Abstract
Megalencephalic leukoencephalopathy with subcortical cysts (MLC) is an autosomal recessive disorder characterized by macrocephaly, deterioration of motor functions with ataxia, and spasticity, eventuating in mental decline. The brain appears swollen on magnetic resonance imaging, with diffuse white-matter abnormalities and the invariable presence of subcortical cysts. MLC was recently localized on chromosome 22qtel. We have narrowed down the critical region by linkage analysis of 11 informative families with MLC to a region of ∼250 kb, containing four known genes. One family with two patients who were siblings did not display linkage between the MLC phenotype and any of the analyzed microsatellite markers on chromosome 22qtel, suggesting genetic heterogeneity and the existence of at least a second MLC locus. The maximum two-point LOD score for the 11 families was 6.6 at recombination fraction .02. Twelve different mutations in seven informative and six uninformative families were found in one of the candidate genes, KIAA0027, which we renamed “MLC1.” The gene encodes a putative membrane protein with eight predicted transmembrane domains. The patients of one family were compound heterozygotes for mutations that both introduced stop codons. The mutations further included frameshifts, splice-acceptor mutations, a putative splice-donor mutation, and amino acid substitutions of residues in predicted transmembrane domains. These data provide strong evidence that mutations of MLC1 cause the disease.
Introduction
Vacuolating megalencephalic leukoencephalopathy with subcortical cysts (MLC [MIM 604004]) is a neurologic disorder characterized by macrocephaly within the first year and delayed onset of a slow deterioration of motor functions with ataxia and spasticity (Van der Knaap et al. 1995; Singhal et al. 1996; Topçu et al. 1998). The cognitive functions are relatively spared. Magnetic resonance imaging of the brain is diagnostic and shows diffusely abnormal, mildly swollen cerebral white matter and, invariably, subcortical cysts—in particular, in the anterior-temporal region. A brain biopsy showed spongiform white-matter changes related to the presence of numerous vacuoles between the outer lamellae of myelin sheaths. The findings suggest either splitting of the outer myelin lamellae along the intraperiod line or incomplete compaction (Van der Knaap et al. 1996). The mode of inheritance is autosomal recessive. An MLC gene locus has been recently assigned, by a genomewide linkage analysis, to a 2-Mb interval on chromosome 22qtel (Topçu et al. 2000). Although the study involved families with MLC from a seemingly homogeneous Turkish population, a founder effect was not apparent from the patient haplotypes on chromosome 22qtel. In our study, the critical region on chromosome 22qtel was narrowed down by linkage analysis of a heterogeneous group of MLC families with microsatellite markers. Recombination events in individual families that border the most likely location of the disease gene were positioned more precisely by the analysis of single-nucleotide polymorphisms (SNPs). Candidate genes that were located in the critical region were screened by sequencing of patient DNA, and we found mutations in one gene, KIAA0027, that are responsible for MLC. We propose to rename KIAA0027 as “MLC1.”
Material and Methods
Samples
All patients with MLC were diagnosed by one of us (M.S.v.d.K.), and, with informed consent, blood samples were collected. Sixteen patients with MLC were from 11 informative families of different ethnic origins. Six of these families were of Turkish background. The other five families were from Croatia, Japan, the United Kingdom (with Indian background), France, and Germany. In nine families, the parents were consanguineous. The informative families included five affected sib pairs and 12 healthy siblings. The estimated maximum LOD score in these families was 13.4. We also studied 19 families who were uninformative for linkage. These families, with single patients and a total of 20 healthy siblings, were of different white and Asian origins. Five of these families were from Croatia.
Marker Analysis
Genomic DNA fragments were amplified as described elsewhere (Leegwater et al. 1999). The markers D22S1161 and D22S1169 were described by Dib et al. (1996), and the oligonucleotide sequences for markers D22S445 and UT580 were derived from The Genome Database. The microsatellite markers ARSA and N66c4 were described by Topçu et al. (2000). The positioning of these markers, except for D22S1161, was based on the DNA sequence of chromosome 22 (Dunham et al. 1999). The positioning of D22S1161 was based on the combination of the chromosome 22 DNA sequence and genetic data (Dib et al. 1996). The DNA sequences of the oligonucleotides for the marker 355c18 were 5′-GTGTCCTGTGGGTATTCCAG-3′ and 5′-GAACCAGGGTGCAGTTCTTG-3′. These primers amplify a polymorphic CA repeat around position 26300 of the RP3 PAC clone 355c18. One of the primers for each of the mentioned microsatellite markers had a fluorescent label 6-FAM, HEX or TET at the 5′-end. The PCR products of microsatellite DNA were analyzed with an Applied Biosystems Genetic Analyzer 310. GS-500 TAMRA (Applied Biosystems) was used as a size standard and DNA from CEPH individual 1347-02 was analyzed as a reference. The alleles correspond to the length of the DNA fragments in base pairs. For the markers UT580, 355c18, ARSA, and N66c4, the lengths were derived from the GENESCAN measurements. The primers for PCR amplification of the SNPs rs4469 and rs4624 were as described in the Web site A Database of Single Nucleotide Polymorphisms. SNP rs8238 was amplified with oligonucleotides 5′-ACCGTCTGCCTGCAGGGAT-3′ and 5′-GAACCCCATGTTCACACGTC-3′. The DNA sequence of the SNP fragments was determined by cycle sequencing with one of the PCR primers and Big Dye terminators (Applied Biosystems), according to the instructions of the manufacturer. The alleles correspond to the base at the polymorphic position, as in A Database of Single Nucleotide Polymorphisms.
Mutation Analysis
The DNA sequences of oligonucleotides for PCR amplification of the MLC1 exons and surrounding intron sequences are available, on request, by e-mail. PCR products of MLC1 exons from patients of all MLC families were analyzed for SSCP, as described elsewhere (Wijker et al. 1999). Exon fragments with banding patterns that were different from control fragments were further analyzed by cycle sequencing, as described above. The PCR products of exon 2 from the heterozygous patient M31 were subcloned in pGEMT vector (Promega), as recommended by the supplier. The DNA inserts of subclones were amplified by PCR and sequenced.
Analysis of Brain MLC1 cDNA
Adult whole-brain cDNA was obtained from Clontech (Palo Alto). Overlapping fragments of MLC1 cDNA were amplified by PCR, using standard conditions with an annealing temperature of 55°C. The primer combinations were 114F (5′-CCATGACCCAGGAGCCATTC)-782R (5′-CGATTACCTCGACGACTGAGTAAGA), 618F (5′-AGGAGGACTGCAAGAAAAAGAAGG)-1362R (5′-CTCACACAAGGGAAAAGAGGTGTTA), and 114F-1362R. The primer numbers correspond to the position on Genbank accession number D25217. The PCR products were sequenced with the PCR primers, as described.
Results
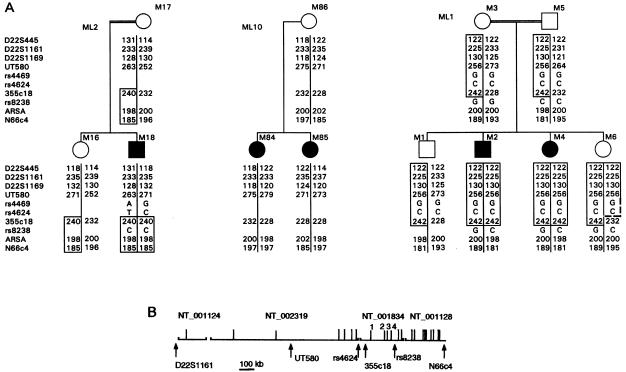
Using linkage analysis, we investigated the location of the MLC gene on chromosome 22qtel in 11 informative families with MLC. One family, ML10, did not display linkage between MLC and chromosome 22qtel. This family included two sibling patients who had different genotypes for all seven microsatellite markers analyzed (fig. 1A). All other informative families displayed linkage between markers on chromosome 22qtel and MLC (data not shown). The newly developed marker 355c18 was the only microsatellite marker that showed overlap between the critical regions of the families with linkage between MLC and chromosome 22qtel. The maximum two-point LOD score for linkage between this marker and MLC was 6.6 at recombination fraction .02. The LOD score was calculated with the data for the 11 informative families, and it proves that an MLC gene on chromosome 22qtel is involved in these families. Analysis of SNPs adjacent to 355c18 narrowed down the critical region (fig. 1). The two sibling patients, M2 and M4, of family ML1 in which the parents are first cousins, are homozygous from D22S445 down to SNP rs8238. The healthy sibling M6 was identical to these patients between D22S445 and UT580, but was a recombinant between markers UT580 and 355c18 (fig. 1A). This family, by itself, placed the most likely location of the MLC gene in a 0.7-Mb interval between markers UT580 and rs8238. Patient M18, whose parents are also first cousins, was homozygous in the chromosomal region with the four most distal markers starting with 355c18. The frequency of the homozygous haplotype was estimated between .02 and .005, indicating that the region was identical by descent in patient M18. Together, families ML1 and ML2 placed the most likely location of the MLC gene on chromosome 22qtel, in a 250-kb region between SNPs rs4624 and rs8238.
Figure 1.
Localization of the MLC gene on chromosome22qtel. A, Chromosome 22qtel haplotypes of three informative families with MLC. Microsatellite and SNP markers are listed on the left, according to their order. The alleles are depicted as PCR fragment length (microsatellites) or DNA sequence at the polymorphic position (SNPs). Haplotypes that are identical by descent in individual patients are indicated by the boxes. In family ML1, there are two patients with parents who are first cousins. The healthy sibling, M6, is a recombinant between markers UT580 and 355c18, indicated by the broken line. Patient M18 also has parents who are first cousins. Together, the two families limit the critical region for MLC between rs4624 and rs8238. The two sibling patients in family ML10 do not share a genotype in the MLC region. B, Physical map of the MLC region. The map is based on the completed and annotated DNA sequence of chromosome 22. Contig numbers and the position of markers are indicated. The gaps between contigs are of unknown length. Vertical lines depict known ESTs and genes. The four candidate EST clusters and genes are numbered: (1) KIAA0027 (“MLC1”); (2) dJ402G11.8 similar to MOV10; (3) dJ402G11.9 similar to MRS1; and (4) dJ402G11.4.
Four genes have been described in the annotated DNA sequence of chromosome 22, between the SNPs rs4624 and rs8238 (fig. 1B). We analyzed, by SSCP analysis and DNA sequencing, two of the three coding exons of dJ402G11.9 and all 11 coding exons of MLC1, of patients from all families with MLC. We found evidence that MLC is caused by mutations in MLC1 (Genbank accession number D25217) (fig. 2 and table 1).
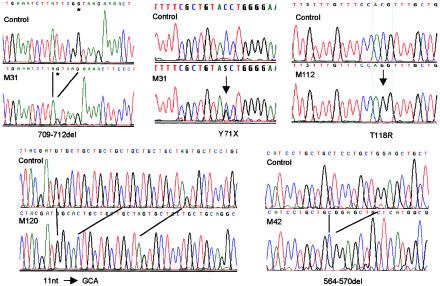
Figure 2.
MLC1 mutations in genomic DNA of patients. Patient M31 was a compound heterozygote for a 4-bp deletion at the 3′ end of exon 7 and a point mutation that led to a stop codon. The allele with the deletion was sequenced after subcloning of the PCR products. The GT sequence of the splice donor is marked by an asterisk in this sequence. Lines indicate homologous residues of control and deletion mutant DNA sequences. The electropherograms were obtained by automated DNA sequencing with PCR products from control and patient DNA templates.
Table 1.
MLC1 Mutations in DNA from MLC Patients
| Patient | Mutationa | Effectb | Exon | State | Origin | Family Type |
| M62 | 954C→T | S280L | 10 | Homozygous | Japan | Informativec |
| M66 | 393C→T | S93L | 4 | Homozygous | Japan | Uninformative |
| M75 | 393C→T | S93L | 4 | Homozygous | Turkey | Uninformative |
| M73 | 393C→T | S93L | 4 | Heterozygous | Japan | Uninformatived |
| M73 | IVS11-2A→G | Aberrant splicing of exon 12 | 12 | Heterozygous | Japan | Uninformativee |
| M2-M4 | IVS10-2A→G | Aberrant splicing of exon 11 | 11 | Homozygous | Turkey | Informativec |
| M112 | 468C→G | T118R | 5 | Homozygous | Turkey | Informativec |
| M14 | IVS5+3insT | Aberrant splicing of exon 5 | 5 | Heterozygous | India | Uninformativee |
| M14 | 749G→A | G212R | 8 | Heterozygous | India | Uninformatived |
| M20 | 564-570del | L149 frameshift | 6 | Homozygous | Turkey | Informativec |
| M39-M42 | 564-570del | L149 frameshift | 6 | Homozygous | Middle East | Informativec |
| M81 | 575insGGAGC | E153 frameshift | 6 | Heterozygous | Yugoslavia | Uninformative |
| M23 | 709-712del | Y198 stop | 7 | Heterozygous | Croatia | Uninformative |
| M31-M32 | 709-712del | Y198 stop | 7 | Heterozygous | Croatia | Informatived |
| M31-M32 | 328C→G | Y71 stop | 3 | Heterozygous | Croatia | Informativee |
| M120 | 1023-1033del+3 | V303 frameshift | 11 | Homozygous | Turkey | Informativef |
Numbers correspond to KIAA0027 cDNA (Genbank accession number D25217).
Numbers correspond to translated KIAA0027, starting with the first methionine codon.
Parents are first cousins.
Maternal allele.
Paternal allele.
Parents are second cousins.
The 12 different mutations in MLC1, found in 13 families with MLC, included formation of stop codons, frameshifts, splice-site mutations, and amino acid substitutions of conserved residues and in predicted transmembrane domains (fig. 2 and table 1). The affected sib-pair M31 and M32 was compound heterozygous for mutations that both introduced stop codons in MLC1 (fig. 2). One stop codon was created in exon 3 by mutation of a C→G residue at codon 71 of the open reading frame (ORF). The mutation that led to a stop codon in the second allele was a 4-bp deletion exactly at the 3′ end of exon 7. The deletion was predicted, by the program SPLICEVIEW (Rogozin and Milanesi 1997), not to affect the efficiency of the exon 7/intron 7 splice donor. The deletion directly led to a stop codon at the exon 7/exon 8 junction. Patient M23, who comes from the same country, was found to be heterozygous for this allele (table 1). Patient M20 and the sib pair M39 and M42 were homozygous for another microdeletion of 7 bp in exon 6 (fig. 2). The deleted 7 bp were part of an 8-bp direct repeat of the DNA sequence TCCTGCTG, separated by a single C residue (fig. 2). The repeats coded for leucine residues of the first of two polyleucine motifs of the MLC1 protein product. Both polyleucine motifs are part of predicted transmembrane domains. Patient M120 was homozygous for a substitution of 11 bp by 3 bp, resulting in a frameshift near the 5′-end of exon 11 (fig. 2). This rearrangement occurred in the region of seven TGC repeats, which encoded part of the second polyleucine motif. Interestingly, the replacement sequence GCA was the reverse complement of the repeat sequence. Patient M81 was heterozygous for an insertion of 5 bp of the sequence GGAGC, next to an inverted repeat with the same sequence in exon 6. The 5-bp insertion occurred at the end of the domain that encoded the first polyleucine motif.
The sibling patients M2 and M4 were homozygous for a mutation of the splice-acceptor AG sequence of exon 11 to GG. The next predicted splice acceptor was 51 nucleotides (nt) downstream from the mutated site. Use of this site would result in a deletion of 17 amino acids that constitute the major part of a predicted transmembrane domain. Similarly, patient M73 was heterozygous for mutation of the AG of the splice-acceptor site of exon 12 to GG. The next predicted splice acceptor was located downstream of the stop codon. If this site were used, the 24 amino acids encoded by exon 12 would be replaced by 27 unrelated residues. Patient M14 was heterozygous for an insertion of a T residue at position +3 of intron 5 (fig. 2). The insertion probably caused incorrect splicing of this intron. The SPLICEVIEW prediction diminished the score from 84% for the wild-type DNA sequence to 0% for the insertion mutant (Rogozin and Milanesi 1997). The next splice donor in the DNA sequence was located 94 bp downstream from the mutated junction. The intron 5 DNA sequence contained an in-frame stop codon and the mutation in patient M14 probably led to a truncated MLC1 gene product of 141 residues, with an addition of 26 unrelated residues at the C-terminal end.
Four additional mutations were found in other patients with MLC that led to amino acid substitutions in the MLC1 gene product (table 1). The serine residue at position 93, which was mutated in three patients from Japan and Turkey (table 1), was conserved between Xenopus and man (fig. 3A). This mutation was not present in 100 chromosomes from unrelated Japanese and 106 chromosomes from unrelated Turkish individuals. The T118R mutation in patient M112, from Turkey, was not present in 106 chromosomes of Turkish origin. This mutation shifted the predicted position of the third transmembrane domain of the protein by 10 residues according to the TMpred program (Hofmann and Stoffel 1993). Patient M62, from Japan, was homozygous for a mutation of the serine residue, at position 280, to a leucine residue. Again, this mutant allele was not present in 100 chromosomes of Japanese origin. Patient M14, of Indian background, was compound heterozygous for the splice-donor mutation described above and for mutation of the glycine residue at position 212 to an arginine residue (table 1). This mutation was not present in 102 chromosomes from unrelated individuals from the same region. The four amino acid substitutions found in the MLC1 gene segregate with the disease. The amino acids concerned are located in predicted transmembrane regions.
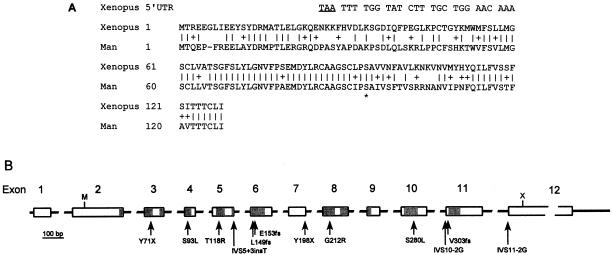
Figure 3.
Organization of MLC1. A, Conservation of the N-terminal end of the MLC1 gene product. The translated DNA sequences of the Xenopus EST and MLC1 (Genbank accession numbers AW640978 and D25217, respectively) are compared. Vertical lines indicate identical residues, and the plus sign (+) indicates conservative changes. The top line shows part of the 5′ end of the Xenopus EST with a stop codon (underlined) in frame with the coding region. The stop codon that is 132 nt upstream of and in frame with the first methionine of MLC1 in EST AU120203 is not shown. The asterisk (*) indicates the conserved serine residue that was mutated in patients M66, M73, and M75. B, Genomic structure of MLC1. The exons are shown as boxes; introns, as solid lines. The gray parts indicate the eight regions that encode transmembrane domains of the protein. The positions of the mutations that were found in DNA of patients with MLC are shown by arrows. Interrupted lines indicate deviations from scale. M = start codon, X = stop codon, and fs = frameshift.
MLC1 is expressed in the brain and other tissues (Nomura et al. 1994). We confirmed the identity of MLC1 transcripts by PCR amplification and DNA sequence analysis of overlapping fragments from adult brain cDNA. PCR amplification with three primer combinations each resulted in single fragments that overlapped. The fragments were of expected size. The DNA sequence was completely identical to the KIAA0027 clone, sequenced earlier (data not shown). These results indicated that alternative splicing of MLC1 does not occur in the brain.
Discussion
In this article, we describe the identification of the gene for MLC. Of the 11 informative families studied, 10 displayed linkage between markers on chromosome 22qtel and MLC. It is possible that, in family ML10, without apparent linkage, one of the sibling patients is a double recombinant between markers UT580 and 355c18 or between 355c18 and ARSA, or that both patients are recombinants between one of these marker pairs, so that they may share a genotype in a small portion of chromosome 22qtel. However, we have not yet been able to detect a mutation in the patients, and the most likely explanation for these results is the involvement of another MLC locus in family ML10.
Recombination events that border the critical region could be pinpointed by the use of SNP markers. Eventually, mutations in one of four candidate genes, MLC1, proved to be responsible for the disorder. The four amino acid substitutions investigated thus far are very unlikely to be polymorphisms, since the mutations were not represented in significant groups of control chromosomes matched for ethnic background. Furthermore, the substituted residues were conserved or located in predicted transmembrane domains, suggesting that the affected amino acids are important for MLC1 protein function. One mutation probably led to deletion of a transmembrane domain, and 8 of the 12 mutations led to disruption and termination of the MLC1 ORF. Interestingly, the two DNA deletions and the duplication involved direct and inverted repeats, respectively. The deletion that occurred in patient M20 and the sib pair M39 and M42 was probably the result of slipped-strand mispairing in the region of the repeat prior to DNA replication (Krawczak and Cooper 1991). Such a mechanism could not explain the rearrangement of the TGC repeat region in patient M120. In this case, a deletion of 8 bp was apparently accompanied by an inversion of one repeat unit. The direct duplication of half an inverted repeat in patient M81 may have been the result of strand slippage by hairpin formation during DNA replication.
We have not yet identified all disease-causing mutations in our group of patients with MLC. Not all mutations alter the mobility of DNA in SSCP analysis, and large deletions in the heterozygous state can be missed, because such alleles may fail to be amplified by PCR (Morgan et al. 1999). Furthermore, we did not analyze the promoter, the noncoding exon 1, complete introns, and the 3′ UTR, all of which may influence the expression of the gene. Also, we cannot exclude that some of the patients carry the gene for MLC at another locus.
The MLC1 cDNA was cloned from the immature myeloid cell line KG-1, and the expression in these cells proved to be high in comparison with the expression in a variety of human tissues (Nomura et al. 1994). Of the tissues examined, the expression of MLC1 was relatively high in the brain. It was also expressed in peripheral white blood cells and spleen, and the expression in ovary, prostate, placenta, thymus, and lung was relatively low. No expression was observed in HeLa cells, heart, liver, skeletal muscle, kidney, pancreas, testis, small intestine, and colon. The northern blot for MLC1, which can be viewed at the HUGE protein database, shows a single band of the same size in these tissues. We confirmed that MLC1 mRNA, with the same ORF, is present in the brain, and we did not detect alternative splice products. In the light of the expression pattern, it is understandable that, if MLC1 exerts an essential function, disruption of the gene would mainly affect the brain. The fact that a gene for a neurological disorder is not exclusively expressed in the nervous system is not uncommon.
The ORF of the MLC1 cDNA has been described as incomplete, because it started at the 5′-end and lacked an apparent start codon (Nomura et al. 1994). However, a strong argument that the MLC1 ORF is complete can be found in the following facts. First, a recently published expressed sequence tag (EST) (Genbank accession number AU120203) from the gene extended exon 1 in the 5′ direction by 20 nt. This EST has a stop codon near the 5′ end in-frame with the MLC1 ORF and the first ATG codon in the extended cDNA sequence corresponds with the first methionine codon in the MLC1 ORF. Second, this methionine is apparently evolutionarily conserved between man and Xenopus. A translated EST (Genbank accession number AW640978) from unfertilized Xenopus egg cells is similar to the N-terminal part of the MLC1 gene product, starting with the first methionine residue. The methionine codon of the Xenopus EST is in frame with an upstream stop codon, indicating that this ATG is the start codon of the Xenopus MLC1 gene (fig. 3A).
The genomic structure of MLC1 contains 12 exons with the start codon in exon 2 and an untranslated 3′ end of 2.2 kb (fig. 3B). The TMpred program predicts the MLC1 gene product to have eight transmembrane domains. A BLAST search did not show homology of the MLC1 product with proteins of known function, although there was a low similarity to the human voltage-gated potassium channel KV1.1. A PROSITE profile search indicated that the amino acid sequence contained a signature motif of the ribosomal protein S14 subunit, but this match is listed by PROSITE as a false-positive because the S14 ribosomal subunit gene obviously has been identified in higher eukaryotes. The MLC1 amino acid sequence also matched the signature of ABC-2 type transporters and of sodium:galactoside symporters with low significance. Our working hypothesis is that MLC1 encodes a membrane protein that may have a transport function for a specific, as yet unknown, substrate.
Acknowledgments
We thank the patients and their families, for cooperation. B.Q.Y. is a scholar supported by China Scholarship Council. This work was supported by the Dutch Foundation for Scientific Research (NWO). We thank Drs. C. A. Catsman, H. Stroink, W. F. M. Arts, P. G. Barth, E. de Vries, F. Goutières, J. Motte, M. Bataillard, I. Baric, V. Diklic, R. A. H. Surtees, T. Balsev, E. B. Skriver, J. Riedel, W. Koehler, A. Haehnelt, A. Fiumara, L. G. Epstein, L. Garcia, J. Clarke, V. Kalra, H. Hattori, T. Koeda, J. Takanashi, and M. A. M. Salih, for referral of patients and collection of material. We thank Dr. J. M. Powers for critical reading of the manuscript.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Database of Single Nucleotide Polymorphisms, A, http://www.ncbi.nlm.nih.gov/SNP
- Genbank, http://www.ncbi.nlm.nih.gov/Genbank/index.html
- Genome Database, The, http://www.gdb.org
- HUGE, A Database of Human Unidentified Gene-Encoded Large Proteins Analyzed by Kazusa cDNA Project, http://www.kazusa.or.jp/huge
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim (for MLC [MIM 604004])
- PROSITE, Database of protein families and domains, http://www.expasy.ch/prosite
- TMpred program, http://www.ch.embnet.org/software/TMPRED_form.html
References
- Dib C, Fauré S, Fizames C, Samson D, Drouot N, Vignal A, Millasseau P (1996) A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature 380:152–154 [DOI] [PubMed] [Google Scholar]
- Dunham I, Shimizu N, Roe BA, Chissoe S, Hunt AR, Collins JE, Bruskiewich R, et al (1999) The DNA sequence of human chromosome 22. Nature 402:489–495 [DOI] [PubMed] [Google Scholar]
- Hofmann K, Stoffel W (1993) TMbase—a database of membrane-spanning proteins segments. Biol Chem Hoppe-Seyler 347:166 [Google Scholar]
- Krawczak M, Cooper DN (1991) Gene deletions causing human genetic disease: mechanisms of mutagenesis and the role of the local DNA sequence environment. Hum Genet 86:425–441 [DOI] [PubMed] [Google Scholar]
- Leegwater PAJ, Könst AAM, Kuyt B, Sandkuijl LA, Naidu S, Oudejans CBM, Schutgens RBH, Pronk JC, Van der Knaap MS (1999) The gene for leukoencephalopathy with vanishing white matter is located on chromosome 3q27. Am J Hum Genet 65:728–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan NV, Tipping AJ, Joenje H, Mathew CG (1999) High frequency of large intragenic deletions in the Fanconi anemia group A gene. Am J Hum Genet 65:1330–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura N, Miyajima N, Sazuka T, Tanaka A, Kawarabayasi Y, Sato S, Nagase T, Seki N, Ishikawa K, Tabata S (1994) Prediction of the coding sequences of unidentified human genes. I. The coding sequences of 40 new genes (KIAA0001–KIAA0040) deduced by analysis of randomly sampled cDNA clones from human immature myeloid cell line KG-1. DNA Res 1:27–35 [DOI] [PubMed] [Google Scholar]
- Rogozin IB, Milanesi L (1997) Analysis of donor splice signals in different organisms. J Mol Evol 45:50–59 [DOI] [PubMed] [Google Scholar]
- Singhal BS, Gursahani RD, Udani VP, Biniwale AA (1996) Megalencephalic leukodystrophy in an Asian Indian ethnic group. Pediatr Neurol 14:291–296 [DOI] [PubMed] [Google Scholar]
- Topçu M, Garitoux C, Ribierre F, Yalcinkaya C, Tokus E, Oztekin N, Beckmann JS, Ozguc M, Seboun E (2000) Vacuoliting megalencephalic leukoencephalopathy with subcortical cysts, mapped to chromosome 22qtel. Am J Hum Genet 66:733–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topçu M, Saatic I, Topcuoglu MA, Kose G, Kunak B ( 1998) Megalencephaly and leukodystrophy with mild clinical course: a report on 12 new cases. Brain Dev 20:142–153 [DOI] [PubMed] [Google Scholar]
- Van der Knaap MS, Barth PG, Stroink H, Van Nieuwenhuizen O, Arts WFM, Hoogenraad F, Valk J (1995) Leukoencephalopathy with swelling and a discrepantly mild clinical course in eight children. Ann Neurol 37:324–334 [DOI] [PubMed] [Google Scholar]
- Van der Knaap MS, Barth PG, Vrensen GF, Valk J (1996) Histopathology of an infantile-onset spongiform leukoencephalopathy with a discrepantly mild clinical course. Acta Neuropathol 92:206–212 [DOI] [PubMed] [Google Scholar]
- Wijker M, Morgan NV, Herterich S, Van Berkel CGM, Tipping AJ, Gross HJ, Gille JJP, et al (1999) Heterogeneous spectrum of mutations in the Fanconi anaemia group A gene. Eur J Hum Genet 7:52–59 [DOI] [PubMed] [Google Scholar]