Abstract
We obtained conclusive linkage of Alzheimer disease (AD) with a candidate region of 19.7 cM at 7q36 in an extended multiplex family, family 1270, ascertained in a population-based study of early-onset AD in the northern Netherlands. Single-nucleotide polymorphism and haplotype association analyses of a Dutch patient-control sample further supported the linkage at 7q36. In addition, we identified a shared haplotype at 7q36 between family 1270 and three of six multiplex AD-affected families from the same geographical region, which is indicative of a founder effect and defines a priority region of 9.3 cM. Mutation analysis of coding exons of 29 candidate genes identified one linked synonymous mutation, g.38030G→C in exon 10, that affected codon 626 of the PAX transactivation domain interacting protein gene (PAXIP1). It remains to be determined whether PAXIP1 has a functional role in the expression of AD in family 1270 or whether another mutation at this locus explains the observed linkage and sharing. Together, our linkage data from the informative family 1270 and the association data in the population-based early-onset AD patient-control sample strongly support the identification of a novel AD locus at 7q36 and re-emphasize the genetic heterogeneity of AD.
Alzheimer disease (AD [MIM 104300]) is a common neurodegenerative disorder of the CNS that is characterized by slowly progressing impairment of memory and intellectual functioning that leads to dementia and, ultimately, death. Clinical symptoms of AD most frequently arise after age 65 years (late-onset AD); however, in <1% of the patients, AD develops before age 65 years (early-onset AD) (Ott et al. 1998). A definite diagnosis of AD can be established only when pathological examination of autopsied brain shows parenchymal senile plaques composed largely of amyloid β and of intraneuronal neurofibrillary tangles of hyperphosphorylated tau. Thus far, positional cloning in multigeneration families with AD has identified three genes, coding for the amyloid precursor protein (APP [MIM 104760]) (21q21) (Goate et al. 1991), presenilin 1 (PSEN1 [MIM 104311]) (14q23) (Sherrington et al. 1995), and presenilin 2 (PSEN2 [MIM 600759]) (1q42.3) (Levy-Lahad et al. 1995; Rogaev et al. 1995). Currently, 172 different mutations in APP, PSEN1, and PSEN2 have been documented in 363 families (Cruts and Van Broeckhoven 1998; Alzheimer Disease & Frontotemporal Dementia Mutation Database). In addition, the ɛ4 allele of the apolipoprotein E gene (APOE [MIM 107741]) (19q13) was identified as a risk factor for both early- and late-onset AD (Saunders et al. 1993; Strittmatter et al. 1993; van Duijn et al. 1994a). For ∼7%–20% of patients, AD is attributable to APOE ɛ4 (Slooter et al. 1998; Warwick et al. 2000). Since the genetic cause remains unknown in a significant percentage of patients with familial AD, including large families with well-documented autosomal dominant AD (Pericak-Vance et al. 2000; Ashley-Koch et al. 2005; Jimenez-Escrig et al. 2005), it is clear that additional AD genes remain to be identified.
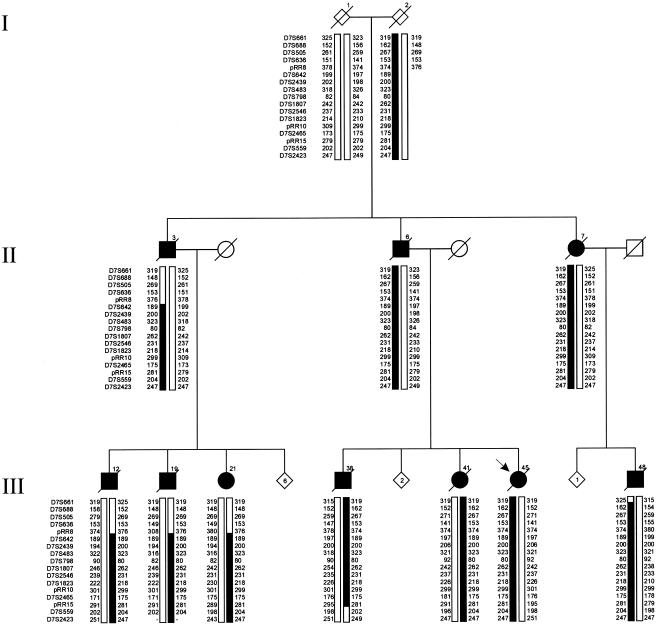
The present study was designed to identify additional loci for AD by positional cloning in multigenerational families. The source for our project was a sample of 110 patients with early-onset AD from a population-based epidemiology study aiming to ascertain all patients with early-onset AD in Rotterdam and the four northern provinces of The Netherlands (Hofman et al. 1989). From this sample, we had previously ascertained 10 multigenerational families for linkage studies (van Duijn et al. 1994b). In two families, a mutation in PSEN1 cosegregated with the disease, whereas mutations in APP and PSEN2 were absent (van Duijn et al. 1991; Cruts et al. 1998). Two other families, 1083 and 1270, were selected for further linkage studies, since simulation linkage data showed that each single family was sufficiently informative to generate significant LOD scores independently. In family 1083, genomewide linkage analysis identified significant linkage at 17q21 (multipoint LOD score 5.41 at D17S951; θ=0.0) (Rademakers et al. 2002). Here, we report on the clinical follow-up and the genetic analyses of the three-generation family 1270. This family was ascertained through its 47-year-old proband with early-onset AD; a total of nine patients and seven at-risk individuals had cooperated, and blood samples had been collected for genetic linkage studies (van Duijn et al. 1994b). A clinical follow-up of family 1270 was performed by neurological examination of incident patients, interviews of first-degree relatives, and review of medical records. Diagnoses of possible or probable AD were made according to National Institute of Neurological and Communicative Diseases and Stroke–Alzheimer's Disease and Related Disorders Association (NINCDS-ADRDA) criteria (McKhann et al. 1984), and that of mild cognitive impairment (MCI) according to Honig and Mayeux (2001). For five patients who were deceased or could not be examined, diagnosis was complemented by information obtained through a family informant. Since the first report (van Duijn et al. 1994b), four family members (III-19, III-21, III-41, and III-43) became symptomatic. The updated pedigree of family 1270 is shown in figure 1; clinical data is summarized in table 1. The overall clinical picture of family 1270 was compatible with AD, with a mean (± SD) age at onset of 66.8±7.4 years (N=13; age range 47–77 years) and a mean duration of disease of 6.7±2.7 years (N=10; range of duration 4–13 years). In proband III-45 and most other patients, the disease initially presented with memory impairment, except for patient III-38, in whom a change of character was the initial complaint, later followed by memory loss. In all patients, the disease progressed into other areas of cognition, such as praxis and speech. Neuroimaging was available for two patients, III-48 at age 74 years and III-21 at age 82 years (table 1), which showed cortical atrophy in both patients. For the only living patient, III-21, who received the diagnosis of possible AD, a recent CT scan (at age 82 years) showed that cortical and subcortical atrophy was most notable in the temporal and frontal regions (fig. 2). No brain pathology of deceased patients was available for family 1270. Since previous studies have already described mutations in the genes encoding the microtubule-associated protein tau (MAPT [MIM 157140]) and the prion protein (PRNP [MIM 176640]) in patients who received a clinical diagnosis of AD but a pathological diagnosis, at autopsy, of frontotemporal dementia (MIM 600274) or Creutzfeld-Jacob disease (MIM 123400) (Dermaut et al. 2000; Rademakers et al. 2003), the identification of the pathology underlying the dementia in family 1270 will be important for the definite classification of the inherited dementia in this family.
Figure 1.
Pedigree of family 1270. A blackened symbol represents a patient, and an unblackened symbol represents an unaffected individual or an at-risk individual with unknown phenotype. The Roman numbers to the left of the pedigree denote generations; Arabic numbers above the symbols denote individuals. All numbering is in accordance with that of van Duijn et al. (1994b). The Arabic numbers below the symbols denote age at onset for patients and either age at last examination or age at death for unaffected individuals and at-risk individuals with unknown phenotype. An asterisk (*) indicates an individual who was included in the linkage analysis. The arrow identifies the proband with early-onset AD (age 47 years).
Table 1.
Clinical Characteristics of Patients of Family 1270
|
Age (in years) at |
|||||
| Patient | Sex | Disease Onset | Death | APOEGenotypea | Diagnosis |
| II-1 | F | 68 | 72 | … | Dementiab |
| II-3 | M | 71 | 77 | … | MCIb |
| II-6 | M | 63 | 74 | … | Dementiab |
| II-7 | F | 65 | 70 | … | Dementiab |
| III-12 | M | 77 | >82 | ɛ3ɛ3 | Probable AD |
| III-19c | M | 75 | 80 | ɛ3ɛ3 | MCIb |
| III-21c | F | 72 | … | ɛ4ɛ4 | Possible AD |
| III-36 | M | 65 | 70 | … | Possible AD |
| III-38 | M | 64 | 72 | ɛ3ɛ4 | Probable AD |
| III-41c | M | 68 | 72 | ɛ3ɛ3 | Probable AD |
| III-43c | F | 65 | 71 | … | Dementiab |
| III-45 | F | 47 | 60 | ɛ4ɛ4 | Probable AD |
| III-48 | M | 69 | >75 | ɛ3ɛ4 | Probable AD |
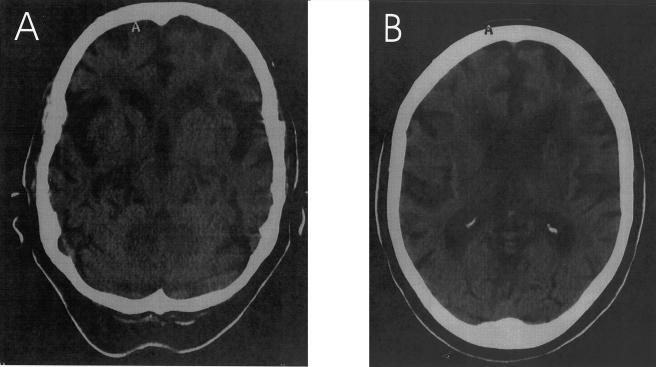
Figure 2.
Brain CT scan of patient III-21 at age 82 years, showing marked cortical and moderate subcortical atrophy, most pronounced in the temporal and frontal regions, with secondary dilatation of the lateral and third ventricles. Mainly frontally localized periventricular leukoencephalomalacia is present as well.
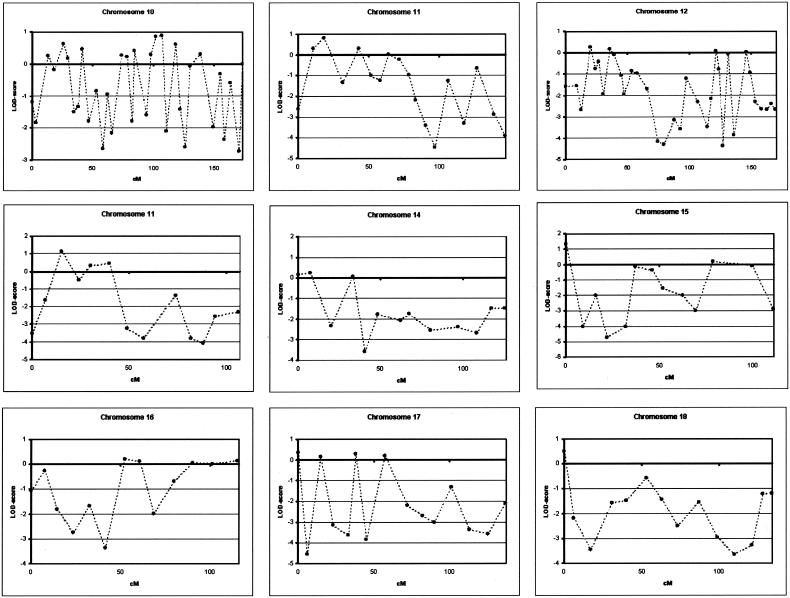
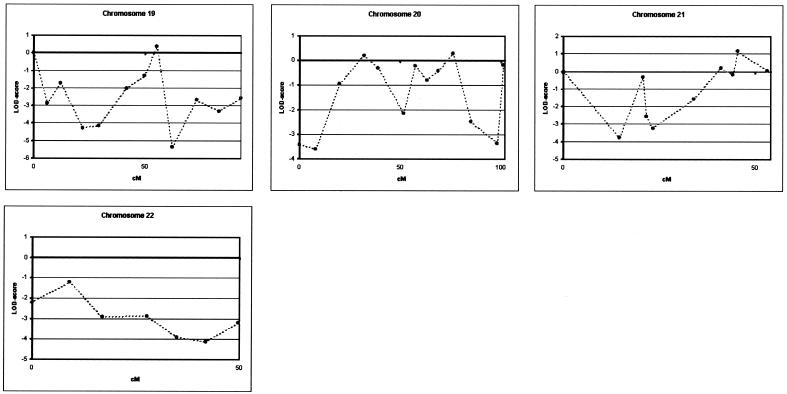
We performed a genomewide linkage analysis of family 1270, using 400 microsatellite markers of the ABI Prism Linkage Mapping Set MD-10 Version 2 (Applied Biosystems). Microsatellite markers were analyzed on an automated ABI 377 or ABI 3700 DNA-Analyzer (Applied Biosystems). For allele identification and scoring, GENESCAN and GENOTYPER were used (Applied Biosystems). We calculated linkage analysis with the assumption of an autosomal dominant mode of inheritance and a disease frequency of 0.1%. A cumulative risk curve was calculated with use of mean age at onset in the family and the assumption of a maximal disease penetrance of 85% at age ⩾85 years, and five disease penetrance classes were defined. Age-dependent phenocopy rates were defined on the basis of the literature (Ott et al. 1995). All patients with AD and MCI were considered affected in the linkage analysis. The phenotypes of individuals I-1 and I-2 were considered unknown. Two-point and multipoint LOD scores were calculated using MLINK and LINKMAP options of the computer package Linkage, version 5.1 (Lathrop et al. 1985), with use of equal recombination rates for males and females. Using this linkage model, we obtained a genomewide conclusive LOD score of 3.39, at θ=0.00, only with marker D7S798 at 7q36 (table 2). The flanking markers, D7S636 and D7S2465, also showed positive LOD scores of 2.69 and 1.70, respectively, at θ=0.00. A summary of all two-point LOD scores obtained in the genomewide scan is provided in figure 3. To fine map the candidate region at 7q36, we analyzed an additional 13 microsatellite markers in a 26.9-cM interval flanked by markers D7S661 and D7S2423, including 9 markers from the Marshfield (Center for Medical Genetics) sex-averaged linkage map, 1 marker from the genetic map of the Cooperative Human Linkage Center, and 3 novel markers—pRR8, pRR10, and pRR15—identified using the Tandem Repeat Finder program (table 2) (Benson 1999). The highest two-point LOD score, 3.41, was obtained with D7S1807 in the absence of recombinants. The maximum multipoint LOD score was 3.47 (data not shown). By use of the genotype data of 17 markers, haplotypes at 7q36 were reconstructed, with consideration of minimal intermarker recombination (fig. 4). In the linkage pedigree, a disease haplotype was present in all patients, and obligate recombinants defined a candidate region of 19.7 cM between pRR8 (individuals III-12, III-19, and III-21) and D7S559 (individual III-38). To estimate the contribution of the 7q36 locus in the Dutch population, we genotyped eight microsatellite markers in 102 patients with AD from the population-based early-onset AD sample, recruited from the same geographical region as family 1270 (Hofman et al. 1989), and 187 age- and sex-matched control individuals, drawn from a large Dutch population-based cohort study of chronic diseases of the elderly (Hofman et al. 1991). Markers D7S1807 and D7S483 were excluded from further analysis: D7S1807 because the linked allele was not observed in any of the patients with early-onset AD, which indicated that the linked allele is extremely rare (frequency <1%) in AD-affected individuals, and D7S483 because >20 different alleles were observed, which is indicative of genetic instability. When we calculated single-marker associations with the remaining six markers in the Dutch early-onset AD patient-control sample with use of Genepop, we identified significant allelic association with D7S798 (P=.03) (table 3). This association could be explained in part by an increase of the rare 80-bp allele, also linked to AD in family 1270, from 1.9% in control individuals to 4.9% in patients with early-onset AD (Pspecific=.04). Examination of individual genotype data of D7S798 revealed that 4 of the 10 patients with early-onset AD who carried the 80-bp allele at D7S798 were probands of families with autosomal dominant AD. We estimated that, in the Dutch patient-control sample, the risk of AD for subjects carrying the 80-bp allele was 2.7 times higher (OR 2.69; 95% CI 1.01–7.17) than for noncarriers.
Table 2.
Two-Point LOD Scores for 7q36 Markers in Family 1270
| LOD at θ= |
||||||||
| Markera | Distanceb(cM) | .00 | .01 | .05 | .10 | .20 | .30 | .40 |
| D7S661 | .0 | −1.31 | −.93 | −.40 | −.15 | .01 | .02 | .01 |
| D7S668 | 4.4 | −.68 | .34 | .97 | 1.15 | 1.09 | .78 | .32 |
| D7S505 | 6.1 | .30 | 1.28 | 1.76 | 1.80 | 1.52 | 1.03 | .43 |
| D7S636 | 7.2 | 2.69 | 2.65 | 2.46 | 2.22 | 1.68 | 1.08 | .43 |
| pRR8c | … | −1.08 | −.09 | .53 | .73 | .70 | .45 | .15 |
| D7S642 | 7.2 | 1.26 | 1.29 | 1.32 | 1.26 | 1.01 | .64 | .24 |
| D7S2439 | 8.6 | 2.88 | 2.83 | 2.62 | 2.34 | 1.74 | 1.09 | .42 |
| D7S483 | 10.1 | 3.32 | 3.27 | 3.04 | 2.74 | 2.08 | 1.34 | .55 |
| D7S798 | 13.9 | 3.39 | 3.33 | 3.11 | 2.81 | 2.14 | 1.38 | .57 |
| D7S1807d | … | 3.41 | 3.36 | 3.14 | 2.84 | 2.16 | 1.40 | .58 |
| D7S2546 | 17.9 | 2.51 | 2.47 | 2.26 | 1.99 | 1.41 | .82 | .28 |
| D7S1823 | 18.6 | 2.72 | 2.69 | 2.52 | 2.29 | 1.74 | 1.12 | .44 |
| pRR10c | … | 1.95 | 1.90 | 1.73 | 1.52 | 1.07 | .61 | .20 |
| D7S2465 | 25.1 | 1.70 | 1.67 | 1.56 | 1.41 | 1.06 | .65 | .23 |
| pRR15c | … | 3.11 | 3.06 | 2.85 | 2.57 | 1.95 | 1.26 | .52 |
| D7S559 | 26.9 | 1.41 | 1.61 | 1.84 | 1.82 | 1.49 | .98 | .38 |
| D7S2423 | 26.9 | −1.04 | −.82 | −.45 | −.28 | −.14 | −.06 | −.02 |
Markers from the 10-cM genomewide scan are in bold italics.
Genetic distances were deduced from the Marshfield (Center for Medical Genetics) sex-averaged linkage map.
pRR8 is located in AC010973.6 starting at nt 93259, pRR10 in AC078834.5 at nt 132119, and pRR15 in AC005534.2 at nt 148849. The number of different alleles observed within family 1270 was three for pRR10, four for pRR8, and five for pRR15.
Selected from the genetic map of the Cooperative Human Linkage Center.
Figure 3.
Genomewide linkage data of the 10-cM density scan of family 1270, with use of the ABI Linkage Mapping Set MD10, version 2, across all chromosomes. Linkage analysis at chromosomes 10, 12, and 21 was performed at a 5-cM density, with use of markers from the ABI Linkage Mapping Set HD-5, version 2. Two-point LOD scores for all markers were calculated using MLINK of the computer package Linkage, version 5.1, and are shown relative to their position on the genetic map (Dib et al. 1996).
Figure 4.
Disguised linkage pedigree of family 1270 (fig. 1). Haplotypes are based on 17 informative markers at 7q36. Haplotypes for individuals from the first and second generations were inferred from genotype data of siblings and offspring. The risk haplotype was arbitrarily set for individual I-2. For confidentiality reasons, haplotypes are shown only for patients; the number of at-risk individuals included in the genotyping are indicated by numbers within diamonds.
Table 3.
Allelic Frequencies and Association Analysis of Dutch Patient-Control Sample of Early-Onset AD, at 7q36
|
Allele Frequencya(%) |
Pb |
|||
| Marker and Allele (bp) | Patientsc(N=204) | Control Individuals (N=372) | Overall | Specific |
| D7S642: | .53 | |||
| 189 | 41.2 | 45.1 | .40 | |
| 191 | 13.7 | 14.2 | ||
| 193 | 1.0 | 2.2 | ||
| 195 | 23.5 | 21.8 | ||
| 197 | 18.6 | 13.2 | ||
| 199 | 2.0 | 3.0 | ||
| 201 | .0 | .3 | ||
| D7S2439: | .12 | |||
| 191 | .0 | .3 | ||
| 193 | .0 | .3 | ||
| 195 | 28.4 | 28.0 | ||
| 197 | 5.9 | 4.8 | ||
| 199 | 24.5 | 19.4 | ||
| 201 | 29.4 | 27.7 | .71 | |
| 203 | 1.5 | 7.0 | ||
| 205 | 2.0 | 4.6 | ||
| 207 | 2.9 | 2.7 | ||
| 209 | 4.9 | 4.0 | ||
| 211 | .5 | .8 | ||
| 213 | .0 | .3 | ||
| 215 | .0 | .3 | ||
| D7S798: | .03 | |||
| 76 | .0 | .8 | ||
| 78 | 6.4 | 5.6 | ||
| 80 | 4.9 | 1.9 | 0.04 | |
| 82 | 10.8 | 14.8 | ||
| 84 | 23.0 | 24.7 | ||
| 86 | .0 | 3.2 | ||
| 88 | 8.8 | 10.8 | ||
| 90 | 8.3 | 8.3 | ||
| 92 | 34.8 | 28.2 | ||
| 94 | 2.5 | 1.3 | ||
| 96 | .5 | .3 | ||
| D7S2546: | .78 | |||
| 228 | .5 | .3 | ||
| 230 | .5 | .0 | ||
| 232 | 18.6 | 18.5 | 1.00 | |
| 234 | 24.5 | 26.6 | ||
| 236 | 9.3 | 11.6 | ||
| 238 | 34.8 | 32.5 | ||
| 240 | 10.3 | 9.7 | ||
| 242 | 1.5 | .5 | ||
| 244 | .0 | .3 | ||
| D7S1823: | .48 | |||
| 201 | .5 | .0 | ||
| 205 | 11.8 | 14.5 | ||
| 209 | 15.2 | 14.5 | ||
| 213 | 3.9 | 4.6 | ||
| 217 | 12.3 | 12.9 | .90 | |
| 221 | 17.2 | 14.5 | ||
| 225 | 18.6 | 21.2 | ||
| 229 | 13.7 | 14.0 | ||
| 233 | 6.4 | 2.7 | ||
| 237 | .5 | 1.1 | ||
| D7S2465: | .25 | |||
| 158 | .0 | .3 | ||
| 160 | .5 | .3 | ||
| 161 | 3.4 | 1.1 | ||
| 164 | 2.0 | 1.6 | ||
| 168 | 1.0 | .0 | ||
| 169 | 1.0 | .5 | ||
| 171 | 3.9 | 5.1 | ||
| 173 | 22.1 | 22.8 | ||
| 174 | .5 | .5 | ||
| 175 | 23.5 | 26.1 | .54 | |
| 176 | 1.0 | .5 | ||
| 177 | 4.9 | 6.5 | ||
| 178 | 17.6 | 12.6 | ||
| 179 | 4.9 | 2.2 | ||
| 180 | 5.4 | 8.6 | ||
| 181 | 1.5 | 1.9 | ||
| 182 | 1.0 | 3.2 | ||
| 183 | 2.0 | 3.0 | ||
| 184 | 3.9 | 3.0 | ||
| 186 | .0 | .3 | ||
N represents the number of chromosomes analyzed.
Association analyses of the linked allele versus all other alleles is shown as specific P values.
In the population with early-onset AD, six patients were probands of families with multiplex AD, which is consistent with autosomal dominant inheritance (van Duijn et al. 1994b); they were not included in the genomewide scan, because none had individually sufficient genetic information to detect conclusive linkage. In all six probands, the onset of AD was at age ⩽65 years (van Duijn et al. 1994b). In three families (1034, 1104, and 1125), the mean age at onset of the relatives was <65 years, whereas the three other families (1027, 1094, and 1242) showed a mean onset of dementia at age >65 years (table 4). The genotypes of the families with multiplex AD were compared for allele sharing with those of family 1270 (table 4). In three families (1034, 1125, and 1242), allele sharing was observed of the rare 80-bp allele at D7S798, as well as the linked alleles at two neighboring markers (D7S2439 and D7S2546). The frequency of the shared haplotype that was calculated on the basis of the frequencies of the linked alleles in Dutch control individuals was 0.6%. Also, all six markers were genotyped in additional patients as well as relatives of families 1034 (2 patients), 1125 (2 patients), and 1242 (1 patient), and segregation analysis of the shared alleles was performed to define haplotypes (fig. 5). In family 1034, all three patients with AD (III-4, III-6, and III-8) inherited the 201-80-232 haplotype. Segregation analysis of family 1242 revealed that both patients (III-14 and III-20) carried the 80-232 haplotype. The lack of sufficient segregation information precluded haplotype determination in family 1125. However, allele sharing in the two additional patients from that family identified the linked 201-80-232 alleles in one patient (III-1), whereas the other patient (III-2) shared the 201-80 alleles. Together, these studies delineated a priority region—between markers D7S2439 and D7S2546, with a maximum genetic distance of 9.3 cM—that is shared by families 1270, 1034, 1242, and 1125. Two-marker sliding-window haplotype analysis in the early-onset AD patient-control sample, with use of the expectation maximization algorithm provided by the Arlequin package, further revealed that the 201-80 haplotype of D7S2439-D7S798 significantly increased from 0.5% in control individuals to 3.4% in patients with AD (Pspecific=.007; OR 6.52; 95% CI 1.34–31.71). The haplotype-sharing data of the four Dutch families with early-onset AD is suggestive of a common founder for AD that is linked to the 7q36 locus in the Dutch population. We documented elsewhere the founder effects within the Dutch population with early-onset AD, since all patients carrying the PSEN1 Ala79Val mutation or the Glu318Gly polymorphism shared an extended haplotype (Cruts et al. 1998; Dermaut et al. 1999). Comparison of the onset ages in the four families with multiplex AD also identified relatively similar mean ages at onset (58.0, 61.3, 66.8, and 72.0 years), with wide intrafamilial ranges (14–30 years) that showed patients with both early- and late-onset AD in each family, which further supported a common phenotype in these families. Previous studies of families with AD have shown that the age at onset can be influenced by the APOE genotype, with APOE ɛ4 carriers having an age at onset earlier than that of noncarriers of APOE ɛ4 (Corder et al. 1993; van Duijn et al. 1994a). However, since ages at onset were 47 and 72 years for two patients homozygous for APOE ɛ4 in family 1270 (table 1), it is unlikely that the broad onset range in the patients with AD in this family is explained by APOE genotypes. Also, in the three families who share a disease haplotype at 7q36, all patients with available DNA were homozygous for APOE ɛ4, whereas ages at onset varied from 51 to 80 years (van Duijn et al. 1994a).
Table 4.
Allele-Sharing Analysis of Microsatellite Markers at 7q36[Note]
|
Genotype (Alleles in bp) of Autosomal Dominant Family |
||||||||
| Marker | Linked Allele in Family 1270 (bp) | Frequency of Linked Allele in Control Individuals(%) | 1027a | 1034b | 1094c | 1104d | 1125e | 1242f |
| D7S642 | 189 | 45.4 | 189–191 | 189–191 | 189–191 | 189–197 | 195–195 | 191–191 |
| D7S2439 | 201 | 29.4 | 201–201 | 201–201 | 195–195 | 199–209 | 199–201 | 199–201 |
| D7S798 | 80 | 1.9 | 82–84 | 80–88 | 84–84 | 90–92 | 80–82 | 80–92 |
| D7S2546 | 232 | 18.6 | 238–238 | 232–236 | 195–195 | 199–209 | 232–238 | 232–232 |
| D7S1823 | 217 | 12.3 | 217–225 | 221–225 | 84–84 | 90–92 | 221–229 | 209–237 |
| D7S2465 | 175 | 23.5 | 180–184 | 173–176 | 169–173 | 175–183 | 173–177 | 173–177 |
Note.— The linked alleles of family 1270 are in bold italics. Ages at onset are taken from the publication by van Duijn et al. (1994b).
Number of patients with dementia=3; mean (± SD) age at onset=72.3±17.9 years; age range 57–92 years.
Number of patients with dementia=4; mean age at onset=61.3±6.1; age range 56–70 years.
Number of patients with dementia=6; mean age at onset=70.5±8.0; age range 61–80 years.
Number of patients with dementia=9; mean age at onset=51.9±9.7; age range 40–67 years.
Number of patients with dementia=5; mean age at onset=58±8.6; age range 49–70 years.
In family 1242, we analyzed allele sharing in patient III-14, since DNA of the proband (III-20) was no longer available (van Duijn et al. 1994b). Number of patients with dementia=4; mean age at onset=72±7.7; age range 63–80 years.
Figure 5.
Linkage pedigrees of Dutch families with early-onset AD who share haplotypes at 7q36. Blackened symbols represent patients with AD; unblackened symbols represent unaffected individuals or at-risk individuals with unknown phenotype. Roman numbers to the left of the pedigree denote generations; Arabic numbers above the symbols denote individuals. All numbers are in accordance with those used in the complete pedigrees published by van Duijn et al. (1994b). For patients, the age (in years) at onset is shown below the symbol. An asterisk (*) indicates that DNA was available for genetic analysis. The arrow identifies the proband. To protect privacy of at-risk individuals, the shared haplotypes are shown only for patients. Haplotypes for individual III-20 in family 1242 and individual III-4 in family 1034 were reconstructed from genotype data obtained from their siblings and offspring. For family 1125, only genotype data are given, since segregation data were not informative for deduction of haplotypes.
We assembled the genomic sequence of the candidate region in family 1270, between pRR8 and D7S559, using publicly available finished and unfinished sequences of large-insert clones. This resulted in two contiguous sequences, of 4.03 Mb and 1.33 Mb, separated by a sequence gap of ∼80 kb located between D7S1823 and pRR10 (table 2) (Hillier et al. 2003). In family 1270, the candidate region segregating the disease gene at 7q36 was therefore estimated at 5.44 Mb. The priority region between D7S2439 and D7S2546 that was shared by the four dominant families with AD corresponded to an estimated physical size of 2.94 Mb. In the course of this study, the complete sequence assembly of 7q36 became available through public genome annotation databases (National Center for Biotechnology Information [NCBI] Map Viewer; University of California–Santa Cruz [UCSC] Genome Bioinformatics). Although two gaps remained in the public genome assembly (NCBI build 34), no major inconsistencies in sequence order were observed with our independently assembled sequence contigs. Within the 5.44-Mb candidate region, we determined the location and genomic organizations of 29 genes by annotating the candidate sequence contigs on the basis of homology with genes and proteins of human and other species, ESTs, and predicted genes and exons (table 5). These included 20 validated, reviewed, and provisional RefSeq genes located in the 150.2–155.8 Mb region of NCBI build 34 of the UCSC Human Genome Browser (UCSC Genome Bioinformatics). We performed mutation analysis of 28 genes from the candidate region by direct genomic DNA (gDNA) sequencing of PCR amplicons of all coding exons in three patients (III-12, III-41, and III-48) and one control individual of family 1270. On the basis of gDNA and cDNA sequence alignment, intronic PCR primers flanking coding exons were developed using SNPbox (Weckx et al. 2005). For the gene encoding myeloid and lymphoid leukemia 3 (MLL3), mutation analysis was performed by cDNA sequencing in two patients (III-38 and III-41) and one control individual. DNA sequences were determined by cycle sequencing with the BigDye Cycle Sequencing kit (Applied Biosystems) and were analyzed on an ABI 3700 and ABI 3730 DNA analyzer (Applied Biosystems). In total, we identified 167 sequence variations: 24 missense and 21 silent mutations in 18 genes, and 122 intronic variations (data not shown). Seven single-nucleotide changes in five different genes segregated in family 1270 on the disease haplotype (table 6). However, analysis of gDNA of 40 Dutch control individuals indicated that six of the seven sequence variants were common polymorphisms, with frequencies of the linked allele varying from 10.7% to 54.0% (table 6). Only g.38030G→C in exon 10 of PAXIP1, which predicted a silent Ala626 mutation, was absent in the 40 control individuals and in an additional 280 Dutch control individuals, which resulted in a C-allele frequency of <0.16% (<1/640 chromosomes). When we examined the expression profile of PAXIP1 on a multiple-human-tissue northern blot, we identified a 4.5-kb transcript of PAXIP1 in all tissues analyzed, except in liver, in which an alternative PAXIP1 transcript of 1.5 kb was expressed (fig. 6A and 6B). Brain expression analysis of PAXIP1 by RT-PCR identified PAXIP1 transcripts in seven different brain regions, including frontal and temporal regions affected in AD-affected brains (fig. 6C). Although g.38030G→C is located 10 bp upstream of the intron 10 splice-donor site, a preliminary RT-PCR analysis of lymphoblast cDNA of two patients (III-38 and III-41) and one control individual of family 1270 did not suggest aberrant PAXIP1 splicing. PAXIP1 g.38030G→C was also not observed in the sample of patients with early-onset AD, including the three probands of the families with autosomal dominant disease who share the priority region between markers D7S2439 and D7S2546. Since PAXIP1 is outside the shared region at 0.44 Mb downstream of the associated D7S2546, the absence of g.38030G→C in the three probands does not contradict a founder effect. Sequencing of PCR amplicons of all coding exons in gDNA of the probands of the six autosomal dominant families further excluded other coding genetic variants in PAXIP1, except for the known g.56231A→G variant (rs3501); this predicts a Met979Val mutation in exon 18, with a frequency of 26.9% for the G-allele, which was present in two heterozygous individuals and one homozygous autosomal dominant proband. Also, expansion of a polyglutamine repeat in PAXIP1 exon 7 was excluded in six patients of family 1270, by Southern blot analysis of PCR-amplified exon 7 genomic fragments. Since PAXIP1 g.38030G→C was absent in the three families with multiplex AD who potentially share a common founder with family 1270 and since we limited our mutation analysis to coding regions, we cannot exclude the possibility that a mutation in 1 of the other 28 positional candidate genes might be the underlying genetic defect in family 1270. Also, a mutation not detected by genomic sequencing of exons—for example, whole-exon deletion—remains plausible. Further, our current annotation efforts might have missed coding sequences. The assembled sequence of the candidate region still contains a gap of ∼80 kb between the genes INSIG1 and EN2 in which additional genes might be present, although the syntenic region in mouse does not contain genes.
Table 5.
Overview of Analyzed Candidate Genes at 7q36
| Gene Symbol | Gene Name | GenBank RefSeq Number |
| ABCF2 | ATP-binding cassette subfamily F, member 2 | NM_005692 |
| ACTR3B | ARP3 actin-related protein 3 homolog B | NM_020445 |
| ASB10 | Ankyrin repeat and SOCS box-containing protein 10 | NM_080871 |
| CENTG3 | Centaurin, gamma 3 | NM_031946 |
| CRYGN | Crystallin, gamma N | NM_144727 |
| CSGlcA-T | Chondroitin sulfate glucuronyltransferase | NM_130797 |
| DPP6 | Dipeptidylpeptidase 6 | |
| EN2 | Engrailed homolog 2 | NM_001427 |
| GALNT11 | UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase | NM_022087 |
| GALNTL5 | UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like | NM_145292 |
| GBX1 | Gastrulation and brain-specific homeobox protein 1 | |
| HTR5A | 5-hydroxytryptamine (serotonin) receptor 5A | NM_024012 |
| INSIG1 | Insulin-induced gene 1 | NM_198337 |
| LOC155100 | Similar to T-complex protein 1 | |
| LOC349136 | ||
| LOC392842 | ||
| LOC392843 | ||
| LOC402628 | Similar to epidermal fatty acid binding protein | |
| MLL3 | Mixed-lineage leukemia 3 | |
| NUB1 | NEDD8 ultimate buster 1 | |
| PAXIP1 | PAX transcription activation domain interacting protein | NM_170606 |
| PRKAG2 | Protein kinase, AMP-activated, gamma 2 | NM_016118 |
| pRR8 | Proline-rich 8 | NM_016203 |
| RBM33 | RNA-binding motif protein 33 | NM_007349 |
| RHEB | Ras homolog enriched in brain | NM_005614 |
| RNF32 | Ring finger protein 32 | NM_030936 |
| SHH | Sonic hedgehog | NM_000193 |
| SMARCD3 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member | NM_003078 |
| XRCC2 | X-ray repair complementing defective repair in Chinese hamster cells 2 | NM_005431 |
Table 6.
Sequence Variations Segregating on the Disease Haplotype in Family 1270
| Genea, GenBankClone Number,and Variant | Predicted Effect on Amino Acid | LinkedAllele | Frequencybin ControlIndividuals(%) | RestrictionEnzymec |
| ABCF2: | ||||
| AC021097.5: | ||||
| g.16531C→T | Leu→Leu | T | 26.2 | … |
| g.19567G→C | … | C | 33.3 | HinfI |
| GALNTL5: | ||||
| AC074257.5: | ||||
| g.176022A→G | Glu→Gly | G | 19.8 | … |
| DPP6: | ||||
| AC006019.2: | ||||
| g.57910C→T | Gly→Gly | T | 18.1 | … |
| PAXIP1: | ||||
| AC093726.3: | ||||
| g.38030G→C | Ala→Ala | C | .0 | Bsh1236I |
| EN2: | ||||
| AC008060.5: | ||||
| g.89332G→A | Pro→Pro | A | 10.7 | SmaI |
| g.89582T→C | Phe→Leu | C | 54.0 | AluI |
ABCF2: ATP-binding cassette subfamily F, member 2; GALNT11: N-acetylgalactosaminyltransferase 11; DPP6: dipeptidylpeptidase 6; PAXIP1: PAX transcription activation domain interacting protein; EN2: engrailed 2.
Allele frequencies for all variations were estimated in 80 control chromosomes. For g.38030G→C in PAXIP1, an additional 560 control chromosomes were tested.
Control individuals were genotyped by a PCR-RFLP assay and electrophoresis of the PCR fragments on a 2% agarose gel or, if the mutation did not affect a restriction enzyme site, by direct sequencing.
Figure 6.
Expression analysis of PAXIP1. A, Northern blot of multiple tissues (FirstChoice Human Blot 1 [Ambion]) hybridized with pooled PAXIP1 cDNA fragments C1 (exons 1–9) and C2 (exons 8–22)—obtained by PCR amplification with use of primers C1F (5′-CGCCGCGGAGCCTCCCGGGCCGCCG-3′), C1R (5′-CAGGTGGCCAGCAGTTGCTTATCAGACATC-3′), C2F (5′-GAAGGCTTCTTATTGGGATG-3′), and C2R (5′-TGAAGGAAGCGCAGCAGCTCC-3′)—in a mixture of cDNA (brain, liver, testis, colon, and lymphoblast). B, β-Actin. Transcript lengths (kb) are specified at the left and were calculated on the basis of the Millennium Marker bands marked on the blot. C, RT-PCR of RNA extracted from seven different human brain regions and lymphoblasts of PAXIP1 C-terminal fragment C2 (exons 8–22). Predicted PCR fragment length of C2 is 1,487 bp.
Together, our linkage data of the informative family 1270, the association data of the population-based patient-control sample of early-onset AD, and the haplotype-sharing data of probands of three additional families with multiplex AD strongly support the locus at 7q36 as a novel locus for AD. Mutation analysis of all 29 known genes in the candidate region identified only an exonic silent mutation in PAXIP1, segregating in family 1270, which was absent from 640 control chromosomes (mutant allele frequency <0.16%). However, whether PAXIP1 has a functional role in expression of AD in family 1270 remains unclear. The novel AD locus at 7q36 provides further evidence that AD is highly genetically heterogeneous, with genes at different loci contributing to the expression of the phenotype. In addition, it can be expected that the identification of the underlying gene defect at 7q36 might largely contribute to our current understanding of the genetic etiology and pathogenesis of AD.
Acknowledgments
We are grateful to the family members for their cooperation in this study. We also acknowledge Gerwin Roks and Hilda Kornman, for their contributions to the genealogy and ascertainment of family 1270; Sebastiaan Engelborghs, for expert second opinion on the clinical diagnoses; and the Flanders Interuniversity Institute for Biotechnology (VIB8) Genetic Service Facility, for genetic analyses. The research was funded by the Special Research Fund of the University of Antwerp; the Fund for Scientific Research Flanders (FWO-F); Interuniversity Attraction Poles program P5/19 of the Belgian Science Policy Office; the International Alzheimer Research Foundation, Belgium; EU contract LSHM-CT-2003-503330 (APOPIS); and the Netherlands Organization for Scientific Research. R.R., M.C., and J.T. are postdoctoral fellows of the FWO-F.
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- Arlequin, http://lgb.unige.ch/arlequin/ (for estimation of haplotype frequencies)
- Alzheimer Disease & Frontotemporal Dementia Mutation Database, http://www.molgen.ua.ac.be/ADMutations/
- Center for Medical Genetics, http://research.marshfieldclinic.org/genetics/ (for the Marshfield Medical Research Foundation and marker position and extra markers in region of interest)
- Cooperative Human Linkage Center, http://gai.nci.nih.gov/CHLC/ (for extra marker in region of interest)
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for PAXIP1 [accession numbers AC093726.3 and NM_007349.6], ABCF2 [accession number AC021097.5], GALNT11 [accession number AC074257.5], DPP6 [accession number AC006019.2], and EN2 [accession number AC008060.5])
- Genepop, http://wbiomed.curtin.edu.au/genepop/ (for single SNP association analysis)
- NCBI Map Viewer, http://www.ncbi.nlm.nih.gov/mapview/ (for gene position as well as links to sequence and protein function information)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for AD, APP, PSEN1, PSEN2,APOE, MAPT, PRNP, frontotemporal dementia, and Creutzfeld-Jacob disease)
- SNPbox, http://www.snpbox.org/
- UCSC Genome Bioinformatics, http://genome.ucsc.edu/ (for gene position and sequence information)
- VIB8 Genetic Service Facility, http://www.vibgeneticservicefacility.be/
References
- Ashley-Koch AE, Shao Y, Rimmler JB, Gaskell PC, Welsh-Bohmer KA, Jackson CE, Scott WK, Haines JL, Pericak-Vance MA (2005) An autosomal genomic screen for dementia in an extended Amish family. Neurosci Lett 379:199–204 [DOI] [PubMed] [Google Scholar]
- Benson G (1999) Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res 27:573–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak-Vance MA (1993) Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science 261:921–923 [DOI] [PubMed] [Google Scholar]
- Cruts M, Van Broeckhoven C (1998) Presenilin mutations in Alzheimer’s disease. Hum Mutat 11:183–190 [DOI] [PubMed] [Google Scholar]
- Cruts M, van Duijn CM, Backhovens H, Van den BM, Wehnert A, Serneels S, Sherrington R, Hutton M, Hardy J, George-Hyslop PH, Hofman A, Van Broeckhoven C (1998) Estimation of the genetic contribution of presenilin-1 and -2 mutations in a population-based study of presenile Alzheimer disease. Hum Mol Genet 7:43–51 [DOI] [PubMed] [Google Scholar]
- Dermaut B, Croes EA, Rademakers R, Van den BM, Cruts M, Hofman A, van Duijn CM, Van Broeckhoven C (2003) PRNP Val129 homozygosity increases risk for early-onset Alzheimer’s disease. Ann Neurol 53:409–412 [DOI] [PubMed] [Google Scholar]
- Dermaut B, Cruts M, Backhovens H, Lubke U, Van Everbroeck B, Sciot R, Dom R, Martin JJ, Van Broeckhoven C, Cras P (2000) Familial Creutzfeldt-Jakob disease in a patient carrying both a presenilin 1 missense substitution and a prion protein gene insertion. J Neurol 247:364–368 [DOI] [PubMed] [Google Scholar]
- Dermaut B, Cruts M, Slooter AJ, Van Gestel S, De Jonghe C, Vanderstichele H, Vanmechelen E, Breteler MM, Hofman A, van Duijn CM, Van Broeckhoven C (1999) The Glu318Gly substitution in presenilin 1 is not causally related to Alzheimer disease. Am J Hum Genet 64:290–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dib C, Faure S, Fizames C, Samson D, Drouot N, Vignal A, Millasseau P, Marc S, Hazan J, Seboun E, Lathrop M, Gyapay G, Morissette J, Weissenbach J (1996) A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature 380:152–154 [DOI] [PubMed] [Google Scholar]
- Goate A, Chartier-Harlin MC, Mullan M, Brown J, Crawford F, Fidani L, Giuffra L, Haynes A, Irving N, James L (1991) Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer’s disease. Nature 349:704–706 [DOI] [PubMed] [Google Scholar]
- Hillier LW, Fulton RS, Fulton LA, Graves TA, Pepin KH, Wagner-McPherson C, Layman D, et al (2003) The DNA sequence of human chromosome 7. Nature 424:157–164 [DOI] [PubMed] [Google Scholar]
- Hofman A, Grobbee DE, de Jong PT, van den Ouweland FA (1991) Determinants of disease and disability in the elderly: the Rotterdam Elderly Study. Eur J Epidemiol 7:403–422 [DOI] [PubMed] [Google Scholar]
- Hofman A, Schulte W, Tanja TA, van Duijn CM, Haaxma R, Lameris AJ, Otten VM, Saan RJ (1989) History of dementia and Parkinson’s disease in 1st-degree relatives of patients with Alzheimer’s disease. Neurology 39:1589–1592 [DOI] [PubMed] [Google Scholar]
- Honig LS, Mayeux R (2001) Natural history of Alzheimer’s disease. Aging (Milano) 13:171–182 [DOI] [PubMed] [Google Scholar]
- Jimenez-Escrig A, Gomez-Tortosa E, Baron M, Rabano A, Arcos-Burgos M, Guillermo PL, Yusta A, Anta P, Perez I, Hierro M, Munoz DG, Barquero S (2005) A multigenerational pedigree of late-onset Alzheimer’s disease implies new genetic causes. Brain 128:1707–1715 [DOI] [PubMed] [Google Scholar]
- Lathrop GM, Lalouel JM, Julier C, Ott J (1985) Multilocus linkage analysis in humans: detection of linkage and estimation of recombination. Am J Hum Genet 37:482–498 [PMC free article] [PubMed] [Google Scholar]
- Levy-Lahad E, Wasco W, Poorkaj P, Romano DM, Oshima J, Pettingell WH, Yu CE, Jondro PD, Schmidt SD, Wang K (1995) Candidate gene for the chromosome 1 familial Alzheimer’s disease locus. Science 269:973–977 [DOI] [PubMed] [Google Scholar]
- McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM (1984) Clinical diagnosis of Alzheimer’s disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer’s Disease. Neurology 34:939–944 [DOI] [PubMed] [Google Scholar]
- Ott A, Breteler MM, van Harskamp F, Claus JJ, van der Cammen TJ, Grobbee DE, Hofman A (1995) Prevalence of Alzheimer’s disease and vascular dementia: association with education: the Rotterdam study. BMJ 310:970–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott A, Breteler MM, van Harskamp F, Stijnen T, Hofman A (1998) Incidence and risk of dementia: the Rotterdam Study. Am J Epidemiol 147:574–580 [DOI] [PubMed] [Google Scholar]
- Pericak-Vance MA, Grubber J, Bailey LR, Hedges D, West S, Santoro L, Kemmerer B, Hall JL, Saunders AM, Roses AD, Small GW, Scott WK, Conneally PM, Vance JM, Haines JL (2000) Identification of novel genes in late-onset Alzheimer’s disease. Exp Gerontol 35:1343–1352 [DOI] [PubMed] [Google Scholar]
- Rademakers R, Cruts M, Dermaut B, Sleegers K, Rosso SM, Van den BM, Backhovens H, Van Swieten J, van Duijn CM, Van Broeckhoven C (2002) Tau negative frontal lobe dementia at 17q21: significant finemapping of the candidate region to a 4.8 cM interval. Mol Psychiatry 7:1064–1074 [DOI] [PubMed] [Google Scholar]
- Rademakers R, Dermaut B, Peeters K, Cruts M, Heutink P, Goate A, Van Broeckhoven C (2003) Tau (MAPT) mutation Arg406Trp presenting clinically with Alzheimer disease does not share a common founder in Western Europe. Hum Mutat 22:409–411 [DOI] [PubMed] [Google Scholar]
- Rogaev EI, Sherrington R, Rogaeva EA, Levesque G, Ikeda M, Liang Y, Chi H, Lin C, Holman K, Tsuda T (1995) Familial Alzheimer’s disease in kindreds with missense mutations in a gene on chromosome 1 related to the Alzheimer’s disease type 3 gene. Nature 376:775–778 [DOI] [PubMed] [Google Scholar]
- Roks G, Dermaut B, Heutink P, Julliams A, Backhovens H, Van den Broeck M, Serneels S, Hofman A, Van Broeckhoven C, van Duijn CM, Cruts M (1999) Mutation screening of the tau gene in patients with early-onset Alzheimer’s disease. Neurosci Lett 277:137–139 [DOI] [PubMed] [Google Scholar]
- Saunders AM, Strittmatter WJ, Schmechel D, George-Hyslop PH, Pericak-Vance MA, Joo SH, Rosi BL, Gusella JF, Crapper-MacLachlan DR, Alberts MJ (1993) Association of apolipoprotein E allele epsilon 4 with late-onset familial and sporadic Alzheimer’s disease. Neurology 43:1467–1472 [DOI] [PubMed] [Google Scholar]
- Sherrington R, Rogaev EI, Liang Y, Rogaeva EA, Levesque G, Ikeda M, Chi H, Lin C, Li G, Holman K (1995) Cloning of a gene bearing missense mutations in early-onset familial Alzheimer’s disease. Nature 375:754–760 [DOI] [PubMed] [Google Scholar]
- Slooter AJ, Cruts M, Kalmijn S, Hofman A, Breteler MM, Van Broeckhoven C, van Duijn CM (1998) Risk estimates of dementia by apolipoprotein E genotypes from a population-based incidence study: the Rotterdam Study. Arch Neurol 55:964–968 [DOI] [PubMed] [Google Scholar]
- Strittmatter WJ, Saunders AM, Schmechel D, Pericak-Vance M, Enghild J, Salvesen GS, Roses AD (1993) Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc Natl Acad Sci USA 90:1977–1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Duijn CM, de Knijff P, Cruts M, Wehnert A, Havekes LM, Hofman A, Van Broeckhoven C (1994a) Apolipoprotein E4 allele in a population-based study of early-onset Alzheimer’s disease. Nat Genet 7:74–78 [DOI] [PubMed] [Google Scholar]
- van Duijn CM, Hendriks L, Cruts M, Hardy JA, Hofman A, Van Broeckhoven C (1991) Amyloid precursor protein gene mutation in early-onset Alzheimer’s disease. Lancet 337:978 [DOI] [PubMed] [Google Scholar]
- van Duijn CM, Hendriks L, Farrer LA, Backhovens H, Cruts M, Wehnert A, Hofman A, Van Broeckhoven C (1994b) A population-based study of familial Alzheimer disease: linkage to chromosomes 14, 19, and 21. Am J Hum Genet 55:714–727 [PMC free article] [PubMed] [Google Scholar]
- Warwick DE, Payami H, Nemens EJ, Nochlin D, Bird TD, Schellenberg GD, Wijsman EM (2000) The number of trait loci in late-onset Alzheimer disease. Am J Hum Genet 66:196–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weckx S, De Rijk P, Van Broeckhoven C, Del Favero J (2005) SNPbox, a modular software package for large-scale primer design. Bioinformatics 21:385–387 [DOI] [PubMed] [Google Scholar]