, and Israel. In addition, table 1 provides the number of markers that were genotyped from each study, as well as the number of markers that were used for the analysis (see the “Genotyping, Genetic Markers, and Map” section). Collection of blood and family history information for each study was done with informed consent and approval of the respective institutional review boards (or equivalent). We used raw genotype data that corresponded to the initial genomewide scans from each data set (avoiding fine-mapping data, etc.), with the exception of the NIMH Wave 3, in which case we used the data available via the Web repository (NIMH Human Genetics Initiative Web Site).
Table 1.
Overview of Data Sets
|
No. of Subjects with BPb |
|||||||||||||||||||
|
No. of Genetic Markersa |
Diagnostic and Ascertainment Criteria |
Narrow |
Broad |
||||||||||||||||
| Data Set | Reference | Population | Race | No. of Pedigrees | No. ofGenotyped Individuals | Genotyped | Mapped | DiagnosticCriterion(a) | Diagnosisof Proband | Schemec | Diagnosis ofAffected Relatived | Total | ARP | FS | IF | Total | ARP | FS | IF |
| Bonn | Cichon et al. 2001 | Germany, Israel, Italy | White | 75 | 387 | 389 | 386 | DSM-IIIR | BPI | 1SB | BPI, SAB, BPNOS, RUD | 103 | 62 | 45 | 27 | 124 | 98 | 69 | 36 |
| Columbia | Liu et al. 2003 | Israel, U.S. | White | 40 | 358 | 334 | 333 | RDC | BPI, SAB | 2FD+ | BPI, SAB | 115 | 97 | 38 | 31 | 200 | 309 | 92 | 35 |
| Johns Hopkins 1 | McInnis et al. 2003b | U.S. | Mixed | 63 | 562 | 823 | 802 | RDC | BPI | 2FD+ | BPI, BPII, SAB, RUD | 125 | 73 | 65 | 32 | 216 | 243 | 169 | 55 |
| Johns Hopkins 2 | Fallin et al. 2004 | U.S. | White | 40 | 175 | 381 | 380 | DSM-IV | BPI | 1FD+ | BPI, BPII | 73 | 33 | 19 | 22 | 95 | 53 | 38 | 40 |
| NIMH Wave 1 | NIMH Genetics Initiative Bipolar Group 1997 | U.S. | Mixed | 95 | 525 | 357 | 351 | DSM-IIIR, RDC | BPI | 1FD | BPI, SAB | 226 | 164 | 94 | 64 | 298 | 294 | 164 | 81 |
| NIMH Wave 2 | Dick et al. 2002; McInnis et al. 2003a; Willour et al. 2003; Zandi et al. 2003 | U.S. | Mixed | 55 | 348 | 465 | 458 | DSM-IIIR, RDC | BPI | 1FD | BPI, SAB | 135 | 85 | 54 | 39 | 174 | 170 | 110 | 46 |
| NIMH Wave 3 | Dick et al. 2003; NIMH Human Genetics Initiative Web Site | U.S. | Mixed | 220 | 982 | 372 | 372 | DSM-IV | BPI | 1SB | BPI, SAB | 489 | 310 | 254 | 189 | 534 | 370 | 288 | 193 |
| NIMH Wave 4 | NIMH Human Genetics Initiative Web Site | U.S. | Mixed | 274 | 1,053 | 384 | 384 | DSM-IV | BPI | 1SB | BPI, SAB | 620 | 416 | 338 | 234 | 665 | 482 | 380 | 243 |
| Portuguese | Pato et al. 2004 | Portugal | White | 16 | 102 | 346 | 342 | DSM-IV | BPI | 1FD | BPI, BPII, SAB | 43 | 35 | 27 | 15 | 44 | 37 | 27 | 15 |
| UCSD | Kelsoe et al. 2001 | Canada, U.S. | White | 20 | 163 | 331 | 324 | DSM-IIIR | BPI, BPII | 2FD+ | BPI, BPII, SAB, RUD | 27 | 11 | 7 | 7 | 41 | 21 | 15 | 12 |
| Wellcome Trust | Bennett et al. 2002 | Great Britain, Ireland | White | 151 |
509 |
380 |
378 |
DSM-IV | BPI | 1SB | BPI, BPII, BPNOS, RUD | 288 |
159 |
150 |
109 |
313 |
195 |
186 |
122 |
| Total | 1,067 | 5,179 | 4,562 | 4,510 | 2,251 | 1,445 | 1,091 | 769 | 2,423 | 2,272 | 1,538 | 878 | |||||||
Genotyped=number of markers genotyped (autosomal only). Mapped=number of markers mapped to the common marker map (autosomal only).
ARP=all possible ARPs; FS=all possible full-sibling pairs ; IF=families informative for nonparametric allele-sharing linkage analysis.
Ascertainment scheme: 1SB=at least one affected sibling; 1FD=at least one affected first-degree relative; 1FD+=at least one affected first- or greater-degree relative; 2FD+=at least two affected first- or greater-degree relatives.
Diagnosis of the relative(s) of proband (satisfying ascertainment scheme). BPNOS=bipolar disorder–not otherwise specified.
Family Ascertainment and Assessment
Detailed descriptions of family ascertainment, clinical assessment, and diagnostic criteria are available in the respective primary references for each data set (table 1). With the exception of the Columbia and University of California at San Diego (UCSD) data sets, all studies included a proband with a Diagnostic and Statistical Manual of Mental Disorders, 3rd edition (DSM-IIIR), Research Diagnostic Criteria (RDC), or DSM-IV diagnosis of BPI but differed with respect to the number, type of relationship, and diagnosis of additional family members used for ascertainment conditions. A brief overview of pertinent information is provided in table 1.
Genotyping, Genetic Markers, and Map
Detailed information about specimen collection, DNA extraction, and genotyping methods for each data set can be found in the primary references for each data set (table 1). In all 11 data sets, variable-repeat microsatellite genetic markers were genotyped for family members with available DNA for each of the 22 autosomes. Genotypes from the X chromosome were not available from all data sets; therefore, they were excluded from the analysis. The number of markers genotyped for each data set was relatively consistent (∼350–400), with the noted exception of the Johns Hopkins 1 data set (n=823).
A unique feature available to meta-analyses of linkage studies that use original genotype data is the ability to construct a standardized genetic map. We mapped respective markers from each data set to one common sex-averaged map, using the Rutgers Combined Linkage–Physical Map of the Human Genome (Kong et al. 2004; Rutgers Combined Linkage–Physical Map of the Human Genome Web Site) as the backbone. If the genetic location (in cM) of a particular marker was found on the Rutgers map, then that location was used. Otherwise, if the genetic location from the Rutgers map was not available for a particular marker, we first determined the physical location (in bp) of that marker, using the National Center for Biotechnology Information (NCBI) Build 35.1 (University of California–Santa Cruz [UCSC] Genome Bioinformatics). We then identified the physical locations of two flanking markers that were found on the Rutgers map. The resultant genetic location of the unknown marker location was interpolated under the assumption that the ratio of the distances between markers on the physical map was the same as the ratio of the distances on the genetic map. If we could not identify a marker either on the Rutgers map or via the NCBI, that marker was discarded. We were able to map the vast majority of markers from each data set to the standardized, common map (table 1).
Pooling Procedure
We combined the raw genotype data from the 11 studies into one large pooled data set. To accommodate the variation in allele coding across different studies, we created unique marker names for each data set, such that no two data sets shared an identical marker name (even if the same marker had been genotyped across more than one study). We then pooled the raw genotypes from each data set, generating missing genotypes for each occurrence of a marker from one of the other data sets. Pooling linkage samples in this way enabled us to estimate allele frequencies within study, which minimized any bias introduced through estimation of allele frequencies across the entire, potentially heterogeneous, pooled sample.
Error Detection
We used a variety of programs to check for Mendelian errors, including family-based association test (FBAT) (Laird et al. 2000) and PEDSTATS (Abecasis et al. 2002). Any incompatible genotypes were set to “unknown” for the entire family for that locus. In addition, we used the software package MERLIN (Abecasis et al. 2002) for its error-detection feature to identify unlikely genotypes (P<.05 from the r-statistic). Any unlikely genotypes detected by MERLIN were set to “unknown.” Relationship errors were assessed using graphical relationship representation (Abecasis et al. 2001). Individuals with genotype data inconsistent with their pedigree relationships, including MZ twins, were excluded from the analysis. Finally, individuals with no genotype or phenotype information who were not needed to define relationships between other individuals in a pedigree were removed using MERLIN’s trim option.
Affection-Status Models
Two hierarchical definitions for BP affection status were used: (1) a narrow model that included individuals who received a diagnosis of BPI only and (2) a broad model that included individuals who received a diagnosis of either BPI or BPII. Relatives who received a diagnosis of disorders other than BPI or BPII were coded as “unknown.” The use of multiple affection models is common practice in linkage analysis of BP, primarily because diagnostic boundaries are ill defined and the underlying genetic model for BP is unknown. Table 1 provides an overview of the diagnostic criteria used for each data set, as well as the number of affected relative pairs (ARPs) that have complete phenotype and genotype information for both the narrow and broad BP definitions; we have the original genotype data for 1,445 ARPs for the narrow phenotype definition and 2,272 ARPs for the broad phenotype definition. Note that families ascertained through non-BPI–affected probands but that met criteria for the BP broad phenotype model were included in the analysis (i.e., BPII-affected probands from UCSD).
ARP LOD-Score Methods
The analytic objective of the project was to conduct nonparametric multipoint linkage analysis with use of the pooled sample for each of the two phenotype definitions. Marker-allele frequencies were estimated within study by use of founder genotypes. If there were no available founder genotypes for a particular family, a random family member with genotype information was used. Use of the default allele frequency calculation in MERLIN (all genotyped individuals) did not alter the results (data not shown). We used MERLIN’s implementation of the Whittemore and Halpern (1994) algorithm to test for allele sharing among all affected individuals, and we generated the nonparametric LOD score via the Kong and Cox (1997) linear model. We also estimated the linkage information at each analysis position, using MERLIN’s measure of entropy. At 1-cM intervals, across all 22 autosomal chromosomes, we analyzed each of the 11 data sets individually, as well as the pooled data set comprising the 11 data sets combined.
Genomewide Significance Thresholds
We followed the guidelines of Lander and Kruglyak (1995) for genomewide significant and suggestive linkage thresholds. We estimated critical values for the LOD score from the pooled analysis, using the method described by Bacanu (2005); to compute genomewide significant and suggestive critical values, this method estimates the correlation between Gaussian statistics at adjacent map points. The threshold for genomewide significance was established at a LOD score of 3.03. Genomewide suggestive thresholds were established at a LOD score of 1.75. The thresholds were Bonferroni adjusted to account for the fact that two phenotypes were analyzed. In our study, the correlation between narrow and broad BP linkage statistics was 0.85. Our simulations (data not shown) suggest that, for this magnitude of correlation, Bonferroni-adjusted thresholds are very close to their empirical counterparts. Consequently, for the present analysis, we report only the Bonferroni adjustment for multiple testing. For comparison, we also calculated simulated thresholds (1,000 genomewide replicates) and confirmed that these were similar to the critical values obtained by the Bacanu (2005) method.
Assessment of Heterogeneity
The identical-by-descent (IBD) sharing probabilities of each affected sibling pair (ASP) generated in MERLIN was used to calculate the maximum-likelihood estimates of sharing 0, 1, and 2 alleles IBD via the expectation-maximization algorithm. All possible ASPs for each family were used and weighted equally. The bootstrap variance estimation procedure (described below) automatically adjusts for correlation between sib pairs among families. Using a custom-written program, we estimated the IBD proportions (treating each sibling pair as independent) from MERLIN’s IBD output by allowing the maximum-likelihood IBD estimates to converge without triangle constraints (Holmans 1993). We chose to use the unconstrained estimates, since imposing the triangle constraints in the presence of heterogeneity may be less powerful (Dizier et al. 2000). In addition, combination of estimates over studies is more meaningful with the unconstrained estimates. Note that our estimation procedure is identical to that of GENEHUNTER (Kruglyak et al. 1996), with the exception of the triangle-constraint restriction. Following a more traditional meta-analytic approach, we used estimates of mean IBD (IBDm) sharing and its variance to quantify heterogeneity among the 11 data sets at selected regions along the genome. Using the unconstrained estimates of sharing 0, 1, or 2 alleles IBD (ibd0, ibd1, ibd2) for ASPs, we calculated the IBDm (0.5×ibd1+ibd2) separately for each study. Note that use of the unconstrained probabilities allows the estimated IBDm for a study to be <0.5, whereas use of the constraints forces IBDm to be at least 0.5. To derive the variance of the IBDm, we used a bootstrap procedure developed for estimating the variance of IBDm sharing among ASPs in linkage analysis samples (M.B.M. and N.M.L., unpublished data). The Q-statistic was utilized to provide a formal test of heterogeneity (Laird and Mosteller 1990). Then, continuing along the traditional meta-analytic path, we pooled the study-specific estimates of IBDm, using a random-effects model (Laird and Mosteller 1990) that allows for the incorporation of between-study heterogeneity and therefore a more realistic summary measure of IBDm, as well as more accurate CIs. Gu et al. (1998) provide a detailed description of this general approach in the context of ASP linkage samples.
Results
LOD-Score Results
Overall, the average information content across all chromosomes of the pooled analysis was ∼0.58 (range 0.49–0.63), which is consistent with genomewide scans that used ∼10-cM marker density. Only one individual genome scan yielded a LOD score >3; eight other LOD scores >2 were found in individual scans. Some of these will be discussed later, in the context of the pooled results; additional results are available on request. The number of families informative for narrow and broad BP per data set is listed in table 1.
Table 2 displays the results for the pooled analysis. The largest Kong and Cox (1997) LOD score observed was with use of the narrow phenotype, which achieved genomewide significance (LOD>3.03) on chromosome 6, at 115 cM (LOD 4.19). With the use of this same phenotype definition, no other chromosome achieved genomewide significance, although we observed genomewide suggestive linkage (LOD>1.75) signals on chromosomes 9 (LOD 2.04), 8 (LOD 1.99), and 20 (LOD 1.91). Table 2 also shows the corresponding results from analysis of the broad phenotype. The LOD score found on chromosome 8 at 151 cM (LOD 3.40) obtained using this phenotype definition exceeds the genomewide significance threshold. Chromosome 9 revealed genomewide suggestive thresholds (LOD 2.04) in the location identical or near to that found with the narrow BP phenotype analysis. Note that the LOD score for chromosome 6 (LOD 1.74) was substantially lower for broad BP compared with narrow BP. In summary, two regions on chromosomes 6 (narrow BP) and 8 (broad BP) achieved genomewide significance. These two regions were selected for more-detailed analysis and presentation.
Table 2.
Results from the Pooled Analysis
| Narrow BP |
Broad BP |
|||||
| Chromosome | GeneticLocationa(cM) | PhysicalLocationb(Mb) | LOD | GeneticLocationa (cM) | PhysicalLocationb(Mb) | LOD |
| 1 | 200 | 185.0 | .41 | 79 | 44.9 | .59 |
| 2 | 92 | 68.0 | .97 | 92 | 68.0 | 1.10 |
| 3 | 1 | .6 | .19 | 69 | 44.5 | .14 |
| 4 | 152 | 154.0 | .39 | 154 | 154.5 | .56 |
| 5 | 79 | 67.0 | .31 | 78 | 66.0 | .11 |
| 6 | 115 | 108.5 | 4.19c | 115 | 108.5 | 1.74 |
| 7 | 187 | 157.1 | .57 | 187 | 157.1 | .70 |
| 8 | 152 | 135.4 | 1.99d | 151 | 134.5 | 3.40c |
| 9 | 46 | 24.5 | 2.04d | 48 | 25.6 | 2.06d |
| 10 | 85 | 70.2 | .07 | 50 | 25.8 | .20 |
| 11 | 72 | 60.0 | .54 | 72 | 60.0 | .57 |
| 12 | 155 | 126.5 | .40 | 155 | 126.5 | .13 |
| 13 | 44 | 42.4 | .62 | 50 | 46.4 | .46 |
| 14 | 79 | 86.5 | .54 | 79 | 86.5 | .19 |
| 15 | 21 | 29.4 | .95 | 25 | 31.2 | .73 |
| 16 | 30 | 12.1 | .18 | 35 | 13.4 | .85 |
| 17 | 98 | 64.3 | 1.36 | 98 | 64.3 | .91 |
| 18 | 70 | 44.9 | 1.47 | 87 | 58.5 | 1.05 |
| 19 | 73 | 51.5 | .33 | 37 | 14.6 | .13 |
| 20 | 12 | 4.2 | 1.91d | 12 | 4.2 | 1.71 |
| 21 | 60 | 43.0 | .06 | 48 | 39.2 | .03 |
| 22 | 2 | 15.0 | .12 | 9 | 16.0 | .03 |
Genetic location from the unified marker map.
Physical location (approximate) from the Rutgers Combined Linkage-Physical Map.
Genomewide significant (LOD>3.03).
Genomewide suggestive (LOD>1.75).
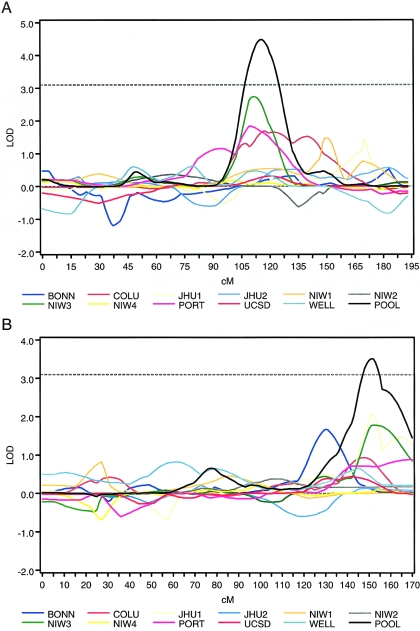
Figures 1A and 1B display the LOD scores from chromosomes 6 (narrow BP) and 8 (broad BP), respectively. In particular, we plotted the LOD score from the pooled data set superimposed on the plot of the LOD scores from each of the 11 component data sets (figs. 1A and 1B). For chromosome 6, the NIMH Wave 3 data set is the only data set that achieves a LOD score of >2.0, despite the fact that the pooled signal surpassed a LOD score of 4.0 (fig. 1A). There were three signals in this region that appear to achieve the highest LOD scores under the pooled signal (NIMH Wave 3, Portuguese, and Columbia). For chromosome 8, we found that no individual data set reached a LOD score of 2.0, despite the fact that the pooled signal exceeded 3.0 (fig. 1B). There were also clear overlapping signals from component data sets (e.g., Johns Hopkins 1 and NIMH Wave 3). Removal of the nonwhite families (and re-estimation of allele frequencies) from the NIMH samples (n=38) did not impact the pooled linkage signal for either chromosome 6 or 8.
Figure 1.
Relative contribution of component data sets to the pooled linkage signals. The LOD scores from the pooled analysis (solid black line) are overlaid with the LOD scores from the data set–specific analysis (the horizontal dotted line indicates the genomewide significance threshold). A, Narrow BP phenotype, chromosome 6. B, Broad BP phenotype, chromosome 8. BONN=Bonn; COLU=Columbia; JHU1=Johns Hopkins 1; JHU2=Johns Hopkins 2; NIW1=NIMH Wave 1; NIW2=NIMH Wave 2; NIW3=NIMH Wave 3; NIW4=NIMH Wave 4; PORT=Portuguese; UCSD=UCSD; WELL=Wellcome Trust; POOL=pooled sample.
Assessment of Heterogeneity Results
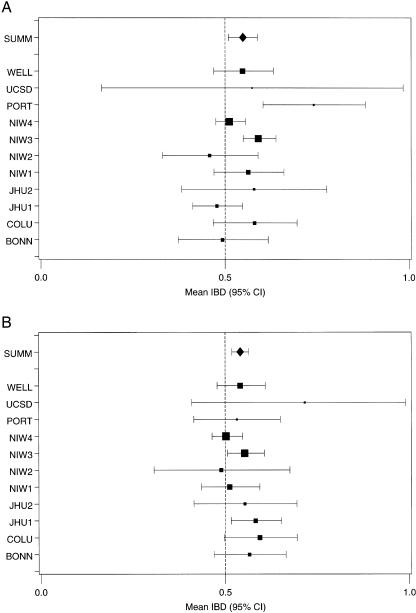
In the region that encompasses the significant linkage signals on chromosome 6 and 8, there was no evidence of heterogeneity among data sets as tested via the Q-statistic (P>.10; results not shown). Using the location from each chromosome that reveals the peak LOD score, we provide a forest plot of the IBDm and 95% CIs for each of the component data sets, as well as the summary IBDm estimate and 95% CI (figs. 2A and 2B). We selected the 115-cM position for chromosome 6 (fig. 2A). Here, we see that only two data sets (NIMH Wave 3 and Portuguese) display significant excess allele sharing, whereas all others have 95% CIs that include 0.50. As seen in figure 2A, the combined estimate of IBDm, which is the random-effects summary estimate that uses each component data set's IBDm estimate and variance, suggests an excess in allele sharing at the position for the studies combined (IBDm=0.55; 95% CI 0.51–0.59). Figure 2B displays chromosome 8 at the 151-cM position. Here, two data sets display significant excess sharing (NIMH Wave 3 and Johns Hopkins 1) and another (Columbia) displays borderline excess sharing. The summary estimate of IBDm also demonstrates excess sharing at this position, despite nine of the component data sets’ 95% CIs including 0.50 (IBDm=0.54; 95% CI 0.52–0.56). Note that the summary IBDm CIs are not adjusted for genomewide comparison.
Figure 2.
IBDm and 95% CIs for the component data sets, as well as the random-effects summary point estimate. A, Narrow BP phenotype, chromosome 6 at 115 cM. B, Broad BP phenotype, chromosome 8 at 151 cM. BONN=Bonn; COLU=Columbia; JHU1=Johns Hopkins 1; JHU2=Johns Hopkins 2; NIW1=NIMH Wave 1; NIW2=NIMH Wave 2; NIW3=NIMH Wave 3; NIW4=NIMH Wave 4; PORT=Portuguese; UCSD=UCSD; WELL=Wellcome Trust; SUMM=random-effects summary IBDm.
Although global tests for heterogeneity are often underpowered, the IBDm procedure was able to detect statistically significant among-study heterogeneity at regions throughout the genome, including regions outside the linkage signals on chromosomes 6 and 8. In particular, the 6pter region and the 35–45-cM region on chromosome 6 have substantial heterogeneity (Q-statistic P<.001) as does the 40–50-cM region of chromosome 8 (Q-statistic P<.001).
There is mounting evidence that unselected sibling pairs tend to share more than half of their alleles IBD (Elston et al. 2005). To provide reassurance that the observed allele sharing among ASPs in the present study is attributable to affection status, we evaluated discordant sibling pairs as well. Estimation of IBDm in unaffected siblings (excluding all psychiatric diagnoses, if available) paired with affected siblings resulted in either null sharing or slightly less than null sharing (not significant) at the locations of the reported linkage signals for each of the data sets that provided adequate diagnostic information (data not shown).
Discussion
To our knowledge, this is the largest and most comprehensive collaboration involving the original genotype data for BP linkage analysis. We identified loci on chromosomes 6q and 8q as meeting genomewide significance and loci on 9p and 20p as meeting suggestive linkage. Our results show that the extra expense and effort required to obtain original genotype data from genome scans has substantial benefits. Our observations from pooling data for the present analysis are consistent with an increase in power, since linkage signals from the pooled sample were generally higher than the linkage signal from any one component data set. Utilizing the more traditional meta-analytic approach, we constructed a summary measure of IBDm to assess the among-study heterogeneity.
Heterogeneity
In general, replication of linkage signals from complex diseases such as BP is likely to be complicated by known sources of heterogeneity among linkage studies. In particular, sources of heterogeneity include different populations under investigation, varied number of affected individuals (with varying diagnoses) per family, varied family size, different ascertainment criteria, varied instruments of assessment and diagnostic criteria, different sets of marker genotypes and corresponding marker maps, as well as dissimilar analytic methods. We restricted our analyses to uniform disease classifications and the use of common methods, including a standardized common marker map. Both within- and between-study heterogeneity likely remains, since subjects were ascertained, were assessed, and received a diagnosis with considerable variation. The presence of genetic heterogeneity may also limit the utility of pooling original data geared toward the identification of candidate loci. Despite these issues, the regions of significant or suggestive linkage lacked evidence of substantial heterogeneity. However, tests of heterogeneity are often underpowered, and acceptance does not preclude heterogeneity in these regions. On the other hand, substantial heterogeneity would likely limit the extent to which signals could be detected; such regions may be worthy of further study.
Phenotype Definition
Our results provide some support for the view that genetic influences on BPI and BPII may be distinguishable. Our most significant result was seen for narrow BP (BPI-only phenotype) on chromosome 6q. When the analysis was expanded to include BPII, the linkage signal on chromosome 6q was attenuated, despite the increase in the number of ARPs. In contrast, removal of the individuals with BPII from the analysis reduced the evidence of linkage on chromosome 8q. It has been argued that the diagnostic reliability of BPII may be less robust than that of BPI, although this may not be true when careful diagnostic procedures are applied (Simpson et al. 2002). Genetic epidemiologic data suggest that BPII is probably a genetically heterogeneous entity in which some cases are genetically distinct from BPI, whereas others are part of a spectrum that includes BPI and/or unipolar depression (Smoller and Finn 2003).
It could be argued from our results that a locus on chromosome 6q is linked specifically to BPI, so that the addition of subjects with BPII resulted in “phenotypic noise,” effectively pulling the estimates of allele sharing toward the null. On the other hand, inclusion of subjects with BPII enhanced the linkage signal on chromosome 8q, suggesting that a locus in this region may influence a broader bipolar spectrum phenotype. It should be noted that, because we did not analyze a BPII-only phenotype, we cannot address the question of whether the 8q locus or loci elsewhere in the genome have more-specific effects on BPII itself.
Prior Meta-Analyses in Context
Table 3 provides an overview of the variations of different data sets included in each of the two prior meta-analyses as well as the present combined analysis. Overall, the MSP meta-analysis included 8 data sets (Badner and Gershon 2002), the GSMA (Segurado et al. 2003) included 18 data sets, and the present combined analysis included 11 data sets (table 3). Only four data sets (Bonn, NIMH Wave 1, UCSD, and Wellcome Trust) were common across all three meta-analyses. There were additional data sets that were common to one or more analyses; however, different variations of the data were often included. For example, the Johns Hopkins 1 data set was included in all three analyses; however, only the GSMA and the present combined analysis used the full sample (65 pedigrees). Much of the variability in the data sets—as well as data-set versions used for each meta-analysis—is largely explained through what was available at the time of each of the meta-analyses. The Johns Hopkins 2; NIMH Waves 2, 3, and 4; and Portuguese data sets became available after the MSP and GSMA analyses. Therefore, as is true in any meta-analysis context, updating analyses to include new information as it becomes available is essential to the confirmation and identification of genomic regions that may harbor disease-susceptibility loci.
Table 3.
Data Sets Included in the Three Recent Meta-Analyses of BP Linkage Scans
|
Meta-Analysisa |
||||
| Study | Comment | MSP | GSMA | Combined |
| Antwerp 1 (Segurado et al. 2003) | Unpublished data | Antwerp 1 | ||
| Antwerp 2 (Segurado et al. 2003) | Unpublished data | Antwerp 2 | ||
| Bonn (Cichon et al. 2001) | Cichon | Bonn | Bonn | |
| Columbia (Liu et al. 2003) | Columbia | Columbia | ||
| Costa Rica (McInnes et al. 1996) | Two-point analysis | Costa Rica | ||
| Costa Rica (Garner et al. 2001) | Markov chain Monte Carlo analysis | Garner | ||
| Edinburgh (Blackwood et al. 1996) | 1 pedigree | Blackwood | ||
| Edinburgh (Blackwood et al. 1996; Segurado et al. 2003) | 7 pedigrees (includes unpublished data) | Edinburgh | ||
| Finland (Ekholm et al. 2003) | Finland | |||
| Johns Hopkins 1 (Friddle et al. 2000) | 50 pedigrees (initial sample) | Friddle | ||
| Johns Hopkins 1 (McInnis et al. 2003b) | 65 pedigrees (expanded sample) | Hopkins/Dana | Johns Hopkins 1 | |
| Johns Hopkins 2 (Fallin et al. 2004) | Ashkenazi population | Johns Hopkins 2 | ||
| NIMH Intramural (Detera-Wadleigh et al. 1999) | Includes 1 Old Order Amish pedigree | Detera-Wadleigh | NIMH-IM | |
| NIMH Wave 1 (NIMH Genetics Initiative Bipolar Group 1997) | NIMH | NIMH | NIMH Wave 1 | |
| NIMH Wave 2 (Dick et al. 2002; McInnis et al. 2003a; Willour et al. 2003; Zandi et al. 2003) | NIMH Wave 2 | |||
| NIMH Wave 3 (Dick et al. 2003; NIMH Human Genetics Initiative Web site) | NIMH Wave 3 | |||
| NIMH Wave 4 (NIMH Human Genetics Initiative Web site) | NIMH Wave 4 | |||
| Ottawa (Turecki et al. 2001) | Ottawa | |||
| Portugal (Pato et al. 2004) | 16 pedigrees (microsatellite scan) | Portuguese | ||
| Quebec (Morissette et al. 1999) | Morissette | Quebec | ||
| Sydney (Badenhop et al. 2002) | 13 pedigrees | Sydney 1 | ||
| Sydney (Badenhop et al. 2002; Segurado et al. 2003) | 15 pedigrees (includes unpublished data) | Sydney 2 | ||
| Turkey (Radhakrishna et al. 2001) | Radhakrishna | |||
| University College (UC) London (Curtis et al. 2003) | UC London | |||
| UCSD (Kelsoe et al. 2001) | Kelsoe | UCSD | UCSD | |
| Utah (Coon et al. 1993) | Coon | Utah | ||
| Wellcome Trust (Bennett et al. 2002) | Bennett | U.K./Irish | Wellcome Trust | |
When a study was included in a meta-analysis, the variable entry for that meta-analysis is listed as it was referenced in each meta-analysis.
Chromosomes 6 and 8
The strongest support derived from this analysis under the narrow BP phenotype definition is for chromosome 6q and under the broad BP phenotype definition, chromosome 8q. Because of practical considerations, an exhaustive review of the BP linkage analysis literature will not be the focus of this discussion (for more detailed review, see Baron [2002]). Rather, a brief discussion of each region follows.
Linkage signals of varying degrees on chromosome 6 have been reported from data sets used in the present analysis as well as other data sets not included here. Figure 1A provides a graphical representation of the relative contribution from each of the component data sets to the signal on 6q. The NIMH Wave 3, Portuguese, and Columbia data sets appear to be the most influential. From the estimated IBDm, all studies in this analysis—with the exception of Bonn, Johns Hopkins 1, and NIMH Wave 2—showed excess allele sharing in ASPs at the pooled linkage peak (fig. 2A). The NIMH Human Genetics Initiative also noted linkage at this region (at or near marker D6S1021) through a linkage analysis incorporating an interaction effect with 6p (Schulze et al. 2004). The study of the Portuguese sample reported a linkage signal to chromosome 6q22 in a follow-up marker scan with use of high-density genotyping (Middleton et al. 2004). In addition, the Wellcome Trust study reported a signal on 6q through a follow-up analysis that included larger sample size and additional marker genotypes (Lambert et al. 2005). Further, in a Danish study that was not a part of the present meta-analysis, Ewald et al. (2002) reported a maximum LOD score of 3.8 for the marker (D6S1021) closest to the maximum LOD score in our pooled sample analysis. Another study that used families from northern Sweden also reported a signal from parametric linkage analysis of chromosome 6q; however, that signal appears nearer to the smaller pooled linkage signal observed in the present meta-analysis (Venken et al. 2005). Moreover, although not statistically significant, Segurado et al. (2003) reported bin 6.4 as meeting a PAveRnk of <.10 for their model 1 (BPI or schizoaffective disorder-bipolar type [SAB]). Badner and Gershon (2002) did not report linkage to BP on 6q.
The pooled signal from chromosome 8 appears to be driven by the Bonn, Johns Hopkins 1, and NIMH Wave 3 data (fig. 1B). All studies in our analysis, aside from NIMH Waves 2 and 4, show excess allele sharing among ASPs at the linkage peak (fig. 2B). In addition, a genomewide scan of psychosis in the Columbia data set provided evidence of linkage to 8q24 (Park et al. 2004). Aside from previous reports from data sets included in this collaboration, we are unaware of any linkage signals that have been reported for 8q from studies outside the collaboration. Segurado et al. (2003) reported that bin 8.6 failed to reach genomewide significance; however, they report that 8q might contain a locus that is “weakly linked” to BP (PAveRnk <.05) for their model 2 (BPI, BPII, and SAB). Badner and Gershon (2002) did not report linkage to BP on 8q.
Conclusions
The present analysis is the first comprehensive collaboration of BP linkage studies to use original genotype data that is aimed at identifying potential candidate regions for future investigation. Using the pooled data, we demonstrated genomewide significant linkage to chromosomes 6q and 8q and genomewide suggestive linkage to chromosomes 9p and 20p. The finding on chromosome 6q, which is based on the BPI phenotype, is supported by some of the individual studies analyzed herein, as well as by other studies that we could not include in our analysis. When subjects with BPII are included in the analysis, the results for chromosome 6q are diminished, and a locus on 8q becomes significant. Evidence for the 8q locus appears limited to studies included in this analysis. Effect size estimates from the ASP analysis suggest relative risks (λs) of 1.34 and 1.26 attributable to loci on chromosomes 6q and 8q, respectively. Thus, the majority of the genetic constituents underlying BP remain unidentified, which suggests that their discovery may require more-elaborate and -focused investigations of bipolar-spectrum phenotypes, as well as more-refined methods for gene mapping.
We have demonstrated that combining data from disparate genome scans provides an effective mechanism for summarizing quantitative information and that lack of consistent findings in individual scans does not preclude finding a significant region in the combined data. There are particular strengths for use of the original data, in that it allows control of many potential sources of variability in the original studies. Fortunately, original genome-scan data are becoming increasingly available, such as those from the NIMH Human Genetics Initiative, and our results suggest these efforts should be supported. Our approach offers a powerful methodology for the identification of linkage regions underlying complex diseases, such as BP, for which there are likely multiple disease-susceptibility loci.
Acknowledgments
The principal investigators of the Genetic Determinants of Bipolar Disorder Project (B.D., S.V.F., N.M.L., V.L.N., P.S., and J.W.S) and M.B.M. were supported by NIMH grants R01-MH063445, MH63420, and MH0667288; M.B.M. was additionally supported by NIMH grant T32-MH017119. Collection of data and biomaterials from the Bonn study was supported by the Deutsche Forschungsgemeinschaft. Data and biomaterials from the four NIMH studies were collected in four projects that participated in the NIMH Bipolar Disorder Genetics Initiative. In 1991–1998, the principal investigators and coinvestigators were: J.I.N., Marvin J. Miller, H.J.E., T.F., and Elizabeth S. Bowman, grant U01 MH46282 to Indiana University, Indianapolis; Theodore Reich, Allison Goate, and J.P.R., grant U01 MH46280 to Washington University, St. Louis; J.R.D., Sylvia Simpson, and Colin Stine, grant U01 MH46274 to Johns Hopkins University, Baltimore; Elliot Gershon, Diane Kazuba, and Elizabeth Maxwell, NIMH Intramural Research Program, Clinical Neurogenetics Branch, Bethesda. Data and biomaterials were collected as part of 10 projects that participated in the NIMH Bipolar Disorder Genetics Initiative. In 1999–2003, the principal investigators and coinvestigators were: J.I.N., Marvin J. Miller, Elizabeth S. Bowman, N. Leela Rau, P. Ryan Moe, Nalini Samavedy, Rif El-Mallakh, (at University of Louisville, Louisville), Husseini Manji and Debra A. Glitz (at Wayne State University, Detroit), Eric T. Meyer, Carrie Smiley, T.F., Leah Flury, Danielle M. Dick, H.J.E., grant R01 MH59545 to Indiana University, Indianapolis; J.P.R., Theodore Reich, Allison Goate, Laura Bierut, grant R01 MH059534 to Washington University, St. Louis; M.G.M., J.R.D., Dean F. MacKinnon, Francis M. Mondimore, J.B.P., P.P.Z., Dimitrios Avramopoulos, and Jennifer Payne, grant R01 MH59533 to Johns Hopkins University, Baltimore; W. Berrettini, grant R01 MH59553 to University of Pennsylvania, Philadelphia; W. Byerley and Mark Vawter, grant R01 MH60068 to University of California at Irvine, Irvine; W.C. and Raymond Crowe, grant R01 MH059548 to University of Iowa, Iowa City; Elliot Gershon, Judith Badner, F.J.M., Chunyu Liu, Alan Sanders, Maria Caserta, Steven Dinwiddie, Tu Nguyen, and Donna Harakal, grant R01 MH59535 to University of Chicago, Chicago; J.R.K. and Rebecca McKinney, grant R01 MH59567 to UCSD, San Diego; W.S., Howard M. Kravitz, Diana Marta, Annette Vaughn-Brown, and Laurie Bederow, grant R01 MH059556 to Rush University, Chicago; F.J.M., Layla Kassem, Sevilla Detera-Wadleigh, Lisa Austin, D.L.M., grant 1Z01MH002810-01 to NIMH Intramural Research Program, Bethesda. Genotyping services were provided in part by the Center for Inherited Disease Research (CIDR). CIDR is fully funded through federal contract N01-HG-65403 from the National Institutes of Health to Johns Hopkins University. Data and biomaterials for the Columbia data set were collected and supported by NIMH grant R01 MH59602 (to M.B.) and by funds from the Columbia Genome Center and the New York State Office of Mental Health. The main contributors to the Columbia project were M.B. (principal investigator), Jean Endicott (coprincipal investigator), Jo Ellen Loth, John Nee, Richard Blumenthal, Lawrence Sharpe, Barbara Lilliston, Melissa Smith, and Kristine Trautman, all from Columbia University Department of Psychiatry, New York. A small subset of the sample was collected in Israel in collaboration with B.L. and Kyra Kanyas, from the Hadassah–Hebrew University Medical Center, Jerusalem. We are grateful to the patients and their family members, for their cooperation and support, and to the treatment facilities and other organizations that collaborated with us in identifying families.
Author affilations.—Department of Epidemiology (M.B.M. and D.B.) and Department of Biostatistics (N.M.L., C.L., and K.V.S.), Harvard School of Public Health, and Psychiatry and Neurodevelopmental Genetics Unit (P.S. and J.W.S.) and Gerontology Research Unit (D.B.), Massachusetts General Hospital, Harvard Medical School, Boston; Broad Institute of Harvard and Massachusetts Institute of Technology (P.S.) and Whitehead Institute (M.J.D.), Cambridge, MA; Department of Psychiatry (B.D. and V.L.N.), Department of Human Genetics (V.L.N.), and Western Psychiatric Institute and Clinic (W.X.), University of Pittsburgh School of Medicine, Pittsburgh; Medical Genetics Research Center and Department of Psychiatry and Behavioral Sciences (S.V.F.) and Center for Neuropsychiatric Genetics (C.N.P. and M.T.P.), State University of New York–Upstate Medical University, Syracuse; Veterans Administration Medical Center, Washington, D.C. (C.N.P. and M.T.P.); Institute of Human Genetics (R.A.J. and J.S.), Department of Genomics, Life and Brain Center (S.C. and M.M.N.), Department of Psychiatry (W.M.), and Institute of Human Genetics (P.P.), University of Bonn, Bonn; Mental State Hospital, Haar, Germany (M.A.); GlaxoSmithKline Research and Development, Research Triangle Park, NC (S.-A.B.); Department of Psychiatry (M.B.) and Department of Medical Genetics (M.B. and S.-H.J.), Columbia University, New York; Department of Psychiatry, UCSD (T.B.B. and J.R.K.), and San Diego VA Healthcare System (T.B.B.), La Jolla; Department of Psychiatry, University of Pennsylvania, Philadelphia (W. Berrettini); Department of Psychiatry, University of California at San Francisco, San Francisco (W. Byerley); Department of Psychiatry, University of Iowa, Iowa City (W.C.); Department of Psychological Medicine, Cardiff University, Cardiff (N.C., I.J., and M.H.); Department of Psychiatry and Behavioral Sciences (J.R.D., J.D.M., J.B.P., and A.E.P.) and Department of Mental Health (P.P.Z.), Bloomberg School of Public Health, and Institute of Genetic Medicine (M.G.M.), Johns Hopkins School of Medicine, Baltimore; Institute of Psychiatric Research and Department of Psychiatry (J.I.N.), Department of Biochemistry and Molecular Biology (H.J.E.), and Department of Medicine and Molecular Genetics (T.F.), Indiana University School of Medicine, Indianapolis; Department of Psychiatry (M.G., D.L., and R.S.) and Department of Genetics (D.L. and R.S.), Trinity College, Dublin; Department of Human Genetics, University of Chicago (T.C.G.), and Department of Psychiatry, Rush University (W.S.), Chicago; Division of Neuroscience, University of Birmingham, Birmingham, United Kindom (L.J.); Department of Psychiatry, Hadassah–Hebrew University Medical Center, Jerusalem (B.L.); Genome Institute of Singapore, Singapore (J.L.); Mood and Anxiety Program (F.J.M.) and Laboratory of Clinical Science (D.L.M.), NIMH, U.S. Department of Health and Human Services, Bethesda; Department of Psychiatry, Washington University, St. Louis (J.P.R.); Division of General Epidemiology in Psychiatry, Central Institute of Mental Health, Mannheim, Germany (M.R.).
Web Resources
The URLs for data presented herein are as follows:
- NIMH Human Genetics Initiative, http://zork.wustl.edu/nimh/NIMH_initiative/NIMH_initiative_link.html
- Rutgers Combined Linkage–Physical Map of the Human Genome, http://compgen.rutgers.edu/maps/
- UCSC Genome Bioinformatics, http://genome.ucsc.edu/
References
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR (2001) GRR: graphical representation of relationship errors. Bioinformatics 17:742–743 10.1093/bioinformatics/17.8.742 [DOI] [PubMed] [Google Scholar]
- ——— (2002) MERLIN—rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 30:97–101 10.1038/ng786 [DOI] [PubMed] [Google Scholar]
- APA (1994) Diagnostic and statistical manual of mental disorders, 4th ed. The American Psychiatric Association, Washington, DC [Google Scholar]
- Bacanu (2005) Robust estimation of critical values for genome scans to detect linkage. Genet Epidemiol 28:24–32 10.1002/gepi.20030 [DOI] [PubMed] [Google Scholar]
- Badenhop RF, Moses MJ, Scimone A, Mitchell PB, Ewen-White KR, Rosso A, Donald JA, Adams LJ, Schofield PR (2002) A genome screen of 13 bipolar affective disorder pedigrees provides evidence for susceptibility loci on chromosome 3 as well as chromosomes 9, 13 and 19. Mol Psychiatry 7:851–859 10.1038/sj.mp.4001114 [DOI] [PubMed] [Google Scholar]
- Badner JA, Gershon ES (2002) Meta-analysis of whole-genome linkage scans of bipolar disorder and schizophrenia. Mol Psychiatry 7:405–411 10.1038/sj.mp.4001012 [DOI] [PubMed] [Google Scholar]
- Baron M (2002) Manic-depression genes and the new millennium: poised for discovery. Mol Psychiatry 7:342–358 10.1038/sj.mp.4000998 [DOI] [PubMed] [Google Scholar]
- Bennett P, Segurado R, Jones I, Bort S, McCandless F, Lambert D, Heron J, Comerford C, Middle F, Corvin A, Pelios G, Kirov G, Larsen B, Mulcahy T, Williams N, O’Connell R, O’Mahony E, Payne A, Owen M, Holmans P, Craddock N, Gill M (2002) The Wellcome Trust UK-Irish bipolar affective disorder sibling-pair genome screen: first stage report. Mol Psychiatry 7:189–200 10.1038/sj.mp.4000957 [DOI] [PubMed] [Google Scholar]
- Blackwood DH, He L, Morris SW, McLean A, Whitton C, Thomson M, Walker MT, Woodburn K, Sharp CM, Wright AF, Shibasaki Y, St. Clair DM, Porteous DJ, Muir WJ (1996) A locus for bipolar affective disorder on chromosome 4p. Nat Genet 12:427–430 10.1038/ng0496-427 [DOI] [PubMed] [Google Scholar]
- Cichon S, Schumacher J, Muller DJ, Hurter M, Windemuth C, Strauch K, Hemmer S, et al (2001) A genome screen for genes predisposing to bipolar affective disorder detects a new susceptibility locus on 8q. Hum Mol Genet 10:2933–2944 10.1093/hmg/10.25.2933 [DOI] [PubMed] [Google Scholar]
- Coon H, Jensen S, Hoff M, Holik J, Plaetke R, Reimherr F, Wender P, Leppert M, Byerley W (1993) A genome-wide search for genes predisposing to manic-depression, assuming autosomal dominant inheritance. Am J Hum Genet 52:1234–1249 [PMC free article] [PubMed] [Google Scholar]
- Curtis D, Kalsi G, Brynjolfsson J, McInnis M, O’Neill J, Smyth C, Moloney E, Murphy P, McQuillin A, Petursson H, Gurling H (2003) Genome scan of pedigrees multiply affected with bipolar disorder provides further support for the presence of a susceptibility locus on chromosome 12q23-q24, and suggests the presence of additional loci on 1p and 1q. Psychiatr Genet 13:77–84 10.1097/00041444-200306000-00004 [DOI] [PubMed] [Google Scholar]
- Detera-Wadleigh SD, Badner JA, Berrettini WH, Yoshikawa T, Goldin LR, Turner G, Rollins DY, Moses T, Sanders AR, Karkera JD, Esterling LE, Zeng J, Ferraro TN, Guroff JJ, Kazuba D, Maxwell ME, Nurnberger JI Jr, Gershon ES (1999) A high-density genome scan detects evidence for a bipolar-disorder susceptibility locus on 13q32 and other potential loci on 1q32 and 18p11.2. Proc Natl Acad Sci USA 96:5604–5609 10.1073/pnas.96.10.5604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick DM, Foroud T, Edenberg HJ, Miller M, Bowman E, Rau NL, DePaulo JR, McInnis M, Gershon E, McMahon F, Rice JP, Beirut LJ, Reich T, Nurnberger J Jr (2002) Apparent replication of suggestive linkage on chromosome 16 in the NIMH Genetics Initiative bipolar pedigrees. Am J Med Genet 114:407–412 10.1002/ajmg.10380 [DOI] [PubMed] [Google Scholar]
- Dick DM, Foroud T, Flury L, Bowman ES, Miller MJ, Rau NL, Moe PR, et al (2003) Genomewide linkage analyses of bipolar disorder: a new sample of 250 pedigrees from the National Institute of Mental Health Genetics Initiative. Am J Hum Genet 73:107–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dizier MH, Quesneville H, Prum B, Selinger-Leneman H, Clerget-Darupoux F (2000) The triangle test statistic (TTS): a test of genetic homogeneity using departure from the triangle constraints in IBD distribution among affected sib-pairs. Ann Hum Genet 64:433–442 10.1046/j.1469-1809.2000.6450433.x [DOI] [PubMed] [Google Scholar]
- Ekholm JM, Kieseppa T, Hiekkalinna T, Partonen T, Paunio T, Perola M, Ekelund J, Lonnqvist J, Pekkarinen-Ijas P, Peltonen L (2003) Evidence of susceptibility loci on 4q32 and 16p12 for bipolar disorder. Hum Mol Genet 12:1907–1915 10.1093/hmg/ddg199 [DOI] [PubMed] [Google Scholar]
- Elston RC, Song D, Iyengar SK (2005) Mathematical assumptions versus biological reality: myths in affected sib pair linkage analysis. Am J Hum Genet 76:152–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald H, Flint T, Kruse TA Mors O (2002) A genome-wide scan shows significant linkage between bipolar disorder and chromosome 12q24.3 and suggestive linkage to chromosomes 1p22–21, 4p16, 6q14-22, 10q26 and 16p13.3. Mol Psychiatry 7:734–744 10.1038/sj.mp.4001074 [DOI] [PubMed] [Google Scholar]
- Fallin MD, Lasseter VK, Wolyneic PS, McGrath JA, Nestadt G, Valle D, Liang K-Y, Pulver AE (2004) Genomewide linkage scan for bipolar-disorder susceptibility loci among Ashkenazi Jewish families. Am J Hum Genet 75:204–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friddle C, Koskela R, Ranade K, Hebert J, Cargill M, Clark CD, McInnis M, Simpson S, McMahon F, Stine OC, Meyers D, Xu J, MacKinnon D, Swift-Scanlan T, Jamison K, Folstein S, Daly M, Kruglyak L, Marr T, DePaulo JR, Botstein D (2000) Full-genome scan for linkage in 50 families segregating the bipolar affective disease phenotype. Am J Hum Genet 66:205–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner C, McInnes LA, Service SK, Spesny M, Fournier E, Leon P, Freimer NB (2001) Linkage analysis of a complex pedigree with severe bipolar disorder, using a Markov chain Monte Carlo method. Am J Hum Genet 68:1061–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu C, Province M, Todorov A, Rao DC (1998) Meta-analysis methodology for combining non-parametric sibpair linkage results: genetic homogeneity and identical markers. Genet Epidemiol 15:609–626 [DOI] [PubMed] [Google Scholar]
- Holmans P (1993) Asymptotic properties of affected-sib-pair linkage analysis. Am J Hum Genet 52:362–374 [PMC free article] [PubMed] [Google Scholar]
- Kelsoe JR, Spence MA, Loetscher E, Foguet M, Sadovnick AD, Remick RA, Flodman P, Khristich J, Mroczkowski-Parker Z, Brown JL, Masser D, Ungerleider S, Rapaport MH, Wishart WL, Luebbert H (2001) A genome survey indicates a possible susceptibility locus for bipolar disorder on chromosome 22. Proc Natl Acad Sci USA 98:585–590 10.1073/pnas.011358498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler RC, McGonagle KA, Zhao S, Nelson CB, Hughes M, Eshleman S, Wittchen HU, Kendler KS (1994) Lifetime and 12-month prevalence of DSM-III-R psychiatric disorders in the United States: results from the National Comorbidity Survey. Arch Gen Psychiatry 51:8–19 [DOI] [PubMed] [Google Scholar]
- Kong A, Cox NJ (1997) Allele-sharing models: LOD scores and accurate linkage tests. Am J Hum Genet 61:1179–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong X, Murphy T, Raj T, He C, White PS, Matise C (2004) A combined linkage-physical map of the human genome. Am J Hum Genet 75:1143–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES (1996) Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet 58:1347–1363 [PMC free article] [PubMed] [Google Scholar]
- Laird N, Horvath S, Xu X (2000) Implementing a unified approach to family based tests of association. Genetic Epidemiol Suppl 19: S36–S42 [DOI] [PubMed] [Google Scholar]
- Laird NM, Mosteller F (1990) Some statistical methods for combining experimental results. Int J Technol Assess Health Care 6:5–30 [DOI] [PubMed] [Google Scholar]
- Lambert D, Middle F, Hamshere ML, Segurado R, Raybould R, Corvin A, Green E, O’Mahony E, Nikolov I, Mulcahy T, Haque S, Bort S, Bennett P, Norton N, Owen MJ, Kirov G, Lendon C, Jones L, Jones I, Holmans P, Gill M, Craddock N (2005) Stage 2 of the Wellcome Trust UK-Irish bipolar affective disorder sibling-pair genome screen: evidence for linkage on chromosomes 6q16-q21, 4q12-q21, 9p21, 10p14-p12 and 18q22. Mol Psychiatry (http://www.nature.com/mp/journal/vaop/ncurrent/abs/4001684a.html) (electronically published May 17, 2005; accessed August 2, 2005) [DOI] [PubMed] [Google Scholar]
- Lander E, Kruglyak L (1995) Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 11:241–247 10.1038/ng1195-241 [DOI] [PubMed] [Google Scholar]
- Levinson DF, Levinson MD, Segurado R, Lewis CM (2003) Genome scan meta-analysis of schizophrenia and bipolar disorder, part I: methods and power analysis. Am J Hum Genet 73:17–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Juo SH, Dewan A, Grunn A, Tong X, Brito M, Park N, Loth JE, Kanyas K, Lerer B, Endicott J, Penchaszadeh G, Knowles JA, Ott J, Gilliam TC, Baron M (2003) Evidence for a putative bipolar disorder locus on 2p13-16 and other potential loci on 4q31, 7q34, 8q13, 9q31, 10q21-24, 13q32, 14a21 and 17q11-12. Mol Psychiatry 8:333–342 10.1038/sj.mp.4001254 [DOI] [PubMed] [Google Scholar]
- McInnes LA, Escamilla MA, Service SK, Reus VI, Leon P, Silva S, Rojas E, Spesny M, Baharloo S, Blankenship K, Peterson A, Tyler D, Shimayoshi N, Tobey C, Batki S, Vinogradov S, Meza L, Gallegos A, Fournier E, Smith LB, Barondes SH, Sandkuijl LA, Freimer NB (1996) A complete genome screen for genes predisposing to severe bipolar disorder in two Costa Rican pedigrees. Proc Natl Acad Sci USA 93:13060–13065 10.1073/pnas.93.23.13060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInnis MG, Dick DM, Willour VL, Avramopoulos D, MacKinnon DF, Simpson SG, Potash JB, et al (2003a) Genome-wide scan and conditional analysis in bipolar disorder: evidence for genomic interaction in the National Institute of Mental Health Genetics Initiative bipolar pedigrees. Biol Psychiatry 54:1265–1273 10.1016/j.biopsych.2003.08.001 [DOI] [PubMed] [Google Scholar]
- McInnis MG, Lan TH, Willour VL, McMahon FJ, Simpson SG, Addington AM, MacKinnon DF, Potash JB, Mahoney AT, Chellis J, Huo Y, Swift-Scanlan T, Chen H, Koskela R, Stine OC, Jamison KR, Holmans P, Folstein SE, Ranade K, Friddle C, Botstein D, Marr T, Beaty TH, Zandi P, DePaulo JR (2003b) Genome-wide scan of bipolar disorder in 65 pedigrees: supportive evidence for linkage at 8q24, 18q22, 4q32, 2p12 and 13q12. Mol Psychiatry 8:288–298 10.1038/sj.mp.4001277 [DOI] [PubMed] [Google Scholar]
- Middleton FA, Pato MT, Gentile KL, Morley CP, Zhao X, Eisener AF, Brown A, Petryshen TL, Kirby AN, Medeiros H, Carvalho C, Macedo A, Dourado A, Coelho I, Valente J, Soares MJ, Ferreira CP, Lei M, Azevedo MH, Kennedy JL, Daly MJ, Sklar P, Pato CN (2004) Genomewide linkage analysis of bipolar disorder by use of a high-density single-nucleotide-polymorphism (SNP) genotyping assay: a comparison with microsatellite marker assays and finding of significant linkage to chromosome 6q22. Am J Hum Genet 74:886–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morissette J, Villeneuve A, Bordeleau L, Rochette D, Laberge C, Gagne B, Laprise C, Bouchard G, Plante M, Gobeil L, Shink E, Weissenbach J, Barden N (1999) Genome-wide search for linkage of bipolar affective disorders in a very large pedigree derived from a homogeneous population in Quebec points to a locus of major effect on chromosome 12q23-q24. Am J Med Genet 88:567–587 [DOI] [PubMed] [Google Scholar]
- NIMH Genetics Initiative Bipolar Group (1997) Genomic survey of bipolar illness in the NIMH Genetics Initiative Pedigrees: a preliminary report. NIMH Genetics Initiative Bipolar Group. Am J Med Genet 74:227–237 [DOI] [PubMed] [Google Scholar]
- NIMH Genetics Workgroup (1999) Report of the NIMH’s Genetics Workgroup: genetics and mental disorders. Biol Psychiatry 45:559–602 [PubMed] [Google Scholar]
- Park N, Juo SH, Cheng R, Liu J, Loth JE, Lilliston B, Nee J, Grunn A, Kanyas K, Lerer B, Endicott J, Gilliam TC, Baron M (2004) Linkage analysis of psychosis in bipolar pedigrees suggests novel putative loci for bipolar disorder and shared susceptibility loci with schizophrenia. Mol Psychiatry 9:1091–1099 10.1038/sj.mp.4001541 [DOI] [PubMed] [Google Scholar]
- Pato CN, Pato MT, Kirby A, Petryshen TL, Medeiros H, Carvalho C, Macedo A, Dourado A, Coelho I, Valente J, Soares MJ, Ferreira CP, Lei M, Verner A, Hudson TJ, Morley CP, Kennedy JL, Azevedo MH, Daly MJ, Sklar P (2004) Genome-wide scan in Portuguese Island families implicates multiple loci in bipolar disorder: fine mapping adds support on chromosomes 6 and 11. Am J Med Genet B Neuropsychiatr Genet 127:30–34 10.1002/ajmg.b.30001 [DOI] [PubMed] [Google Scholar]
- Radhakrishna U, Senol S, Herken H, Gucuyener K, Gehrig C, Blouin JL, Akarsu NA, Antonarakis SE (2001) An apparently dominant bipolar affective disorder (BPAD) locus on chromosome 20p11.2-q11.2 in a large Turkish pedigree. Eur J Hum Genet 9:39–44 10.1038/sj.ejhg.5200584 [DOI] [PubMed] [Google Scholar]
- Schulze TG, Buervenich S, Badner JA, Steele CJM, Detra-Wadleigh SD, Dick D, Foroud T, Cox NJ, MacKinnon DF, Potash JB, Berrettini WH, Byerley W, Coryell W, DePaulo JR, Gershon ES, Kelsoe JR, McInnis MG, Murphy DL, Reich T, Scheftner W, Nurnberger JI, McMahon FJ (2004) Loci on chromosomes 6q and 6p interact to increase susceptibility to bipolar affective disorder in the National Institute of Mental Health Genetics Initiative pedigrees. Biol Psychiatry 56:18–23 10.1016/j.biopsych.2004.04.004 [DOI] [PubMed] [Google Scholar]
- Segurado R, Detera-Wadleigh SD, Levinson DF, Lewis CM, Gill M, Nurnberger JI Jr, Craddock N, et al (2003) Genome scan meta-analysis of schizophrenia and bipolar disorder, part III: bipolar disorder. Am J Hum Genet 73:49–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson SG, McMahon FJ, McInnis MG, MacKinnon DF, Edwin D, Folstein SE, DePaulo JR (2002) Diagnostic reliability of bipolar II disorder. Arch Gen Psychiatry 59:736–740 10.1001/archpsyc.59.8.736 [DOI] [PubMed] [Google Scholar]
- Smoller JW, Finn CT (2003) Family, twin, and adoption studies of bipolar disorder. Am J Med Genet C Semin Med Genet 123:48–58 10.1002/ajmg.c.20013 [DOI] [PubMed] [Google Scholar]
- Tsuang MT, Faraone SV (2000) The genetic epidemiology of bipolar disorder. In: Marneros A, Angst J (eds) Bipolar disorders: 100 years after manic-depressive insanity. Kluwer Academic, Zurich, pp 231–242 [Google Scholar]
- Turecki G, Grof P, Grof E, D’Souza V, Lebuis L, Marineau C, Cavazzoni P, Duffy A, Betard C, Zvolsky P, Robertson C, Brewer C, Hudson TJ, Rouleau GA, Alda M (2001) Mapping susceptibility genes for bipolar disorder: a pharmacogenetic approach based on excellent response to lithium. Mol Psychiatry 6:570–578 10.1038/sj.mp.4000888 [DOI] [PubMed] [Google Scholar]
- Venken T, Claes S, Sluijs S, Paterson D, van Duijn C, Adolfsson R, Del-Favero J, Van Broekhoven C (2005) Genomewide scan for affective disorder susceptibility loci in families of a northern Swedish isolated population. Am J Hum Genet 76:237–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman M, Bruce M, Leaf PJ, Florio LP, Holzer C (1991) Psychiatric disorders in America. In: Robins L, Regier D (eds) Affective disorders. Free Press, New York, pp 53–81 [Google Scholar]
- Weissman MM, Bland RC, Canino GJ, Faravelli C, Greenwald S, Hwu HG, Joyce PR, Karam EG, Lee CK, Lellouch J, Lepine JP, Newman SC, Rubio-Stipec M, Wells JE, Wickramaratne PJ, Wittchen H, Yeh EK (1996) Cross-national epidemiology of major depression and bipolar disorder. JAMA 276:293–299 10.1001/jama.276.4.293 [DOI] [PubMed] [Google Scholar]
- Whittemore AS, Halpern J (1994) A class of tests for linkage using affected pedigree members. Biometrics 50:118–127 [PubMed] [Google Scholar]
- Willour VL, Zandi PP, Huo Y, Diggs TL, Chellis JL, MacKinnon DF, Simpson SG, McMahon FJ, Potash JB, Gershon ES, Reich T, Foroud T, Nurnberger JI Jr, DePaulo JR Jr, McInnis MG (2003) Genome scan of the fifty-six bipolar pedigrees from the NIMH genetics initiative replication sample: chromosomes 4, 7, 9, 18, 20 and 21. Am J Med Genet B Neuropsychiatr Genet 121:21–27 10.1002/ajmg.b.20051 [DOI] [PubMed] [Google Scholar]
- Wise LH, Lanchbury JS, Lewis CM (1999) Meta-analysis of genome searches. Ann Hum Genet 63:263–272 10.1046/j.1469-1809.1999.6330263.x [DOI] [PubMed] [Google Scholar]
- Zandi PP, Willour VL, Huo Y, Chellis J, Potash JB, MacKinnon DF, Simpson SG, McMahon FJ, Gershon E, Reich T, Foroud T, Nurnberger J Jr, DePaulo JR Jr, McInnish MG (2003) Genome scan of a second wave of NIMH Genetics Initiative bipolar pedigrees: chromosomes 2, 11, 13, 14 and X. Am J Med Genet B Neuropsychiatr Genet 119:69–76 10.1002/ajmg.b.10063 [DOI] [PubMed] [Google Scholar]