Abstract
In this study, enterobacterial repetitive intergenic consensus PCR (ERIC-PCR) and randomly amplified polymorphic DNA PCR (RAPD-PCR) were optimized for characterization of Arcobacter butzleri, Arcobacter cryaerophilus, and Arcobacter skirrowii. In addition, a simple and rapid DNA extraction method was tested for use in both typing procedures. Both methods had satisfactory typeability and discriminatory power, but the fingerprints generated with ERIC-PCR were more reproducible and complex than those obtained with RAPD-PCR. The use of nondiluted boiled cell suspensions as DNA templates was found to be very useful in ERIC-PCR. Characterization of large numbers of Arcobacter isolates is therefore preferably performed by the ERIC-PCR procedure. Isolates for which almost identical ERIC fingerprints are generated may subsequently be characterized by RAPD-PCR, although adjustment and standardization of the amount of the DNA template are necessary. In the second part of this study, the genotypic diversity of arcobacters present on broiler carcasses was assessed by using both typing methods. A total of 228 cultures from 24 samples were examined after direct isolation and enrichment. The isolates were identified by using a multiplex PCR as A. butzleri (n = 182) and A. cryaerophilus (n = 46). A total of 131 types (91 A. butzleri types and 40 A. cryaerophilus types) were discerned without discordance between the two typing techniques. The analysis of the poultry isolates showed that poultry products may harbor not only more than one species but also multiple genotypes. All genotypes were confined to one poultry sample, and only three genotypes were found after simultaneous enrichment and direct isolation. These results demonstrate that different outcomes can be obtained in epidemiological studies depending on the isolation procedure used and the number of isolates characterized.
The genus Arcobacter belongs to rRNA superfamily VI of the Proteobacteria and currently includes four species (23). Two species, Arcobacter butzleri and A. cryaerophilus, have been associated with human diarrheal illness (11, 21) and bacteremia (7, 16, 30). They have also been isolated from farm animals with clinical symptoms of mastitis (12), diarrhea (9), and reproduction abnormalities (3, 20), as well as from clinically healthy farm animals (28) and poultry (29). A third species, A. skirrowii, has been recovered from fecal samples from experimentally infected poultry (29) and pigs (27) and from lambs with diarrhea (24). Additional isolates have been obtained from the internal organs of porcine, ovine, and bovine aborted fetuses and from preputial sheet washes of bulls. This organism has also been recovered from two broiler carcasses (1). Association of A. skirrowii with human infection has not been reported. The fourth Arcobacter species, A. nitrofigilis, is a nitrogen-fixing bacterium associated with the roots of Spartina alterniflora, a salt marsh plant (14). This organism requires an elevated salt level for growth and has never been associated with animal or human infection. Therefore, this species was not considered in this study.
Examination of human and veterinary clinical specimens for the presence of Arcobacter species is rarely performed, and in most cases suboptimal procedures are used (6).
In addition, little is known about the risk factors for human infection. However, the high rates of Arcobacter contamination of raw poultry products (1, 5) and the distribution of the same biotypes (2; H. Lior and D. L. Woodward, abstract from the International Workshop on Campylobacter, Helicobacter, and Related Organisms, 21 to 23 September 1993, Brussels, Belgium, Acta Gastro-Enterol. Belg. Suppl. 6:28, 1993) and serotypes (H. Lior and D. L. Woodward, Acta Gastro-Enterol. Belg. Suppl. 6:29, 1993) among human and poultry isolates indicate that handling of raw poultry and consumption of undercooked poultry products may be routes of infection. As the precise contribution of poultry to human infection is still unclear, characterization of Arcobacter isolates can contribute to our knowledge of the epidemiological behavior of these organisms. However, few typing studies have been described so far. Methods such as biotyping (Lior and Woodward, Acta Gastro-Enterol. Belg. Suppl. 6:28, 1993), serotyping (Lior and Woodward, Acta Gastro-Enterol. Belg. Suppl. 6:29, 1993), and comparison of cellular protein profiles (24) have been shown to have limited discriminatory capabilities and to rely on phenotypic characteristics that may be unstable and poorly reproducible. Molecular typing methods, such as the randomly amplified polymorphic DNA (RAPD) assay (H. Lior and G. abstract from the International Workshop on Campylobacter, Helicobacter, and Related Organisms, 21 to 23 September 1993, Brussels, Blegium, Wang, Acta Gastro-Enterol Belg. Suppl. 6:29, 1993), ribotyping (10), and repetitive-motif-based polymorphic DNA analysis (22), have been performed with limited numbers of arcobacters, but there has not been a profound evaluation of the performance and discriminatory abilities of these methods for the three Arcobacter species examined in this study. It has been shown that pulsed-field gel electrophoresis of macrorestriction fragments (8; Lior and Wang, Acta Gastro-Enterol. Belg. Suppl. 6:29, 1993) is a well-suited method with respect to its discriminatory power for A. butzleri and A. cryaerophilus, but this procedure is complex, costly, and time-consuming (15).
In this study, we optimized enterobacterial repetitive intergenic consensus PCR (ERIC-PCR) and RAPD-PCR for genetic characterization of A. butzleri, A. cryaerophilus, and A. skirrowii and assessed the performance and discriminatory abilities of these methods by using a well-defined strain collection consisting of members of these three Arcobacter species. In addition, the usefulness of a simple and fast DNA preparation method was evaluated. The genotypic diversity of arcobacters present on broiler carcasses was studied, and the impact of the isolation procedure and the number of isolates characterized on epidemiological investigation results was determined.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
For optimization and evaluation of the typeability, reproducibility, and discriminatory power of the two typing methods, 24 geographically and epidemiologically unrelated Arcobacter strains were obtained from the BCCM/LMG Bacteria Collection (Ghent University, Ghent, Belgium). These collection strains were isolated from humans and animals and are listed in Table 1. The collection used also included three A. butzleri isolates from a well-documented outbreak in a nursery and primary school in Italy (25). The strains were grown on Mueller-Hinton agar plates (CM 337; Oxoid, Basingstoke, United Kingdom) and were incubated for 48 h at 29°C under microaerobic conditions by evacuating 80% of the normal atmosphere and introducing a gas mixture consisting of 8% CO2, 8% H2, and 84% N2 into each jar. For each strain, a cell suspension was prepared in 10 ml of sterile water with an optical density at 660 nm of about 0.074 ± 0.002, which corresponded to a concentration of approximately 108 CFU/ml.
TABLE 1.
Arcobacter strains used in this study
| Species | Isolate | Reference no.a | Source | Country of origin |
|---|---|---|---|---|
| A. butzleri | 1 | LMG 6620 | Human, blood | United Kingdom |
| 2 | LMG 9869 | Piglet, liver | United Kingdom | |
| 3 | LMG 9906 | Porcine fetus, eye | Northern Ireland | |
| 4 | LMG 10220 | Pig, feces | Canada | |
| 5 | LMG 10223 | Bovine, liver | Canada | |
| 6 | LMG 10240 | Horse, feces | Canada | |
| 7 | LMG 10828T | Human, feces | United States | |
| 8 | LMG 11118 | Human, feces | Italy | |
| 9 | LMG 11119 | Human, feces | Italy | |
| 10 | LMG 11120 | Human, feces | Italy | |
| 11 | LMG 14714 | Human, feces | Greece | |
| 12 | LMG 15918 | Human, feces | Hungary | |
| A. cryaerophilus | 13 | LMG 7536T | Ovine fetus | Northern Ireland |
| 14 | LMG 9066 | Porcine fetus, eye | Northern Ireland | |
| 15 | LMG 9905 | Bovine fetus, kidney | Northern Ireland | |
| 16 | LMG 10210 | Bovine fetus | Canada | |
| 17 | LMG 9861 | Bovine fetus, peritoneal | Northern Ireland | |
| 18 | LMG 9864 | Porcine fetus, eye | Northern Ireland | |
| 19 | LMG 9867 | Equine fetus, spleen | Northern Ireland | |
| 20 | LMG 10209 | Bovine fetus | Canada | |
| 21 | LMG 10217 | Porcine fetus, placenta | Canada | |
| A. skirrowii | 22 | LMG 6621T | Lamb, feces | United Kingdom |
| 23 | LMG 9802 | Bull, preputial fluid | Belgium | |
| 24 | LMG 9912 | Bovine fetus, thoracal | Northern Ireland | |
| 25 | LMG 10234 | Porcine fetus, lung | Canada | |
| 26 | LMG 10239 | Bovine, feces | Canada | |
| 27 | LMG 14983 | Bull, preputial fluid | United States |
T = type strain. LMG, Bacteria Collection Laboratory, Ghent University, Ghent, Belgium.
DNA preparation.
Template DNA was extracted from a 1.0-ml cell suspension of each strain by two methods. Before extraction, all of the cell suspensions were centrifuged for 5 min at 13,000 rpm (Eppendorf model 5417-R centrifuge) to pellet the cells and the supernatants were discarded. In the first method, DNA extracts were prepared by resuspending the cell pellets in 500 μl of sterile distilled water and boiling the suspensions for 10 min. After a quick spin to pellet the cell debris, the supernatants were used as DNA templates for subsequent amplifications. In the second method, the bacterial pellets were resuspended in 100 μl of Tris-EDTA buffer and genomic DNA was extracted by the guanidinium thiocyanate method described by Pitcher et al. (18).
Five microliters of each DNA preparation was size separated by electrophoresis to analyze the integrity of the DNA extracted. The concentration of each DNA template was determined spectrophotometrically at A260 and adjusted to 25 ng μl−1. The DNA templates obtained by both methods were stored at −25°C.
Conditions used for RAPD-PCR.
All PCR amplifications were performed by using 25 pmol of primer with Ready-To-Go RAPD analysis beads (Amersham Pharmacia Biotech, Uppsala, Sweden) containing premixed, predispensed Ampli Taq DNA polymerase, as well as all necessary buffer ingredients and nucleotides. The cycling parameters were as follows: denaturing at 95°C for 30 s, annealing at 36°C for 1 min, and extension at 72°C for 2 min for a total of 45 cycles. Prior to cycling, samples were heated at 94°C for 5 min. For each strain, 25, 50, and 100 ng of DNA, obtained by both extraction methods, were tested. In addition, 1 μl of each nondiluted boiled cell suspension was analyzed. For the initial screening, RAPD fingerprints were generated for all 27 strains by using the six arbitrary 10-mer primers included in the RAPD analysis primer set (Amersham Pharmacia Biotech). All amplifications were performed in 25-μl mixtures by using a Perkin-Elmer GeneAmp System 9700 PCR thermocycler.
Conditions used for the ERIC-PCR assay.
Each 50-μl PCR mixture was composed of autoclaved deionized water, 5 μl of 10× PCR buffer (Invitrogen, GmbH), 5 U of Taq polymerase (Invitrogen, GmbH), and a deoxynucleoside triphosphate mixture containing each deoxynucleoside triphosphate (Invitrogen, GmbH) at a final concentration of 0.2 mM. Several concentrations of primers, MgCl2, and DNA templates were tested. Annealing was performed at different temperatures, as indicated below. The ERIC motifs 1R (5′-ATGTAAGCTCCTGGGGATTCAC-3′) and 2 (5′-AAGTAAGTGACTGGGGTGAGCG-3′) designed by Versalovic et al. (26) were each used at concentrations of 25, 50, and 100 pmol/50 μl. The effects of MgCl2 concentrations of 2.5, 3.0, 3.5, and 4.0 mM were evaluated. The DNA concentrations that were used in the RAPD assays, including 1-μl portions of the boiled cell suspensions, were tested. The PCR consisted of 35, 38, 40, or 42 cycles of 94°C for 1 min, 25, 40, or 50°C for 1 min, and 72°C for 2 min. Prior to cycling, samples were heated at 94°C for 5 min.
Reproducibility.
In order to assess the reproducibility of the two typing methods, genomic DNA of the 27 collection strains were extracted by the two extraction procedures and amplified by the two PCR-based typing methods on three nonconsecutive days, and the fingerprints generated were compared.
Poultry isolates.
Neck skin samples from 24 broiler carcasses were collected in four different Belgian poultry slaughterhouses and examined for the presence of Arcobacter. Each sample was taken on a different day and from a different poultry flock. Samples were obtained immediately after chilling and were examined by using the protocol developed previously, including isolation with and without a primary enrichment step (5). Briefly, 10 g of neck skin was added to 10 ml of Arcobacter enrichment broth consisting of 28 g of Arcobacter broth (CM 965;Oxoid) per liter, 5% lysed horse blood, and the previously developed selective supplement (5) and homogenized in a stomacher blender for 1 min at the normal speed. One hundred microliters of each neck skin homogenate was inoculated by the spiral plating method onto an Arcobacter selective agar plate containing 28 g of Arcobacter broth per liter, 12 g of Agar Technical no. 3 (L13; Oxoid Ltd., Basingstoke, Hampshire, England) per liter, and the previously developed selective supplement (5) and incubated at 28°C for 24 to 72 h. To detect low Arcobacter contamination levels, the neck skin homogenates were incubated for 48 h at 28°C in a microaerobic atmosphere and then inoculated onto Arcobacter selective agar plates. Following incubation, all colonies obtained by direct isolation were examined. In addition, 2 to 10 randomly sampled colonies, obtained after enrichment, were picked from the agar plates. Subsequently, all isolates were identified by using a species-specific multiplex PCR assay (4) and were characterized further by the two typing methods.
Analysis of PCR products.
The PCR products were size separated by electrophoresis of 8-μl portions of the reaction mixtures in ethidium bromide-stained 2% agarose gels with 1× Tris-borate-EDTA buffer for 2.5 h at 100 V. The DNA profiles were visualized by UV transillumination and photographed. The gels were analyzed both by visual inspection and by computer-aided methods. DNA patterns that differed in one or more DNA fragments were considered patterns that represented different types. Whenever type differences relied on only one band, a repeat analysis was performed (including a repeat DNA extraction) to confirm the reproducibility of the fingerprint. To interpret the profiles of the poultry isolates, computer-based normalization and interpolation of the DNA profiles and numerical analysis using the Pearson product moment correlation coefficient with 1% position tolerance were performed using the GelCompar 4.2 software package (Applied Maths, Kortrijk, Belgium). Dendrograms were constructed by using the unweighted pair group linkage analysis method. For convenience, the correlation levels were expressed as percentages of similarity.
RESULTS
DNA preparation.
DNA templates of all 27 Arcobacter collection strains were prepared on three occasions by using both extraction procedures. DNA was detected in all extracts; the concentrations ranged from 35 to 600 ng of DNA μl−1, and there was no apparent effect of extraction procedure, Arcobacter species, or day of extraction. When the integrity of the DNA extracts was checked on agarose gels, clear-cut DNA bands were observed when the extraction procedure of the Pitcher et al. (18) was used. When boiled lysates were analyzed, smears of DNA appeared in the agarose gel.
RAPD-PCR.
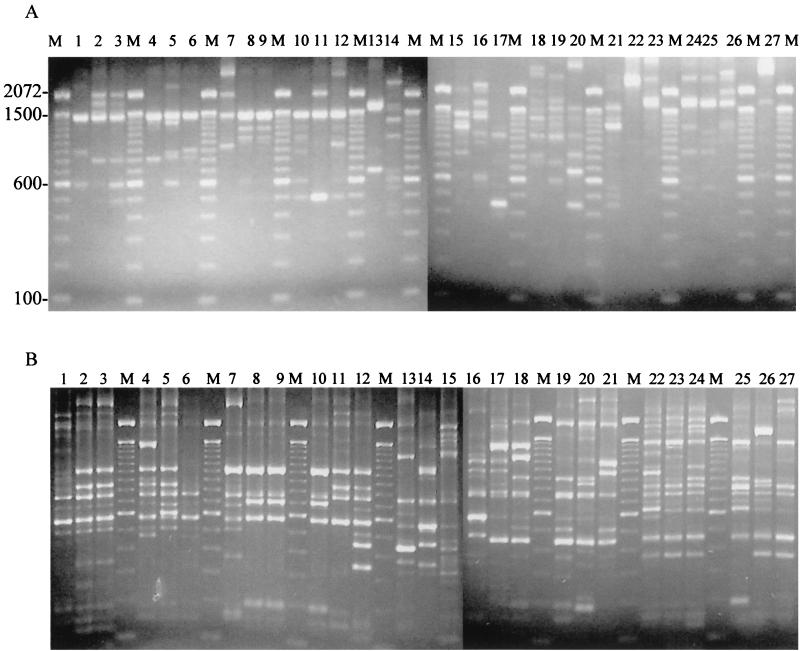
Of the six random primers tested, primer 1 (5′-GGTGCGGGAA-3′) gave the clearest and most distinctive band pattern for each of the 27 strains examined. When 25 pmol of this primer was used, unique profiles were obtained for all of the Arcobacter strains except the three outbreak isolates (Fig. 1A). The profiles were composed of two to nine fragments with different intensities. The sizes of the DNA fragments generated by the amplification process and used for determination ranged from 300 to 2,072 bp. When 100 ng of DNA or 1 μl of nondiluted boiled cell suspension was used, few or no DNA fragments were amplified, while nondiscriminatory profiles consisting of two or three fragments were obtained when 50 ng of DNA was utilized (data not shown). When the fingerprints generated with 25-ng DNA templates obtained by the two extraction methods were compared, similar degrees of discrimination were observed, although the profiles generated were not identical (data not shown).
FIG. 1.
RAPD (A) and ERIC (B) fingerprints of the 27 Arcobacter collection strains. The lane numbers correspond to the isolate numbers shown in Table 1. Lanes M contained a reference marker.
ERIC-PCR.
To enhance the discriminatory power of ERIC-PCR fingerprinting, the optimal annealing temperature and optimal concentrations of MgCl2, DNA template, and primers were determined. The Mg2+ concentration and the annealing temperature were observed to have profound effects on the complexity of the band pattern (data not shown). Only the use of 4 mM MgCl2 and an annealing temperature of 25°C produced strain-specific band patterns. While an MgCl2 concentration of 2.5 or 3 mM and annealing at 25°C resulted in poor amplification, higher annealing temperatures generated species-specific profiles with all MgCl2 concentrations tested. The fingerprints, however, did not vary significantly when different amounts of DNA template or primers were used (irrespective of the DNA preparation procedure), although slight differences in band intensity were noticed.
An annealing temperature of 25°C with 4 mM MgCl2 and 25 pmol of each primer generated reproducible fingerprints with a sufficient number of amplimers for typing purposes. The profiles were specific for unrelated strains and identical for the three outbreak Arcobacter isolates (Fig. 1B). The number of amplimers in the ERIC-PCR profiles ranged from 3 to 15. The band patterns used to determine the ERIC-PCR types comprised DNA fragments between 100 and 2,072 bp long. For optimal results, 40 amplification cycles were used, as fewer cycles resulted in fewer amplimers and a poor yield of some fragments (data not shown).
Reproducibility.
Twenty-five nanograms of DNA of each collection strain, obtained by the two extraction methods, was examined with both typing methods in triplicate. One microliter of nondiluted boiled cell suspension was tested only with the ERIC-PCR, as using 1 μl of a nondiluted boiled lysate in RAPD-PCR resulted in amplification of few or no DNA fragments.
Excellent reproducibility was obtained for ERIC-PCR with all three types of DNA templates and for RAPD-PCR with 25-ng DNA templates obtained by the guanidinium thiocyanate or boiled cell method. In some fingerprints, the major bands were less intense and the minor bands were difficult to visualize, but the presence or absence of DNA fragments was very consistent.
Poultry isolates.
Of the 24 broiler neck skin samples, 20 and 9 obtained after enrichment and by direct isolation, respectively, were found to be positive for the presence of Arcobacter strains. The numbers of colonies randomly collected from the agar plates after enrichment and the total numbers of colonies obtained after direct isolation are shown in Table 2. For ERIC-PCR and multiplex PCR, the poultry isolates were examined by using boiled cell suspensions as DNA templates. Based on the results obtained by comparing different amounts of DNA from the collection strains, a portion of each supernatant was adjusted so that the DNA concentration was 25 ng μl−1, and 1 μl was used as the DNA template in RAPD-PCR.
TABLE 2.
Numbers of Arcobacter isolates analyzed
| Abattoir | Broiler | After enrichment
|
Direct isolation
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
A. butzleri
|
A. cryaerophilus
|
A. butzleri
|
A. cryaerophilus
|
||||||||||
| No. of colonies | No. of ERIC types | No. of RAPD types | No. of colonies | No. of ERIC types | No. of RAPD types | No. of colonies | No. of ERIC types | No. of RAPD types | No. of colonies | No. of ERIC types | No. of RAPD types | ||
| A | 1 | 0a | — | — | 5 | 4 | 3 | 0 | — | — | 9 | 7 | 7 |
| A | 2 | 0 | — | — | 5 | 5 | 5 | 0 | — | — | 0 | — | — |
| A | 3 | 5 | 4 | 4 | 0 | — | — | 0 | — | — | 0 | — | — |
| A | 4 | 5 | 2 | 2 | 0 | — | — | 0 | — | — | 0 | — | — |
| A | 5 | 5 | 2 | 2 | 0 | — | — | 0 | — | — | 9 | 9 | 9 |
| B | 6 | 0 | — | — | 0 | — | — | 0 | — | — | 0 | — | — |
| B | 7 | 2 | 1 | 1 | 0 | — | — | 21 | 8 | 6 | 2 | 2 | 2 |
| B | 8 | 2 | 1 | 1 | 0 | — | — | 7 | 3 | 3 | 2 | 2 | 2 |
| B | 9 | 0 | — | — | 0 | — | — | 0 | — | — | 0 | — | — |
| B | 10b | 2 | 1 | 1 | 0 | — | — | 7 | 3 | 3 | 1 | 1 | 1 |
| C | 11 | 2 | 1 | 1 | 0 | — | — | 14 | 7 | 6 | 1 | 1 | 1 |
| C | 12 | 2 | 2 | 2 | 0 | — | — | 1 | 1 | 1 | 2 | 2 | 2 |
| C | 13 | 2 | 2 | 2 | 0 | — | — | 0 | — | — | 0 | — | — |
| C | 14 | 0 | — | — | 0 | — | — | 0 | — | — | 0 | — | — |
| C | 15 | 10 | 4 | 4 | 0 | — | — | 0 | — | — | 0 | — | — |
| C | 16 | 0 | — | — | 0 | — | — | 0 | — | — | 0 | — | — |
| C | 17b | 10 | 5 | 5 | 0 | — | — | 8 | 7 | 7 | 2 | 1 | 1 |
| D | 18 | 6 | 4 | 4 | 0 | — | — | 5 | 4 | 4 | 4 | 3 | 2 |
| D | 19 | 10 | 3 | 3 | 0 | — | — | 0 | — | — | 0 | — | — |
| D | 20 | 4 | 1 | 1 | 0 | — | — | 0 | — | — | 0 | — | — |
| D | 21 | 10 | 3 | 3 | 0 | — | — | 0 | — | — | 0 | — | — |
| D | 22 | 10 | 3 | 3 | 0 | — | — | 0 | — | — | 0 | — | — |
| D | 23 | 10 | 3 | 3 | 0 | — | — | 6 | 6 | 6 | 1 | 1 | 1 |
| D | 24 | 10 | 7 | 7 | 0 | — | — | 3 | 3 | 3 | 3 | 2 | 3 |
| Total | 110 | 49 | 49 | 10 | 9 | 8 | 72 | 42 | 40 | 36 | 31 | 31 | |
0, no arcobacters isolated; —, not applicable.
One of the A. butzleri types was found both after enrichment and after direct isolation.
A total of 228 subcultures were identified by multiplex PCR as A. butzleri (182 isolates) or A. cryaerophilus (46 isolates) and were characterized further by the two typing methods. When direct isolation was used, both Arcobacter species were isolated simultaneously from nine broiler samples. When enrichment was used, A. cryaerophilus was isolated only in the absence of A. butzleri. A. cryaerophilus was detected in 11 broiler samples by direct isolation but in only 2 samples by enrichment culture. Furthermore, discordance in the Arcobacter species isolated was observed for broiler 5, from which A. butzleri was isolated by enrichment culture and A. cryaerophilus was isolated by direct isolation.
All 228 isolates were examined by both typing methods. ERIC-PCR generated fingerprints for 197 isolates; DNA smears appeared for the remaining 31 isolates. The supernatants of these 31 isolates were diluted 1/10 in sterile water, and amplification was performed again by using 1-μl portions of the diluted supernatants as the DNA templates. After this second analysis, only three isolates (one A. butzleri isolate and two A. cryaerophilus isolates) remained uncharacterized. RAPD fingerprints were obtained for all but 12 isolates. Genomic DNA was reextracted, the concentration was adjusted to 25 ng μl−1, and 1 μl was used in a new RAPD-PCR. When this was done, six isolates (three A. butzleri isolates and three A. cryaerophilus isolates) remained uncharacterized. DNA were extracted by the guanidinium thiocyanate method from the three isolates that were not characterized by ERIC-PCR and the six isolates that were not characterized by RAPD-PCR, and these DNA were tested by using both PCR methods. ERIC and RAPD fingerprints were obtained for all isolates. However, the latter patterns were not included in the subsequent analysis because of the different DNA extraction method used.
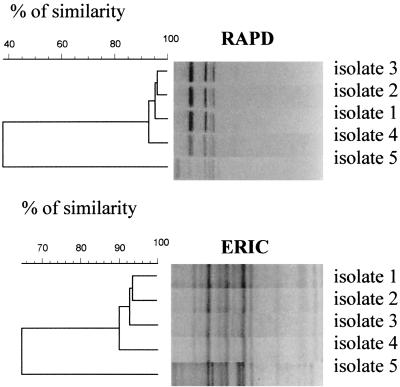
Computer analysis of the patterns allowed rapid comparison of the different gel tracks and construction of dendrograms. For both methods, groups consisting of identical fingerprints or fingerprints with minor differences in band intensity were characterized by Pearson correlation coefficients of 90% or higher. Pearson correlation coefficients lower than 78% were obtained for band patterns that differed by the presence of two or more distinct DNA fragments. For some isolates (for example, isolates 1 and 2 from broiler 1 [Fig. 2]), ERIC fingerprints differed in only one fragment. Such fingerprints were characterized by Pearson correlation coefficients ranging from 78 to 90%. When the corresponding RAPD profiles were analyzed, different patterns (and thus different types) were observed, which were characterized by Pearson correlation coefficients lower than 78%. Cluster analysis of the DNA patterns of all of the isolates tested revealed great heterogeneity among the Arcobacter isolates obtained after direct isolation, as well as among the isolates obtained after enrichment. Ninety-one A. butzleri types and 40 A. cryaerophilus types were determined by ERIC-PCR, and 89 A. butzleri types and 39 A. cryaerophilus types could be distinguished by RAPD-PCR analysis (Table 2). The discriminatory powers of the two methods were identical. Differences in the total number of types obtained by the two typing methods (Table 2) were due to the presence of some isolates for which no fingerprint was obtained by one of the methods used. The same type was never detected in more than one poultry sample, and only three identical A. butzleri types were found after direct isolation and after enrichment in the same sample.
FIG. 2.
Dendrograms based on RAPD and ERIC fingerprints for A. butzleri isolates obtained after enrichment from broiler 4.
DISCUSSION
In the first part of this study, ERIC-PCR and RAPD-PCR were optimized for differentiation of A. butzleri, A. cryaerophilus, and A. skirrowii strains. In addition, a simple and rapid DNA extraction method was tested for use with both PCR methods, in order to obtain maximum resolution with a minimum of effort. In the second part of the study, both typing methods were used to characterize 228 arcobacters isolated from 24 broiler carcasses with and without prior enrichment.
The usefulness of the two typing methods was evaluated by using the three main criteria for typing schemes, namely, typeability, reproducibility, and discriminatory power (19). To do this, a well-defined collection of three related Arcobacter isolates and 24 unrelated Arcobacter isolates that represented the three animal-associated species in this genus was used. With both methods, satisfactory typeability and reproducibility were obtained, as a reproducible profile was obtained for each strain tested. The two typing methods generated identical and unique fingerprints for the outbreak and unrelated strains, respectively, illustrating the discriminatory power of the methods and their suitability for use in epidemiological investigations. The amount of DNA used in the PCR-based methods and the method of DNA extraction seemed to have important effects on the results obtained. Particularly in the RAPD-PCR assay, variations in the quality and amount of DNA template had a major impact on the results obtained. For this assay, the best typing results were obtained with DNA template standardized at a concentration of 25 ng μl−1 and extracted by the guanidinium thiocyanate or boiled cell method. The use of nondiluted boiled cell extract as the DNA template generated useful fingerprints only in ERIC-PCR.
When the 228 Arcobacter poultry isolates were analyzed, 86 and 95% of the isolates could be characterized by ERIC and RAPD fingerprints, respectively. No discordance between the typing results obtained by the two typing methods was observed. Dilution of the DNA templates of 31 isolates for which DNA smears instead of band patterns were obtained after ERIC-PCR increased the typeability of the method to 98%. There was great variation among the DNA patterns of Arcobacter poultry isolates. A total of 131 types (91 A. butzleri types and 40 A. cryaerophilus types) from 24 neck skin samples from 24 broilers were discerned. The diversity of patterns was especially interesting because a particular genotype never occurred in more than one poultry sample. Moreover, only three genotypes were detected after enrichment and after direct isolation. These findings have major implications for epidemiological studies of arcobacters. In order to characterize the source of Arcobacter contamination by genotyping, it is necessary to examine a large number of isolates, and both direct isolation and enrichment should be used in order to recover as many different strains as possible.
The presence of considerable heterogeneity among arcobacters in one animal specimen was suggested previously by biotyping (2; Lior and Woodward, Acta Gastro-Enterol. Belg. Suppl. 6:28, 1993) and serotyping (2; Lior and Woodward, Acta Gastro-Enterol. Belg. Suppl. 6:29, 1993) studies. When DNA-based methods were used, genotypic variation was demonstrated among Arcobacter isolates from a farrow-to-finish swine facility (8) and among A. butzleri isolates from mechanically separated turkey samples (13). The possible explanations for the large amount of heterogeneity include multiple sources of contamination, the presence of multiple parent genotypes for both species in a single animal, and a high degree of genomic recombination among the progeny of parent genotypes. The ability to undergo genomic rearrangements has been demonstrated in Campylobacter species (17) but not in members of the genus Arcobacter. Genomic rearrangements were not detected among 14 outbreak isolates of A. butzleri, as all isolates belonged to the same serotype and had identical phenotypic characteristics and protein and ERIC-PCR profiles (22). Furthermore, in vitro cultivation of Arcobacter isolates over a period of time did not induce genomic rearrangements, as identical fingerprints were obtained in the reproducibility experiments in this study.
In conclusion, ERIC-PCR and RAPD-PCR are both valuable techniques for characterizing A. butzleri, A. cryaerophilus, and A. skirrowii isolates. Both methods have satisfactory typeability and discriminatory power. One-band differences between two fingerprints generated by one method reliably represented different types, as confirmed by the results of the other typing method. The fingerprints generated with ERIC-PCR were more reproducible and complex than the fingerprints generated with RAPD-PCR. Moreover, the use of nonstandardized amounts of boiled cell suspensions was possible only with ERIC-PCR. Therefore, for characterization of a large number of Arcobacter isolates, ERIC-PCR performed with boiled cell suspensions as the DNA templates is the first choice for a characterization method. Isolates for which almost identical ERIC fingerprints are generated may subsequently be characterized by RAPD-PCR, although adjustment and standardization of the amount of DNA template will be necessary. Our analysis of poultry isolates showed that poultry products may harbor not only more than one species but also multiple genotypes of the species. Depending on the isolation procedure used, different Arcobacter species can be isolated. Moreover, a large number of isolates should be characterized in order to recover as many different strains as possible.
Acknowledgments
We thank the BCCM/LMG Bacteria Collection, Ghent University (Ghent, Belgium), for providing the Arcobacter collection strains used in this study. The skilled technical assistance provided by Bieke Verbeke was greatly appreciated.
REFERENCES
- 1.Atabay, H. I., J. E. L. Corry, and S. L. W. On. 1998. Diversity and prevalence of Arcobacter spp. in broiler chickens. J. Appl. Microbiol. 84:1007-1016. [DOI] [PubMed] [Google Scholar]
- 2.de Boer, E., J. Tilburg, D. Woodward, H. Lior, and W. Johnson. 1996. A selective medium for the isolation of Arcobacter from meats. Lett. Appl. Microbiol. 23:64-66. [DOI] [PubMed] [Google Scholar]
- 3.Fernandez, H., X. Rojas, and T. Gajardo. 1995. First isolation in Chile of Arcobacter cryaerophilus from a bovine abortion. Arch. Med. Vet. 27:111-114. [Google Scholar]
- 4.Houf, K., A. Tutenel, L. De Zutter, J. Van Hoof, and P. Vandamme. 2000. Development of a multiplex PCR assay for the simultaneous detection and identification of Arcobacter butzleri, Arcobacter cryaerophilus and Arcobacter skirrowii. FEMS Microbiol. Lett. 193:89-94. [DOI] [PubMed] [Google Scholar]
- 5.Houf, K., L. A. Devriese, L. De zutter, J. Van Hoof, and P. Vandamme. 2001. Development of a new protocol for the isolation and quantification of Arcobacter species from poultry products. Int. J. Food Microbiol. 71:189-196. [DOI] [PubMed] [Google Scholar]
- 6.Houf, K., L. A. Devriese, L. De Zutter, J. Van Hoof, and P. Vandamme. 2001. Susceptibility of Arcobacter butzleri, Arcobacter cryaerophilus, and Arcobacter skirrowii to antimicrobial agents used in selective media. J. Clin. Microbiol. 39:1654-1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hsueh, P., L. Teng, P. Yang, S. Wang, S. Chang, S. Ho, W. Hsieh, and K. Luh. 1997. Bacteremia caused by Arcobacter cryaerophilus 1B. J. Clin. Microbiol. 35:489-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hume, M. E., R. B. Harvey, L. H. Stanker, R. E. Droleskey, T. L. Poole, and H. B. Zhang. 2001. Genotypic variation among Arcobacter isolates from a farrow-to-finish facility. J. Food Prot. 64:645-651. [DOI] [PubMed] [Google Scholar]
- 9.Kiehlbauch, J. A., D. Brenner, M. Nicholson, C. Baker, C. Patton, A. Steigerwalt, and I. Wachsmuth. 1991. Campylobacter butzleri sp. nov. isolated from humans and animals with diarrheal illness. J. Clin. Microbiol. 29:376-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kiehlbauch, J. A., D. N. Cameron, and I. K. Wachmuth. 1994. Evaluation of ribotyping techniques as applied to Arcobacter, Campylobacter and Helicobacter. Mol. Cell. Probes 8:109-116. [DOI] [PubMed] [Google Scholar]
- 11.Lerner, J., V. Brumberger, and V. Preac-Mursic. 1994. Severe diarrhea associated with Arcobacter butzleri. Eur. J. Clin. Microbiol. Infect. Dis. 13:660-662. [DOI] [PubMed] [Google Scholar]
- 12.Logan, E., S. Neill, and D. Mackie. 1982. Mastitis in dairy cows associated with an aerotolerant Campylobacter. Vet. Rec. 110:229-230. [DOI] [PubMed] [Google Scholar]
- 13.Manke, T. R., I. V. Wesley, J. S. Dickson, and K. M. Harmon. 1998. Prevalence and genetic variability of Arcobacter species in mechanically separated turkey. J. Food. Prot. 61:1623-1628. [DOI] [PubMed] [Google Scholar]
- 14.McClung, C. R., D. G. Patriquin, and R. E. Davis. 1983. Campylobacter nitrofigilis sp. nov., a nitrogen fixing bacterium associated with roots of Spartina alterniflora Loisel. Int. J. Syst. Bacteriol. 33:605-612. [Google Scholar]
- 15.Olive, D. M., and P. Bean. 1999. Principles and applications of methods for DNA-based typing of microbial organisms. J. Clin. Microbiol. 37:1661-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.On, S. L. W., A. Stacey, and J. Smyth. 1995. Isolation of Arcobacter butzleri from a neonate with bacteraemia. J. Infect. 31:225-227. [DOI] [PubMed] [Google Scholar]
- 17.On, S. L. W. 1998. In vitro genotypic variation of Campylobacter coli documented by pulsed-filed gel electrophoresis DNA profiling: implications for epidemiological studies. FEMS Microbiol. Lett. 165:341-346. [DOI] [PubMed] [Google Scholar]
- 18.Pitcher, D. G., N. A. Saunders, and R. J. Owen. 1989. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett. Appl. Microbiol. 8:151-158. [Google Scholar]
- 19.Power, E. G. M. 1996. RAPD typing in microbiology—a technical review. J. Hosp. Infect. 34:247-265. [DOI] [PubMed] [Google Scholar]
- 20.Shroeder-Tucker, L., I. V. Wesley, J. A. Kiehlbauch, D. J. Larson, L. A. Thomas, and G. A. Erickson. 1996. Phenotypic and ribosomal RNA characterization of Arcobacter species isolated from porcine aborted fetuses. J. Vet. Diagn. Investig. 8:186-195. [DOI] [PubMed] [Google Scholar]
- 21.Tee, W., R. Baird, M. Dyall-Smith, and B. Dwyer. 1988. Campylobacter cryaerophila isolated from a human. J. Clin. Microbiol. 26:2469-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vandamme, P., B. A. Giesendorf, A. Van Belkum, D. Pierard, S. Lauwers, K. Kesters, J. P. Butzler, H. Goossens, and W. G. Quint. 1993. Discrimination of epidemic and sporadic isolates of Arcobacter butzleri by polymerase chain reaction-mediated DNA fingerprinting. J. Clin. Microbiol. 31:3317-3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vandamme, P., E. Falsen, R. Rossau, B. Hoste, P. Segers, R. Tytgat, and J. De Ley. 1991. Revision of Campylobacter, Helicobacter, and Wolinella taxonomy: emendation of generic descriptions and proposal of Arcobacter gen. nov. Int. J. Syst. Bacteriol. 41:88-103. [DOI] [PubMed] [Google Scholar]
- 24.Vandamme, P., M. Vancanneyt, B. Pot, L. Mels, B. Hoste, D. Dewettinck, L. Vlaes, C. Van Den Borre, R. Higgens, J. Hommez, K. Kesters, J. P. Butzler, and H. Goossens. 1992. Polyphasic taxonomic study of the emended genus Arcobacter with Arcobacter butzleri comb. nov. and Arcobacter skirrowii sp. nov., an aerotolerant bacterium isolated from veterinary specimens. Int. J. Syst. Bacteriol. 42:344-356. [DOI] [PubMed] [Google Scholar]
- 25.Vandamme, P., P. Pugina, G. Benzi, R. Van Etterijck, L. Vlaes, K. Kesters, J. P. Butzler, H. Lior, and S. Lauwers. 1992. Outbreak of recurrent abdominal cramps associated with Arcobacter butzleri in an Italian school. J. Clin. Microbiol. 30:2335-2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Versalovic, J., T. Koeuth, and J. R. Lupski. 1991. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 19:6823-6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wesley, I. V., A. L. Baetz, and D. J. Larson. 1996. Infection of cesarean-derived colostrum-deprived 1-day-old piglets with Arcobacter butzleri, Arcobacter cryaerophilus, and Arcobacter skirrowii. Infect. Immun. 64:2295-2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wesley, I. V., S. J. Wells, K. M. Harmon, A. Green, L. Schroeder-Tucker, M. Glover, and I. Siddique. 2000. Fecal shedding of Campylobacter and Arcobacter spp. in dairy cattle. Appl. Environ. Microbiol. 66:1994-2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wesley, I. V., and A. L. Baetz. 1999. Natural and experimental infections of Arcobacter in poultry. Poultry Sci. 78:536-545. [DOI] [PubMed] [Google Scholar]
- 30.Yan, J. J., W. C. Ko, A. H. Huang, H. M. Chen, Y. T. Jin, and J. J. Wu. 2000. Arcobacter butzleri bacteremia in a patient with liver cirrhosis. J. Formosan Med. Assoc. 99:166-169. [PubMed] [Google Scholar]