Abstract
Angiotensin I–converting enzyme inhibitors (ACEi), which are used to treat common cardiovascular diseases, are associated with a potentially life-threatening adverse reaction known as angioedema (AE-ACEi). We have previously documented a significant association between AE-ACEi and low plasma aminopeptidase P (APP) activity. With eight large pedigrees, we hereby demonstrate that this quantitative trait is partially regulated by genetic factors. We tested APP activity using a variance-component QTL analysis of a 10-cM genomewide microsatellite scan enriched with seven markers over two candidate regions. We found significant linkage (LOD = 3.75) to a locus that includes the XPNPEP2 candidate gene encoding membrane-bound APP. Mutation screening of this QTL identified a large coding deletion segregating in one pedigree and an upstream single-nucleotide polymorphism (C–2399A SNP), which segregates in the remaining seven pedigrees. Measured genotype analysis strongly suggests that the linkage signal for APP activity at this locus is accounted for predominantly by the SNP association. In a separate case-control study (20 cases and 60 controls), we found significant association of this SNP to ACEi-induced AE (P=.0364). In conclusion, our findings provide supporting evidence that the C-2399A variant in XPNPEP2 is associated with reduced APP activity and a higher incidence of AE-ACEi.
Introduction
Angiotensin I–converting enzyme inhibitors (ACEi) are a class of drugs used by >40 million patients worldwide for the treatment of cardiovascular diseases such as hypertension, congestive heart failure, and diabetes (Unger and Gohlke 1994; Brown and Vaughan 1998). Angioedema (AE) associated with ACEi therapy (AE-ACEi) is a potentially fatal adverse event that affects 0.1%–0.7% of white patients (Israili and Hall 1992; Vleeming et al. 1998) and is four to five times more prevalent among African Americans (Brown et al. 1996; Coats 2002). This racial difference suggests that genetic factors modulate AE risk, but environmental factors are also important, because smokers taking ACEi have an increased susceptibility to AE (Coats 2002; Kostis et al. 2004). The incidence of AE-ACEi is likely underestimated because the clinical symptoms can develop years after starting ACEi therapy, thus obscuring the adverse event's relationship with the drug and often leading to misdiagnosis (Agostoni et al. 2004).
A majority of AE-ACEi cases do not respond to antihistamines or corticosteroids, indicating that these cases are not allergic reactions (Agostoni and Cicardi 2001). Currently, there is no effective treatment for AE-ACEi and no method for identifying individuals with increased susceptibility to this adverse reaction. Better understanding of the pathogenetic mechanism underlying this type of AE may also have significant implications for AE associated with other vasopeptidase inhibitors (i.e., Omapatrilat), which block the activities of both ACE and neutral endopeptidase (NEP). The AE risk associated with these drugs is even higher than with ACEi (Coats 2002) and has curtailed the regulatory approval for usage of these vasopeptidase inhibitors to treat cardiovascular diseases.
Previous reports have suggested that bradykinin (BK), a potent vasodilatory and proinflammatory nonapeptide, plays a central role in the pathophysiology of AE-ACEi (Israili and Hall 1992; Nussberger et al. 1998). BK is rapidly degraded in the plasma of healthy individuals by angiotensin I–converting enzyme (ACE) and aminopeptidase P (APP) (Bhoola et al. 1992). Kininase I enzymes normally transform a minute fraction (3.5%) of BK into its active metabolite, des-arginine9-bradykinin (des-Arg9-BK) (Blais et al. 2000). This carboxy-truncated metabolite is, in turn, broken down by APP and ACE (Cyr et al. 2001). In the presence of ACE inhibition, however, kininase I activity is increased (transforms 28% of BK into des-Arg9-BK), and APP acts as the major metabolizing enzyme of both BK and des-Arg9-BK (Blais et al. 2000).
An increase of BK has been measured in the plasma of patients during episodes of AE-ACEi, but, unlike hereditary forms of AE (MIM 106100), there is no increase in cleavage of the BK precursor, high-molecular-weight kininogen (HK) (Nussberger et al. 1998; Agostoni et al. 1999; Cugno et al. 2003). This suggests that impaired BK metabolism, rather than increased BK production, plays an important role in AE-ACEi. We previously reported significantly lower plasma APP activities in patients with a history of AE-ACEi, which is strongly correlated with a significant decrease in des-Arg9-BK degradation in vitro (Blais et al. 1999a; Adam et al. 2002; Molinaro et al. 2002). Blais et al. demonstrated that half of AE-ACEi cases have an enzyme defect involved in des-Arg9-BK metabolism (Blais et al. 1999b). Accumulated levels of des-Arg9-BK have been shown to cause proinflammatory effects in vivo (Blais et al. 1997; Blais et al. 1999a). One study reported that ACEi patients who received subcutaneous injections of the APP inhibitor Apstatin developed local inflammations (Kim et al. 2000). Thus, previous data indicate that reduced plasma APP activity may predict increased risk for AE associated with ACEi therapy.
The aim of this study is to determine whether plasma APP activity is regulated by genetic factors and to identify the QTL or QTLs that confer susceptibility to AE-ACEi. There are two known APP enzymes in humans: membrane-bound (mAPP) and cytosolic (cAPP). The gene encoding the former is XPNPEP2 (MIM 300145), localized to chromosome Xq26.1 (Sprinkle et al. 1998), and the latter is the product of XPNPEPL (MIM 602443) on chromosome 10q25.1 (Sprinkle et al. 2000). Although these represent good candidate genes for the interindividual variability in plasma APP activity, other genetic loci may serve as important regulators. Plasma APP activity has been shown to form a continuous distribution in the general population (Cyr et al. 2001), suggesting that plasma APP activity is a complex quantitative trait likely influenced by multiple genetic loci and nongenetic factors (e.g., smoking and hormone replacement therapy [Gallagher et al. 1999]). Identification of the genetic factors underlying reduced plasma APP activity would provide a better understanding of the pathogenesis of AE-ACEi and could facilitate the development of a clinical assay to detect those individuals with greater AE risk.
Subjects and Methods
Blood and Plasma Samples
The ethics committees from Centre Hospitalier de l’Universite de Montreal, Institut de Cardiologie de Montreal, and McGill University, all in Montreal, reviewed and approved all protocols involving human subjects. Informed consent was obtained from all participants. DNA extraction from blood was performed following a standardized protocol (Gentra Systems).
Participants
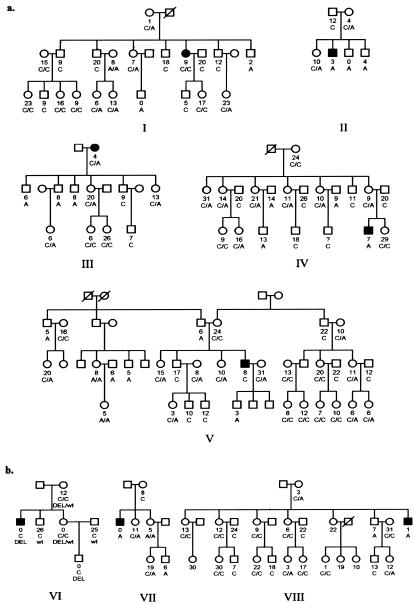
Cohort 1 (fig. 1a), composed of five white pedigrees (88 DNA samples), was collected for a genomewide microsatellite scan, followed by a variance-component linkage analysis for APP activity. Cohort 2 (fig. 1b), consisting of three white pedigrees (35 DNA samples), was subsequently selected for genotyping of 11 microsatellite markers (table 1), which cover four genomic regions that provided LOD scores >1 in the linkage analysis of cohort 1 and the locus including the XPNPEPL candidate gene. Plasma samples were also collected from all 123 participants for characterization of APP activity. The kindreds were selected from Canada (Montreal) and France on the basis of the presence of one individual in each family with a history of AE and/or another acute adverse effect related to ACEi therapy, anaphylactoid reactions (AR-ACEi) (Bright et al. 1999). The clinical diagnosis was not considered for linkage analysis because of lack of power. Instead, plasma APP activity measurements were used as the quantitative trait in a variance-component linkage analysis.
Figure 1.
Cohorts included in variance-component linkage analysis. One member of each kindred, depicted in black, developed AE and/or AR associated with ACEi therapy. All available members were quantified for plasma APP activity, shown as numbers that represent units of arginine released per minute per milliliter of plasma. Genotypes of the C-2399A SNP upstream of XPNPEP2 are shown as C/C, C/A, or A/A for females, and C or A for males. a, DNA samples from cohort 1 were included in a genomewide microsatellite scan, which was analyzed for linkage using a variance-component method (SOLAR program). b, Eleven genome-scanned markers were genotyped in cohort 2 to further evaluate linkage. A large deletion (DEL) in XPNPEP2 segregates in pedigree VI.
Table 1.
Variance-Component Two-Point QTLs for Markers Genotyped in Cohorts 1 and 2[Note]
| LOD |
||||
| Marker | Position(cM) | Chromosome Band | Cohorts 1 and 2 (8 Pedigrees) | Cohort 1 Only |
| D6S1017 | 63.3 | 6p21.1 | .97 | 2.62 |
| D6S1280 | 73.1 | 6p12.3 | .18 | 1.09 |
| D6S1056 | 102.8 | 6q16.1 | 1.32 | 1.68 |
| D8S277 | 8.0 | 8p23.1 | .22 | .69 |
| D8S373 | 164.5 | 8q24.3 | 1.72 | 1.76 |
| D9S925 | 32.2 | 9p22.2 | .16 | 1.71 |
| D9S1122 | 75.9 | 9q21.2 | .13 | .76 |
| Markers flanking candidates: | ||||
| D10S1220 | 70.2 | 10q11.23 | .24 | .74 |
| DXS8057 | 136.4 | Xq25 | 2.10 | 1.82 |
| DXS1047 | 143.2 | Xq25 | 2.04 | .28 |
| XPNPEP2_CA | 146.0 | Xq25 | 3.75 | 2.15 |
Note.— Eleven microsatellite markers that were already part of the analysis in cohort 1 were genotyped in cohort 2 to cover the four loci that provided LOD scores >1 in cohort 1 and the region proximal to the XPNPEPL candidate gene (D10S1220).
DNA from 20 independent AE-ACEi cases (table A1 [online only]) was extracted for mutation screening experiments. All subjects were whites with histories of AE-ACEi and origins in Canada (n=7), Belgium (n=3), or the United States (n=10). Plasma samples were available for all AE subjects except two from the United States. The onset of clinical symptoms ranged from a few hours to 8 years after starting ACEi therapy. In addition, we collected three unrelated white controls to match each AE patient for country of origin and gender (n=60). Controls from Belgium and Canada had no history of ACEi therapy, whereas the medical histories were unknown for US controls. Controls were not matched for age because we did not find age effects in our samples, which is in agreement with previous findings (Cyr et al. 2001).
Quantification of Plasma APP Activity
APP activity was measured in plasma with modification of an assay described elsewhere (Simmons and Orawski 1992; Blais et al. 1999a). Our assay used the synthetic substrate arginine-proline-proline. Plasma APP activity is expressed as nanomoles of arginine released per minute per milliliter of plasma sample (1 unit = 1 nmol/min/ml).
Genome Scan and Microsatellite Genotyping
A 10-cM genomewide microsatellite scan (marker panel SS4 on the ABI-3700 DNA analyzer at the Genome Quebec Innovation Centre, Montreal), enriched for seven markers flanking the two APP candidate genes on chromosomes 10 and X (D10S534, D10S1741, D10S562, DXS1212, DXS8057, XPNPEP2_CA, and DXS1047), was performed on cohort 1 (total of 397 microsatellite markers). Primers for the XPNPEP2_CA repeat marker located upstream of XPNPEP2 (∼4 kb were designed for this study) were sense 5′-GCTCTTTCCCCCTGCTGTGT-3′ and antisense 5′-GGTGCTGTTGGGTGCCTCATC-3′.
Genotyping of the 11 genome-scanned markers in cohort 2 was conducted in our laboratory. Genomic DNA was amplified by radiolabeled (α-35S-dATP) PCR. The products were separated by electrophoresis on 6% denaturing polyacrylamide gels. The alleles were sized by comparison to the M13mp18 sequence ladder, and each individual was assigned a genotype. Marker-allele sizes and frequencies were obtained from the Fondation Jean Dausset CEPH database.
Statistical Analyses
The SOLAR (Sequential Oligogenic Linkage Analysis Routines, v. 2.1.4) (Almasy and Blangero 1998) program was used to estimate heritability of plasma APP activity and to perform a variance-component linkage analysis. Age and sex were considered as covariates, but were not significant (P > .05). APP activity was transformed for normality to avoid convergence failure of SOLAR by using the Box-Cox transformation (Box and Cox 1964), such that T(APP)={[(APP+1)×0.40-1]/0.40}×4. The two-point (all chromosomes) and multipoint (autosomes only) variance-component linkage analyses were conducted for Box-Cox transformed plasma APP activity in cohorts 1 and 2. Empirical LOD adjustment based on 10,000 simulations was used for all linkage analyses. QTL results of the genomewide scan for the five pedigrees in cohort 1 were adjusted with an empirical LOD factor of 0.98220 (results shown in fig. 2). QTL results of cohorts 1 and 2 were corrected with an empirical LOD adjustment factor of 0.74595 (table 1).
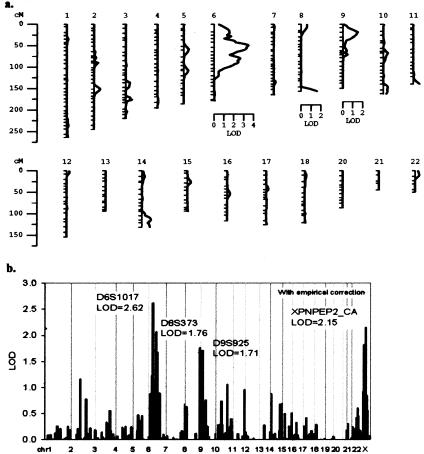
Figure 2.
Results of the variance-component QTL analyses in the five pedigrees of cohort 1. All results were corrected empirically for α ⩽ 0.05. a, String diagram of autosomal multipoint results, LODs >1, are shown next to a LOD scale. b, Two-point linkage analysis results including chromosome X.
A measured genotype approach using variance-component analysis (Soria et al. 2000) in SOLAR was used to test for the C-2399A SNP genotype-specific differences in plasma APP-activity means at the XPNPEP2_CA marker. This approach accounts for the different relationships among family members in performing association on the measured genotypes and quantitative traits (Box-Cox transformed plasma APP activity). The three C-2399A SNP genotypes, CC, CA, and AA, were coded as −1, 0, and 1 in females, respectively, and the C and A hemizygous genotypes in males as −1 and 1, respectively. The measured genotype test was conducted on all families except pedigree VI, which segregates the coding deletion accounting for the APP variation in this family. Other statistical analyses were conducted in SAS (v. 9.1.3). All tests were two-sided.
Mutation Detection
Oligonucleotides for PCR were designed from the genomic sequences for human XPNPEP2 (accession number AL023653) and XPNPEPL (accession number AL354951), obtained from the National Center for Biotechnology Information (NCBI) database. The exons, 5′ and 3′ UTRs of both candidate genes, were amplified using genomic DNA and radiolabeled deoxyadenosine triphosphate for single-stranded conformational polymorphism (SSCP) analysis. Electrophoresis of PCR products was performed on 9.5% polyacrylamide (5% glycerol) and on 50% mutation detection enhancement gels (Biowhittaker Molecular Applications), followed by autoradiography. Samples showing altered migration patterns were selected for sequencing. All sequencing was performed at the Genome Quebec Innovation Centre in Montreal. Variations that altered a restriction enzyme digestion site were genotyped in additional individuals using RFLP assays. The silent variant in exon 6 of XPNPEP2 (SNP rs3747343) was genotyped by EcoNI digestion.
The C-2399A SNP was genotyped using a modification of the allele-specific PCR assay described elsewhere (Germer and Higuchi 1999). This required three PCR primers for each polymorphic site (sequences available upon request): one common and two allele-specific oligonucleotides. The allele-specific primers differed only at the position of the 3′-most nucleotide. Two standard PCRs were performed using the common primer and either one of the allele-specific primers. Products were visualized on 1.5% agarose gels by electrophoresis.
mRNA Analysis
Extraction of total RNA from suspended lymphoblast cells was performed following a standardized protocol (Qiagen). Synthesis of cDNA required two steps: (1) annealing oligoDT primers and random hexamers to the RNA template in an initial reaction mix (3 μg of total RNA, 1 μg each of oligoDT primers and random hexamers, and HPLC-grade water), incubated at 70°C for 3 min; and (2) extension with the addition of a second reaction (1 μl of Moloney murine leukemia virus reverse transcriptase [100 U], 10 μl of 5× First Strand Buffer, 5 μl of 0.1 M DTT, 1 μl of RNase Inhibitor, and 2 μl of dNTP [25 mM]), incubated at 37°C for 1 h. Primer pairs for PCR amplification of cDNA were designed from the transcript sequence of the human XPNPEP2 gene (accession number U90724), obtained from the NCBI database. We applied a nested PCR protocol for amplification of the cDNA to compensate for the low gene expression of XPNPEP2 in lymphoblasts (Venema et al. 1997). Products from the first PCR were diluted 30-fold and then used as template DNA in the second PCR. Amplified products were visualized on 1.5% agarose gels and then sequenced.
Results
Quantification of Plasma APP Activity
The mean plasma APP activity (±SE) among our AE-ACEi cases was 13 ± 3 units (table 2). There was a nonsignificant sex difference (t test, P=.2549) with mean APP values (±SE) of 15 ± 4 units for women and 9 ± 6 units for men. These mean values for AE-ACEi patients differ from those previously reported in healthy whites (24 ± 9 units and 19 ± 7 units in women and men, respectively) (Cyr et al. 2001). Moreover, the majority (67%) of our AE-ACEi cases have APP values of <10 units, which is comparatively more than the 10% reported elsewhere for the general population (Cyr et al. 2001). However, a third of our affected patients have normal or high plasma APP activity (⩾20 units), suggesting that additional factors are likely involved in determining risk to AE-ACEi.
Table 2.
C-2399A SNP Genotypes in ACEi-Associated AE Cases and Matched Controls[Note]
| AE-ACEi Cases(n=20) |
Controls(n=60) |
||||
| C-2399A SNP Genotype | No. of Males | No. of Females | Mean APP ± SE(units) | No. of Males | No. of Females |
| CC or C | 4 | 8 | 18 ± 4 | 18 | 31 |
| CA | − | 4 | 10 ± 5 | − | 8 |
| AA or A | 3 | 1 | 2 ± 2 | 3 | 0 |
| All | 13 ± 3 | ||||
Note.— Mean APP activity is represented as units of arginine released per minute per milliliter of plasma sample.
Mutation Screening of Candidate Genes
DNA from 10 unrelated individuals with plasma APP activities <10 units and histories of AE-ACEi were screened for mutations in XPNPEP2 and XPNPEPL by SSCP. A band variant was detected in exon 6 of XPNPEP2 in four individuals. Sequencing revealed a T→C substitution (SNP rs3747343), which does not result in an amino acid change. Genotyping by RFLP analysis in the family members of these cases and in 20 healthy individuals showed no correlation between this variant and plasma APP activity. No other sequence variants in the candidate genes were detected in these subjects.
Given the absence of a causative mutation, we concluded that, if a functional variant conferring risk to AE-ACEi exists within these candidate loci, it might be located in the regulatory regions affecting transcription, splicing, or message stability. Alternatively, APP activity might be regulated by other genetic loci. For example, variants in genes coding for transcription factors might modulate expression of the APP coding gene(s). Genomewide studies in yeast have shown that 75% of transcripts are regulated by trans-acting elements (Yvert et al. 2003). Previous studies have also shown that expression differences in humans can be explained by genetic variations located elsewhere in the genome (e.g., Wilm’s Tumor [Discenza and Pelletier 2004]). Thus, we conducted a genomewide scan and variance-component linkage analyses, in addition to further mutation screening experiments involving additional AE-ACEi cases, to identify other potential genetic loci regulating APP activity.
Variance-Component Linkage Analysis
Cohort 1 (fig. 1a), included in the genome scan and characterized for plasma APP activities, was tested for variance-component QTL analysis using SOLAR. The mean of the transformed trait in the five pedigrees was 16.8 units, with SD of 6.3 units, skewness of −0.17 units, and kurtosis of −0.14 units. Heritability of APP activity was estimated at 37.5% ± 26.5% (P=.0336), indicating that the observed phenotypic variability within these families partly results from genotypic differences. The genome scan performed on cohort 1 resulted in the identification of three loci with LOD scores >1 in multipoint linkage analysis (fig. 2a) on chromosomes 6, 8, and 9, and one region on chromosome X that included the XPNPEP2 candidate gene with a two-point LOD score of 2.15 (fig. 2b).
We proceeded to genotype cohort 2 (fig. 1b) for 11 genome-scanned microsatellite markers covering the four regions above and the XPNPEPL candidate gene (table 1). Heritability of plasma APP activity for all eight pedigrees combined was estimated at 33.6% ± 25.1% SD, P=.0452. Analyses of the combined genotype data from cohorts 1 and 2 produced a maximum two-point LOD score of 3.75 for the marker XPNPEP2-CA (table 1). Review of the linkage data strongly suggested that the XPNPEP2 locus is a major genetic factor controlling APP activity in our families, despite no mutation having been found in the initial mutation screening experiments. This locus, however, is not the only regulator of APP activity, as multipoint linkage analysis in cohort 1 (fig. 2a) provided strong linkage to chromosome 6 (LOD = 3.47), but the linkage signal was greatly reduced in combined analyses of both cohorts (LOD = 1.43). Our linkage analysis also provided positive linkage to loci on chromosomes 8 and 9. Further investigation of these loci is necessary to either confirm or reject linkage.
Mutation Screening of the XPNPEP2 Locus
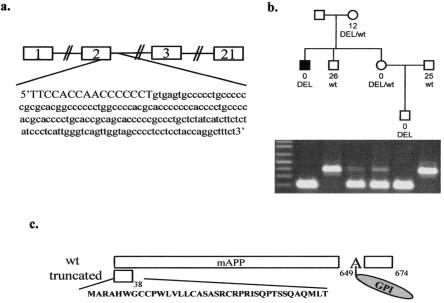
Given evidence of strong linkage to the XPNPEP2 locus, we sequenced the coding regions of this candidate gene in all available AE-ACEi cases with APP activities <10 units. A coding deletion in XPNPEP2 was detected in the proband of pedigree VI (fig. 1b) (not included in the initial mutation screening experiments) who first suffered an anaphylactoid reaction (AR) associated with ACEi during hemodialysis and subsequently suffered an AE-ACEi episode. The 175-bp genomic deletion (g.2953-3127del) includes 16 bp at the 3′ end of exon 2, the donor splice site and a fragment of the downstream intron (fig. 3a). The deleted allele is also present in three relatives of this AE/AR-ACEi patient, who all have negligible or reduced plasma APP activities (fig. 3b). The two men with relatively high plasma APP activities in this family each have a normal copy of this gene. This deletion was not found in other AE-ACEi cases or controls included in this study.
Figure 3.
Coding deletion in XPNPEP2. a, The deleted genomic region of 175 bp consists of 16 bp of exon 2 (in blue), the 3′ donor splice site (in green) and intronic sequences. b, The deleted allele (DEL) is present in four members of this pedigree who have negligible or reduced (in one female heterozygote) plasma APP activity. The two men with normal alleles (wt) both have high APP measurements. c, The deletion results in an abnormally spliced mRNA transcript coding for a premature stop codon that is predicted to translate into a severely truncated protein of 38 amino acids. This short peptide would not be membrane-bound because it lacks a glycosylphosphatidylinsitol (GPI) anchor attached to the alanine (A) at position 650.
Total RNA from cultured lymphoblastoid cells of the AE/AR-ACEi male was used to determine the effects of this deletion on the mRNA. Sequencing of his cDNA revealed an erroneously spliced transcript missing exon 2 and a segment of exon 3 (314_431del). This mutant transcript encodes a premature stop codon, which presumably translates into a truncated protein of 38 amino acids that is significantly shorter than the wild-type mAPP of 674 amino acids (fig. 3c). The first 21 amino acids of the protein constitute the signal peptide for the human mAPP, as previously predicted by the SignalP program, and are consequently cleaved off in the mature protein (Nielsen et al. 1997). The mutant protein lacks the predicted active site residues involved in metal ion coordination and proton shuttling, as well as a glycosylphosphatidylinsitol (GPI) anchor (Molinaro et al. 2004).
Next, we sequenced upstream regions of XPNPEP2 that are highly conserved in the mouse (UCSC; April 2002 freeze) to identify potential regulatory variants. We identified one SNP (C-2399A; rs3788853) that segregates in seven of our eight linkage families (genotypes shown in fig. 1). The A allele is absent only in the pedigree segregating the coding deletion described above. The two probands in pedigrees I and V also lack the A allele, which segregates in their families. Interestingly, only pedigree I contributes to the linkage signal at the XPNPEP2_CA marker (LOD = 0.99). We further sequenced 3 kb upstream, 1 kb downstream, and the coding region of XPNPEP2 in all AE-ACEi patients with the A allele, but did not detect any other sequence variations that might be in linkage disequilibrium with the C-2399A SNP.
The MatInspector program (Genomatix Suite) predicts that this substitution potentially affects the binding sites of two trans-acting elements: hepatic nuclear factor 4 (HNF-4) and peroxisome proliferator-activated receptors/retinoid X receptor (PPAR/RXR) heterodimer. The biological significance of this SNP remains to be determined. We have conducted quantitative RT-PCR and northern blots without success because of the low expression of XPNPEP2 in human lymphoblastoid cells (Venema et al. 1997). We have yet to ascertain more relevant tissues (i.e., kidney and lung) for functional analysis.
Measured Genotype Analysis of the C-2399A SNP
To evaluate the extent to which the C-2399A SNP accounts for the linkage observed at the XPNPEP2 locus, we repeated linkage analysis at the XPNPEP2_CA microsatellite marker after regressing out the effect of the C-2399A SNP association by including it as a covariate in the linkage model (Almasy and Blangero 2004). The trait mean calculated on the seven pedigrees segregating the SNP (fig. 1) was 17.25 units. The standard deviation in the model with the SNP as covariate was 5.55, compared with 6.70 in the model without the SNP as a covariate, and the proportion of variance explained by the covariate SNP was 0.3149. The beta term for the SNP genotypes in the model was −4.67. The SNP upstream of XPNPEP2 was significant as a covariate in the model (P < .0001). When all pedigrees were used except family VI, which segregates the XPNPEP2 coding deletion, the linkage signal diminished substantially in two-point linkage at the microsatellite marker XPNPEP2_CA, from LOD = 3.24 to LOD = 0.54. This marked diminution in LOD score indicates that the linkage signal for plasma APP activity at this locus is mostly accounted for by the C-2399A SNP association.
Association of the C-2399A SNP with AE-ACEi
We genotyped 20 independent AE-ACEi cases and 60 unrelated, matching controls for the C-2399A SNP (table 2). The A allele was present in 8 of the 20 AE patients (table A1 [online only]). By allele counting, the A allele was found at a frequency of 11.1% in our controls and 27.3% in our AE-ACEi cases. Genotype analysis in cases versus controls, where the male hemizygote is considered to be equivalent to the female homozygote, yields a P=.0364 (Armitage trend test) when comparing AE cases to population controls.
Discussion
This report provides evidence that variable plasma APP activity is partially regulated by genetic factors. We estimated that 34% of the phenotypic variation in our linkage families results from genotypic differences. Linkage analysis significantly identified a QTL (LOD = 3.75) near XPNPEP2, a candidate gene encoding mAPP. Despite the absence of mutations detected in the initial gene-screening experiments, further investigation of this locus identified two sequence variations. The genomic deletion segregating in pedigree VI results in an erroneously spliced transcript that is predicted to translate into a severely truncated protein. The C-2399A SNP genotype significantly accounts for the linkage signal at this QTL and is significantly associated with AE-ACEi (P=.0364). No other variants that may be in linkage disequilibrium with the A allele were found at this locus in additional sequencing experiments, further suggesting that this SNP may be responsible for the observed reduction in plasma APP activity.
The above variants were found in 9 of 20 AE-ACEi cases (table A1 [online only]). This suggests that the low plasma APP activity observed in additional AE-ACEi patients may result from other genetic loci or environmental factors. Our multipoint variance-component linkage analysis of cohort 1 provided evidence of significant linkage to a locus on chromosome 6 (LOD = 3.47) and positive linkage to loci on chromosomes 8 and 9. Multipoint linkage analysis including cohort 2, however, reduced linkage to the chromosome 6 locus (LOD = 1.43), as well as the LOD scores for the other two autosomal loci. Genotyping of additional markers at these loci in more families are necessary to either confirm or reject linkage.
Given that a third of our AE patients have normal plasma APP activity (⩾20 units), this quantitative trait clearly explains a fraction of the AE risk associated with ACEi therapy. Individuals who are not genetically predisposed to have low APP activity may develop AE-ACEi due to nonspecific inhibition of APP by certain kinds of ACEi drugs (Hooper et al. 1992) or other unidentified causes. Other potential risk factors include elevated levels of the sensory neuropeptide substance P, which correlate with reduced amounts of dipeptidyl peptidase IV activity (Ferreira et al. 2000; Lefebvre et al. 2002). In addition, clinical trials for new vasopeptidase inhibitors (that inhibit NEP and ACE) result in higher AE incidence than with ACE inhibition alone, suggesting that NEP activity may play a role in AE risk. Furthermore, other, nongenetic factors (i.e., smoking and estrogen replacement) may contribute to AE-ACEi risk.
Our study confirms previously published data showing that reduced plasma APP activity is a relatively common phenotype in the general population (Lefebvre et al. 2002). Other than predisposition to AE-ACEi, no clinical phenotype has been associated with low APP activity to date. This suggests that its physiological role may be nonessential or that there is functional redundancy. Furthermore, certain ACEi patients with low APP activity do not develop AE during therapy. This suggests that factors (i.e., environmental triggers) acting in pathways other than des-Arg9-BK degradation may contribute to AE risk.
Finally, low APP activity may determine risk for other ACEi-associated adverse effects. We previously reported an association between reduced plasma APP activity and anaphylactoid reactions during hemodialysis induced by ACEi therapy (Blais et al. 1999a). Further genetic investigations are necessary to determine if variants in the XPNPEP2 locus, such as those described here, play a role in AR-ACEi risk. Other adverse reactions associated with ACEi therapy include chronic cough, which affects ∼10%–15% of ACEi patients. It would be interesting to further characterize plasma APP activity in these patients and screen XPNPEP2 as a candidate susceptibility gene for these side effects of ACEi.
In conclusion, this report demonstrates that genetic variants at the XPNPEP2 locus are partially responsible for reduced plasma APP activity. Furthermore, we show a significant association between the C-2399A polymorphism and AE-ACEi. Thus, we have successfully mapped a QTL that may, in part, predict susceptibility to a potentially fatal adverse reaction associated with one of the most commonly used drugs worldwide. Our study represents a potential application of pharmacogenomics in health care, a field destined to transform medical care, but in which there have been few successes to date. Our findings, in addition to further studies, may facilitate the rational design of safer drugs for treating cardiovascular diseases, as well as clinical assays for predicting individuals who are genetically more liable to develop AE associated with ACEi or other vasopeptidase inhibitors.
Acknowledgments
We thank all the participants in this study; Dr. André Toulouse, for molecular expertise; Dominique Verlaan, for careful review of this manuscript; Nicole Gervais and Kateri Brisebois, for technical assistance; and the logistical assistance of Hospal France. This work was funded by the National Institutes of Health grant 1-RO1-HL079184. G.A.R. and G.M. are supported by the Canadian Institutes of Health Research. A.A. receives funding from Fonds de Recherche en Santé du Québec.
Appendix A
Table A1.
XPNPEP2 Genotypes in 20 AE-ACEi Patients[Note]
| AE-ACEi Patient No. | APP(units) | Sex | Origin | C-2399A SNP | Other Mutation |
| 1 | 0 | M | Montreal | C | g.3217del3391 |
| 2 | 9 | M | Belgium | C | … |
| 3 | 38 | M | USA | C | … |
| 4 | Unknown | M | USA | C | … |
| 5 | 4 | F | Montreal | C/C | … |
| 6 | 9 | F | USA | C/C | … |
| 7 | 9 | F | USA | C/C | … |
| 8 | 9 | F | USA | C/C | … |
| 9 | 21 | F | USA | C/C | … |
| 10 | 22 | F | Montreal | C/C | … |
| 11 | 24 | F | USA | C/C | … |
| 12 | 48 | F | USA | C/C | … |
| 13 | 5 | F | Montreal | C/A | … |
| 14 | 6 | F | Montreal | C/A | … |
| 15 | 20 | F | Belgium | C/A | … |
| 16 | Unknown | F | USA | C/A | … |
| 17 | 0 | M | USA | A | … |
| 18 | 0 | M | Montreal | A | … |
| 19 | 7 | M | Montreal | A | … |
| 20 | 0 | F | Belgium | A/A | … |
Note.— APP activity is represented as units of arginine released per minute per milliliter of plasma sample. The C-2399A SNP genotype is shown as C/C, C/A, or A/A for females (F), and C or A for males (M). The coding deletion resulting in an erroneously spliced transcript is represented as g.3217del3391.
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- Fondation Jean Dausset CEPH, http://www.cephb.fr/
- MatInspector (Genomatix Suite), http://www.genomatix.de/
- National Center for Biotechnology Information (NCBI), http://www.ncbi.nlm.nih.gov/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/
- University of Santa Cruz (UCSC) Genome Browser, http://genome.ucsc.edu/
References
- Adam A, Cugno M, Molinaro G, Perez M, Lepage Y, Agostoni A (2002) Aminopeptidase P in individuals with a history of angio-oedema on ACE inhibitors. Lancet 359:2088–2089 10.1016/S0140-6736(02)08914-6 [DOI] [PubMed] [Google Scholar]
- Agostoni A, Aygoren-Pursun E, Binkley KE, Blanch A, Bork K, Bouillet L, Bucher C, et al (2004) Hereditary and acquired angioedema: problems and progress: proceedings of the third C1 esterase inhibitor deficiency workshop and beyond. J Allergy Clin Immunol 114:S51–131 10.1016/j.jaci.2004.06.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agostoni A, Cicardi M (2001) Drug-induced angioedema without urticaria. Drug Saf 24:599–606 [DOI] [PubMed] [Google Scholar]
- Agostoni A, Cicardi M, Cugno M, Zingale LC, Gioffre D, Nussberger J (1999) Angioedema due to angiotensin-converting enzyme inhibitors. Immunopharmacology 44:21–25 10.1016/S0162-3109(99)00107-1 [DOI] [PubMed] [Google Scholar]
- Almasy L, Blangero J (1998) Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet 62:1198–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almasy L, Blangero J (2004) Exploring positional candidate genes: linkage conditional on measured genotype. Behav Genet 34:173–177 10.1023/B:BEGE.0000013731.03827.69 [DOI] [PubMed] [Google Scholar]
- Bhoola KD, Figueroa CD, Worthy K (1992) Bioregulation of kinins: kallikreins, kininogens, and kininases. Pharmacol Rev 44:1–80 [PubMed] [Google Scholar]
- Blais C Jr, Couture R, Drapeau G, Colman RW, Adam A (1997) Involvement of endogenous kinins in the pathogenesis of peptidoglycan-induced arthritis in the Lewis rat. Arthritis Rheum 40:1327–1333 [DOI] [PubMed] [Google Scholar]
- Blais C Jr, Marc-Aurele J, Simmons WH, Loute G, Thibault P, Skidgel RA, Adam A (1999a) Des-Arg9-bradykinin metabolism in patients who presented hypersensitivity reactions during hemodialysis: role of serum ACE and aminopeptidase P. Peptides 20:421–430 10.1016/S0196-9781(99)00020-0 [DOI] [PubMed] [Google Scholar]
- Blais C Jr, Marceau F, Rouleau JL, Adam A (2000) The kallikrein-kininogen-kinin system: lessons from the quantification of endogenous kinins. Peptides 21:1903–1940 10.1016/S0196-9781(00)00348-X [DOI] [PubMed] [Google Scholar]
- Blais C Jr, Rouleau JL, Brown NJ, Lepage Y, Spence D, Munoz C, Friborg J, Geadah D, Gervais N, Adam A (1999b) Serum metabolism of bradykinin and des-Arg9-bradykinin in patients with angiotensin-converting enzyme inhibitor-associated angioedema. Immunopharmacology 43:293–302 10.1016/S0162-3109(99)00133-2 [DOI] [PubMed] [Google Scholar]
- Box GEP, Cox DR (1964) An analysis of transformations. J R Stat Soc Ser B (Methodological) 26:211–252 [Google Scholar]
- Bright RA, Torrence ME, Daley WR, McClellan WM (1999) Preliminary survey of the occurrence of anaphylactoid reactions during haemodialysis. Nephrol Dial Transplant 14:799–800 10.1093/ndt/14.3.799 [DOI] [PubMed] [Google Scholar]
- Brown NJ, Ray WA, Snowden M, Griffin MR (1996) Black Americans have an increased rate of angiotensin converting enzyme inhibitor-associated angioedema. Clin Pharmacol Ther 60:8–13 10.1016/S0009-9236(96)90161-7 [DOI] [PubMed] [Google Scholar]
- Brown NJ, Vaughan DE (1998) Angiotensin-converting enzyme inhibitors. Circulation 97:1411–1420 [DOI] [PubMed] [Google Scholar]
- Coats AJ (2002) Omapatrilat—the story of Overture and Octave. Int J Cardiol 86:1–4 10.1016/S0167-5273(02)00389-3 [DOI] [PubMed] [Google Scholar]
- Cugno M, Nussberger J, Cicardi M, Agostoni A (2003) Bradykinin and the pathophysiology of angioedema. Int Immunopharmacol 3:311–317 [DOI] [PubMed] [Google Scholar]
- Cyr M, Lepage Y, Blais C Jr, Gervais N, Cugno M, Rouleau JL, Adam A (2001) Bradykinin and des-Arg(9)-bradykinin metabolic pathways and kinetics of activation of human plasma. Am J Physiol Heart Circ Physiol 281:H275–283 [DOI] [PubMed] [Google Scholar]
- Discenza MT, Pelletier J (2004) Insights into the physiological role of WT1 from studies of genetically modified mice. Physiol Genomics 16:287–300 [DOI] [PubMed] [Google Scholar]
- Ferreira PK, Campos MM, Calixto JB (2000) The role of sensorial neuropeptides in the edematogenic responses mediated by B(1) agonist des-Arg(9)-BK in rats pre-treated with LPS. Regul Pept 89:29–35 [DOI] [PubMed] [Google Scholar]
- Gallagher PE, Li P, Lenhart JR, Chappell MC, Brosnihan KB (1999) Estrogen regulation of angiotensin-converting enzyme mRNA. Hypertension 33:323–328 [DOI] [PubMed] [Google Scholar]
- Germer S, Higuchi R (1999) Single-tube genotyping without oligonucleotide probes. Genome Res 9:72–78 [PMC free article] [PubMed] [Google Scholar]
- Hooper NM, Hryszko J, Oppong SY, Turner AJ (1992) Inhibition by converting enzyme inhibitors of pig kidney aminopeptidase P. Hypertension 19:281–285 [DOI] [PubMed] [Google Scholar]
- Israili ZH, Hall WD (1992) Cough and angioneurotic edema associated with angiotensin-converting enzyme inhibitor therapy. A review of the literature and pathophysiology. Ann Intern Med 117:234–242 [DOI] [PubMed] [Google Scholar]
- Kim KS, Kumar S, Simmons WH, Brown NJ (2000) Inhibition of aminopeptidase P potentiates wheal response to bradykinin in angiotensin-converting enzyme inhibitor-treated humans. J Pharmacol Exp Ther 292:295–298 [PubMed] [Google Scholar]
- Kostis JB, Packer M, Black HR, Schmieder R, Henry D, Levy E (2004) Omapatrilat and enalapril in patients with hypertension: the Omapatrilat Cardiovascular Treatment vs. Enalapril (OCTAVE) trial. Am J Hypertens 17:103–111 [DOI] [PubMed] [Google Scholar]
- Lefebvre J, Murphey LJ, Hartert TV, Jiao Shan R, Simmons WH, Brown NJ (2002) Dipeptidyl peptidase IV activity in patients with ACE-inhibitor-associated angioedema. Hypertension 39:460–464 [DOI] [PubMed] [Google Scholar]
- Molinaro G, Boileau G, Adam A (2004) Aminopeptidase P and vasoactive peptides. In: Hooper NM, Lendeckel (eds) Aminopeptidases in biology and disease. Kluwer Academic/Plenum, New York [Google Scholar]
- Molinaro G, Cugno M, Perez M, Lepage Y, Gervais N, Agostoni A, Adam A (2002) Angiotensin-converting enzyme inhibitor-associated angioedema is characterized by a slower degradation of des-arginine(9)-bradykinin. J Pharmacol Exp Ther 303:232–237 [DOI] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G (1997) Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng 10:1–6 [DOI] [PubMed] [Google Scholar]
- Nussberger J, Cugno M, Amstutz C, Cicardi M, Pellacani A, Agostoni A (1998) Plasma bradykinin in angio-oedema. Lancet 351:1693–1697 [DOI] [PubMed] [Google Scholar]
- Simmons WH, Orawski AT (1992) Membrane-bound aminopeptidase P from bovine lung. Its purification, properties, and degradation of bradykinin. J Biol Chem 267:4897–4903 [PubMed] [Google Scholar]
- Soria JM, Almasy L, Souto JC, Tirado I, Borell M, Mateo J, Slifer S, Stone W, Blangero J, Fontcuberta J (2000) Linkage analysis demonstrates that the prothrombin G20210A mutation jointly influences plasma prothrombin levels and risk of thrombosis. Blood 95:2780–2785 [PubMed] [Google Scholar]
- Sprinkle TJ, Caldwell C, Ryan JW (2000) Cloning, chromosomal sublocalization of the human soluble aminopeptidase P gene (XPNPEP1) to 10q25.3 and conservation of the putative proton shuttle and metal ligand binding sites with XPNPEP2. Arch Biochem Biophys 378:51–56 [DOI] [PubMed] [Google Scholar]
- Sprinkle TJ, Stone AA, Venema RC, Denslow ND, Caldwell C, Ryan JW (1998) Assignment of the membrane-bound human aminopeptidase P gene (XPNPEP2) to chromosome Xq25. Genomics 50:114–116 [DOI] [PubMed] [Google Scholar]
- Unger T, Gohlke P (1994) Converting enzyme inhibitors in cardiovascular therapy: current status and future potential. Cardiovasc Res 28:146–158 [DOI] [PubMed] [Google Scholar]
- Venema RC, Ju H, Zou R, Venema VJ, Ryan JW (1997) Cloning and tissue distribution of human membrane-bound aminopeptidase P. Biochim Biophys Acta 1354:45–48 [DOI] [PubMed] [Google Scholar]
- Vleeming W, van Amsterdam JG, Stricker BH, de Wildt DJ (1998) ACE inhibitor-induced angioedema: incidence, prevention and management. Drug Saf 18:171–188 [DOI] [PubMed] [Google Scholar]
- Yvert G, Brem RB, Whittle J, Akey JM, Foss E, Smith EN, Mackelprang R, Kruglyak L (2003) Trans-acting regulatory variation in Saccharomyces cerevisiae and the role of transcription factors. Nat Genet 35:57–64 [DOI] [PubMed] [Google Scholar]