Abstract
Multicolor karyotyping technologies, such as spectral karyotyping (SKY) (Schröck et al. 1996; Liyanage et al. 1996) and multiplex (M-) FISH (Speicher et al. 1996), have proved to be extremely useful in prenatal, postnatal, and cancer cytogenetics. However, these technologies have inherent limitations that, in certain situations, may result in chromosomal misclassification. In this report, we present nine cases, which fall into five categories, in which multicolor karyotyping has produced erroneous interpretations. Most errors appear to have a similar mechanistic basis.
SKY and M-FISH are molecular cytogenetic technologies that use combinatorially labeled chromosome-specific paints to differentiate and classify nonhomologous human or mouse chromosomes (reviewed by Lee et al. 2000). SKY simultaneously excites multiple fluorochromes and determines the spectral profile at each pixel of an image by means of an interferometer. M-FISH, on the other hand, excites and detects each of the five employed fluorochromes separately with narrow band-pass excitation/emission filters. The presence or absence of each fluorochrome at a given pixel of an image is evaluated; computerized superimposition of the five fluor layers then is performed. Both SKY and M-FISH subsequently identify chromosomal material by comparing the spectral information obtained with the labeling scheme of the probe set used.
These multicolor karyotyping technologies have already been used to detect subtle interchromosomal rearrangements that were otherwise below the resolution level of conventional banding methods (Veldman et al. 1997; Uhrig et al. 1999) and have been effective in determining the chromosomal composition of ambiguous marker chromosomes (Haddad et al. 1998). Because of the increased number and complexity of chromosomal aberrations, neoplastic cells are a particularly attractive target for multicolor karyotyping (Veldman et al. 1997; Sawyer et al. 1998). Although these technologies are becoming more widely applied to cytogenetic studies and clinical diagnoses (Eils et al. 1998; Lindbjerg Anderson et al. 2000; Lu et al. 2000), their limitations are ill defined.
We present nine cases (table 1) in which multicolor classification yielded misleading results. Our collective analyses suggest that most multicolor karyotyping errors have a similar mechanistic basis. Structural rearrangements, which juxtapose nonhomologous chromosome material, frequently result in overlapping fluorescence at the interface of the translocated segments, a phenomenon sometimes referred to as “flaring” (Lu et al. 2000). This flaring effect can obscure or distort the fluorescence pattern of adjacent chromatin, leading to misinterpretation by the multicolor karyotyping systems.
Table 1.
Cases Illustrating Multicolor Classification Errors[Note]
| Case | Diagnosis | G-Band Classification | Multicolor Analysis | Multicolor Classification | Single-Paint Classification |
| 1 | Mouse fibroblast cell line | Not analyzed | SKY | der(15)t(Y;15;19) | der(15)t(15;19) |
| 2 | Plasma cell leukemia | mara | SKY | der(X)t(X;Y;8) | t(X;8) |
| 3 | Dedifferentiated liposarcoma | mar | M-FISH (S) | der(1 or 12)t(1;5;9;12;15)hsr(12) | der(1 or 12)t(1;5;9;12;15)hsr(1;12) |
| 4 | Pleomorphic adenoma | del(8)(q12q21)b | M-FISH (C) | Undetected | der(8)ins(8;14) |
| 5 | Acute lymphoblastic leukemia | Undetected | SKY | der(12)t(12;21) | der(12)t(8;12) |
| 6 | Adenoid cystic carcinoma | add(1)(p34-35)b | M-FISH (C) | der(1)t(1;4)der(1)t(1;8)der(1)t(1;17) | der(1)t(1;8) |
| 7 | Pleomorphic adenoma | marb | M-FISH (C) | der(17)t(13;17)hsr(12) | der(13)ins(13;12)hsr(12) |
| 8 | Mouse tumor | Not analyzed | SKY | der(9)t(9;19) | t(9;19) |
| 9 | Malignant fibrous histiocytoma | +2r | SKY | r(5;12) | r(9;12) |
| M-FISH (C) | r(3;12),r(9;12),r(12;15) |
Metaphase chromosomes were hybridized with commercially available SKY or M-FISH probe sets. SKY paints were analyzed on an SD200 SpectraCube System (Applied Spectral Imaging), and M-FISH probes were analyzed on a Genus/Chromofluor System (Applied Imaging International). To corroborate structural rearrangements, ⩾10 metaphase cells were analyzed for each case followed by one- or two-color whole-chromosome-painting (WCP) procedures. Misclassifications were identified as discrepancies between the multicolor karyotype results and WCP data. The observed discrepancies fell into one of the following contexts:
-
1.
False insertions at the interface of translocated segments. In case 1, two seemingly complex three-way translocations were observed, involving chromosomes Y, 15, and 19; and 13, 15, and 17. The small intercalated segments from chromosome Y, in the former case, and chromosome 15, in the latter, could not be corroborated by WCP (fig. 1a). In case 2, SKY interpreted Y-chromosomal material between 8 and X chromatin in a der(X)t(X;Y;8). Painting only corroborated the presence of 8 and X material. A more complex scenario was seen in case 3, where multiple supernumerary marker chromosomes were shown, by M-FISH, to contain material from chromosomes 5, 9, 12, and 15 (fig. 1e). Although the relatively large segments of material from chromosomes 5, 9, and 12 were corroborated by conventional WCP, small intercalary chromosome-15 segments could not be validated (fig. 1e).
-
2.
Misclassified small insertions. In case 4, a deletion in 8q was observed by G-banding, whereas M-FISH failed to show any chromosome 8 rearrangements (fig. 1b). When WCP14 was performed to confirm a der(14)t(8;14) detected in the same case, chromosome-14 material was also found inserted into 8q.
-
3.
Misclassified small translocations. Case 5 showed no aberrations by conventional banding, but SKY showed a small segment of chromosome-21 material translocated to 12p13. WCP21 could not confirm this finding, and additional WCP experiments revealed that the translocated segment actually originated from chromosome 8. Case 6 exhibited a similar scenario, involving a small segment translocated to 1p35. In different metaphase cells, M-FISH identified this small segment as being from chromosomes 4, 8, or 17 (fig. 1c). Dual-color painting showed that the added material originated from chromosome 8. No material from either chromosome 4 or chromosome 17 could be detected.
-
4.
Rearrangements involving pericentric regions. Case 7 contained a large marker chromosome that could not be resolved by conventional banding. M-FISH classified it as material from chromosome 12, flanked on the telomeric side by material from chromosome 13 and on the centromeric side by a small segment from chromosome 17 (fig. 1d). Painting confirmed the presence of material from chromosomes 12 and 13 but showed that the segment classified as 17 actually originated from chromosome 13. In case 8 (not shown), a murine tumor, analyzed with the SKY mouse probe set, identified a der(9)t(9;19). Painting corroborated this aberration but also detected a der(19)t(9;19) with a breakpoint close to the centromere, indicating that the observed translocation was apparently balanced.
-
5.
Coamplification of material from nonhomologous chromosomes. The large marker chromosomes in case 3 were shown, by M-FISH, to contain mostly chromosome-12 material with occasional chromosome-1 foci at the edges (fig. 1e). Minimal color spotting was observed on the other M-FISH–interpreted chromosome images, indicating an effective hybridization. Subsequent two-color chromosome-painting experiments demonstrated an interspersed distribution of material from chromosomes 1 and 12 in this chromosome region, suggestive of high copy number, coamplification of material from these two chromosomes. In case 9 (not shown), several supernumerary ring chromosomes were observed, which could not be further resolved by G-banding. SKY analysis classified these as r(5;12), whereas M-FISH analysis classified them as either r(3;12), r(9;12), or r(12;15), in different metaphase cells. Painting for these chromosomes consistently showed rings containing material only from chromosomes 9 and 12 in more than 15 analyzed cells.
Figure 1.
a, Partial karyotype of der(15) and der(13) mouse chromosomes from case 1: pseudocolored chromosomes are on the left, inverted DAPI-banded chromosomes are in the center, and classification-colored chromosomes are on the right. The insertions of material from chromosomes Y and 15, interpreted by SKY, could not be corroborated by WCP (Cambio). The Y-chromosome paint was tested earlier and was found to adequately detect the euchromatic portion of the mouse Y chromosome. b, Partial karyotype of chromosome 8s, from case 4: G-banded chromosomes are on the left, M-FISH–painted chromosomes are in the center. A chromosome 8 from the same case after two-color FISH with chromosome-8 (green) and chromosome-14 (red) WCP probes (Vysis) show insertion of chromosome-14 material in one of the chromosome 8s (right). c, Partial karyotype of a der(1), from case 6. The addition of material from dark G-banded chromosome to the terminus of 1p can be seen by G-banding (left). This material was classified variably, by M-FISH, as being from chromosome 4, 8, or 17 (center). WCP experiments showed that the material originated only from chromosome 8 (right). d, Partial karyotype of a marker chromosome from case 7: the marker chromosome appeared to be comprised of three chromatin components (pseudocolored M-FISH image on left). M-FISH classified these components as arising from chromosomes 17, 12, and 13 (center). WCP experiments showed that the proximal and distal components of this chromosome are both material from chromosome 13 (right). No chromosome-17 material was detectable by WCP (Vysis). e, Partial karyotype of a large marker chromosome from case 3: M-FISH classification suggested that this marker chromosome contained mainly material from chromosomes 9, 12, and 5 (left). WCP experiments showed that the region containing material from chromosome 12 was actually comprised of coamplified material from chromosomes 1 (red) and 12 (green). Light-blue intercalated segments, corresponding to chromosome-15 material, could not be validated with a chromosome-15–specific paint probe (Vysis) and were therefore considered artifactual.
It has been well accepted that SKY/M-FISH is incapable of detecting intrachromosomal rearrangements such as duplications, deletions, or inversions (Uhrig et al. 1999). Furthermore, the resolution of insertions/translocations on metaphase spreads from lymphocytes has been estimated at ∼1 Mb for both SKY (Schröck et al. 1996; Fan et al. 2000) and M-FISH (Jalal and Law 1999). However, several factors can result in suboptimal SKY/M-FISH conditions and lead to an increased propensity for chromosome classification errors. Poor chromosome preparations and inadequate pretreatments can result in increased cytoplasm and inferior probe hybridization. Overdenaturation of the chromosomes disrupts the chromatin compaction, leading to a decrease in the signal intensity for a given locus and increased fluorescence flaring. Drying out of the probe during hybridization can reduce hybridization efficiency and increase background. Finally, the analysis of relatively short chromosomes decreases the physical resolution of detecting translocations/insertions.
In this report, we have presented other limitations for multicolor karyotyping systems that appear to have their basis in algorithmic interpretation of fluorescence flaring. Although this fluorescence flaring can also be seen in conventional chromosome-painting experiments, algorithmic interpretation is not required and thus does not result in similar misinterpretations. Fluorescence flaring, at the boundaries of juxtaposed chromosome regions of different origins, can lead to the false interpretation of additional nonhomologous chromosome fragments. Caution is therefore particularly warranted when the “inserted” chromosomal material has a characteristic fluorochrome combination that is a mixture of the fluorochrome profile of the two adjacent chromosome segments.
Fluorescence flaring also appears to be a significant cause for the misclassification of rearrangements involving small chromosomal segments (fig. 2). Both insertions (case 4) and translocations (cases 5 and 6) involving small euchromatic segments have been missed/misclassified. In the former case, the inserted chromosomal segment was labeled with the same fluors as the receiver chromosome but in a different ratio. In the latter cases, the false classifications had a fluor combination containing a mix of fluors from both translocation partners. A contributing factor may also be that small translocations can be lost during the employment of automatic segmentation masking used by certain M-FISH software. This program subroutine attempts to automatically determine the chromosome contours using the DAPI counterstaining. Fluorescent signals beyond these established boundaries are then considered unimportant and are eliminated during automatic background subtraction. Because DAPI staining is sometimes less intense at the ends of human chromosomes, fluorochrome information at these chromosomal regions may be lost or misinterpreted.
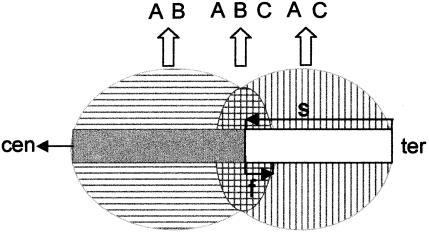
Figure 2.
Fluorescence blending at the breakpoint of a translocation between chromosomes labeled with fluors A + B and A + C, resulting in the false classification of material at the boundary as ABC. The risk of misclassification of the entire translocated segment increases as the size of the translocated segment (s) approaches the range of flaring (f).
Pericentric regions contain highly repetitive DNA sequences, which are shared by several chromosomes (Choo 1997; Lee et al. 1997). Incomplete suppression of these repetitive elements can lead to intense and uninterpretable multicolor fluorescence, capable of blending with the fluorescence profile of the immediately juxtaposed chromosomal region (Eils et al. 1998; Castleman et al. 2000). Mouse chromosomes, with their prominent centromeric heterochromatin, may be more problematic in this regard than human chromosomes.
Detection of coamplified material from nonhomologous chromosomes is difficult, at best, when current multicolor karyotyping systems are used. Depending on the respective size of coamplified segments, two scenarios can be envisioned. Either the array of coamplification will be interpreted as a homogeneous mixture of the fluorochromes used to label the two contributing chromosomes or one of the fluorochrome combinations will prevail during algorithmic data processing. The former scenario leads to misclassification of the entire region, whereas the latter may result in failure to detect one or several amplified components. To our knowledge, there have been no reports of coamplification successfully resolved by multicolor classification alone. Because genomic amplification involving material from at least two chromosomes occurs with significant frequency in tumor cells (Guan et al. 1994), this appears to be an important limitation in multicolor analysis of neoplastic cells.
Multicolor karyotyping generally has a high level of accuracy and therefore is becoming an important tool in the karyotypic analyses of prenatal, constitutional, and tumor cases. It provides an effective screening method for the entire genome in a labor-efficient manner when a priori discrimination of the specific chromosomal aberrations is not available. However, because of these inherent problems of SKY/M-FISH, it is highly recommended that these technologies not be considered as complete on their own and that they be confirmed with other molecular cytogenetic technologies or conventional banding techniques. Careful examination of an associated G-banded/inverse DAPI image is mandatory. Where translocations/insertions are inferred, one- or two-color chromosome-painting experiments should be performed to corroborate the M-FISH/SKY interpretations. This especially applies to interpretations of nonreciprocal rearrangements where two-color painting experiments occasionally demonstrate apparently reciprocal exchanges (cases 4 and 8). If available and warranted, other techniques—such as comparative genomic hybridization or reverse chromosome painting with microdissected/flow-sorted chromosome material (Lu et al. 2000)—could be used to verify the karyotypic information.
Acknowledgments
We are grateful to Dr. J. Wiegant for critical reading of the manuscript. This study was supported by the Swedish Cancer Society, the Swedish Child Cancer Fund, the John and Augusta Persson Foundation, and the Inga Britt and Arne Lundberg Foundation. D.G. was supported by a Young Investigators Award from the American Cancer Society and by grants from the Swedish Medical Society and the Blanceflor Foundation.
References
- Castleman KR, Eils R, Morrison L, Piper J, Saracoglu K, Schulze MA, Speicher MR (2000) Classification accuracy in multiple color fluorescence imaging microscopy. Cytometry 41:139–147 [PubMed] [Google Scholar]
- Choo KHA (1997) The centromere. Oxford University Press, Oxford [Google Scholar]
- Eils R, Uhrig S, Saracoglu K, Sätzler K, Bolzer A, Peterson I, Chassery J-M, Ganser M, Speicher MR (1998) An optimized, fully automated system for fast and accurate identification of chromosomal rearrangements by multiplex-FISH (M-FISH). Cytogenet Cell Genet 82:160–171 [DOI] [PubMed] [Google Scholar]
- Fan Y-S, Siu V, Jung JH, Xu J (2000) Sensitivity of multiple color spectral karyotyping in detecting small interchromosomal rearrangements. Genet Testing 4:9–14 [DOI] [PubMed] [Google Scholar]
- Guan XY, Meltzer PS, Dalton WS, Trent JM (1994) Identification of cryptic sites of DNA sequence amplification in human breast cancer by chromosome microdissection. Nat Genet 8:155–161 [DOI] [PubMed] [Google Scholar]
- Haddad BR, Schröck E, Meck J, Cowan J, Young H, Ferguson-Smith MA, du Manoir S, Ried T (1998) Identification of de novo chromosomal markers and derivatives by spectral karyotyping. Hum Genet 103:619–625 [DOI] [PubMed] [Google Scholar]
- Jalal SM, Law ME (1999) Utility of multicolor fluorescent in situ hybridization in clinical cytogenetics. Genet Med 1:181–186 [DOI] [PubMed] [Google Scholar]
- Jin C, Martins C, Jin Y, Wiegant J, Wennerberg J, Dictor M, Gisselsson D, Strömbeck B, Fonseca I, Mitelman F, Tanke HJ, Höglund M, Mertens F (2001) Characterization of chromosome aberrations in salivary gland tumors by FISH, including multicolor COBRA-FISH. Genes Chromosome Cancer 30:161–167 [PubMed] [Google Scholar]
- Lee C, Rens W, Yang F (2000) Multicolor fluorescence in situ hybridization (FISH) approaches for simultaneous analysis of the entire human genome. In: Dracopoli NC, Haines JL, Korf BR, Morton CC, Seidman CE, Seidman JG, Smith DR (eds) Current protocols in human genetics. John Wiley and Sons, New York, pp 4.9.1–4.9.11. [Google Scholar]
- Lee C, Wevrick R, Fisher RB, Ferguson-Smith MA, Lin CC (1997) Human centromeric DNAs. Hum Genet 100:291–304 [DOI] [PubMed] [Google Scholar]
- Lindbjerg Anderson C, Ostergaard M, Nielsen B, Pedersen B, Koch J (2000) Characterization of three hairy cell leukemia-derived cell lines (ESKOL, JOK-1, and Hair-M) by multiplex-FISH, comparative genomic hybridization, FISH, PRINS, and dideoxyPRINS. Cytogenet Cell Genet 90:30–39 [DOI] [PubMed] [Google Scholar]
- Liyanage M, Coleman A, du Manoir S, Veldman T, McCormack S, Dickson RB, Barlow C, Wynshaw-Boris A, Janz S, Wienberg J, Ferguson-Smith MA, Schröck E, Ried T (1996) Multicolour spectral karyotyping of mouse chromosomes. Nat Genet 14:312–315 [DOI] [PubMed] [Google Scholar]
- Lu YJ, Morris JS, Edwards PA, Shipley J (2000) Evaluation of 24-color multifluor-fluorescence in-situ hybridization (M-FISH) karyotyping by comparison with reverse chromosome painting of the human breast cancer cell line T-47D. Chromosome Res 8:127–132 [DOI] [PubMed] [Google Scholar]
- Nordgren A, Farnebo F, Björkholm M, Sahlén S, Porwit-MacDonald A, Ösby E, Kytölä S, Larsson C, Nordenskjöld M, Blennow E (2000) Detailed characterization of a complex karyotype in a patient with primary plasma cell leukaemia using multicolour spectral karyotyping and micro-FISH. Hemat J 1:95–101 [DOI] [PubMed] [Google Scholar]
- Sawyer JR, Lukacs JL, Munshi N, Desikan KR, Shingal S, Mehta J, Siegel D, Shaughnessy J, Barlogie B (1998) Identification of new nonrandom translocations in multiple myeloma with multicolor spectral karyotyping. Blood 92:4269–4278 [PubMed] [Google Scholar]
- Schröck E, du Manoir S, Veldman T, Schoell B, Wienberg J, Ferguson-Smith MA, Ning Y, Ledbetter DH, Bar-Am I, Soenksen D, Garini Y, Ried T (1996) Multicolor spectral karyotyping of human chromosomes. Science 273:494–497 [DOI] [PubMed] [Google Scholar]
- Speicher MR, Gwyn Ballard S, Ward DC (1996) Karyotyping human chromosomes by combinatorial multi-fluor FISH. Nat Genet 12:368–375 [DOI] [PubMed] [Google Scholar]
- Tanke HJ, Wiegant J, van Gijlswijk RP, Bezrookove V, Pattenier H, Heetebrij RJ, Talman EG, Raap AK, Vrolijk J (1999) New strategy for multi-colour fluorescence in situ hybridization: COBRA: COmbined Binary RAtio labeling. Eur J Hum Genet 7:2–11 [DOI] [PubMed] [Google Scholar]
- Uhrig S, Schuffenhauer S, Fauth C, Wirtz A, Daumer-Haas C, Apacik C, Cohen M, Muller-Navia J, Cremer T, Murken J, Speicher MR (1999) Multiplex-FISH for pre- and postnatal diagnostic applications. Am J Hum Genet 65:448–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldman T, Vignon C, Schröck E, Rowley JD, Ried T (1997) Hidden chromosome abnormalities in hematological malignancies. Nat Genet 15:406–410 [DOI] [PubMed] [Google Scholar]