Abstract
Melanocortin-1 receptor (MC1R) gene variants are associated with fair skin and red hair and, independently of these, with cutaneous malignant melanoma. The association of MC1R gene variants with nonmelanoma skin cancer is largely unknown. A total of 838 subjects were included in the present study: 453 patients with nonmelanoma skin cancer and 385 subjects with no skin cancer. The coding sequence of the human MC1R gene was tested using single-stranded conformation polymorphism analysis followed by sequencing of unknown variants. Risk of skin cancer dependent on the various MC1R gene variants was estimated using the exposure odds ratio. We investigated whether subjects with MC1R variant alleles were at increased risk of developing nonmelanoma skin cancer and, if so, whether this increased risk was mediated by fair skin and red hair. A total of 27 MC1R gene variants were found. The number of carriers of one, two, or three MC1R gene variants was 379 (45.2%), 208 (24.8%), and 7 (0.9%), respectively. A strong association between MC1R gene variants and fair skin and red hair was established, especially the variants Arg151Cys and Arg160Trp (P<.0001). Carriers of two variant alleles were at increased risk for developing cutaneous squamous cell carcinoma (odds ratio 3.77; 95% confidence interval [CI] 2.11–6.78), nodular basal cell carcinoma (odds ratio 2.26; 95% CI 1.45–3.52), and superficial multifocal basal cell carcinoma (odds ratio 3.43; 95% CI 1.92–6.15), compared with carriers of two wild-type alleles. Carriers of one variant allele had half the risk. The highest relative risks of nonmelanoma skin cancer were found in carriers of the Asp84Glu, His260Pro, and Asp294His variant alleles, and the risk was only slightly lower for carriers of the Val60Leu, Val92Met, Arg142His, Arg151Cys, and Arg160Trp variant alleles. When subjects were stratified by skin type and hair color, analysis showed that these factors did not materially change the relative risks. These findings indicate that MC1R gene variants are important independent risk factors for nonmelanoma skin cancer.
Introduction
Cutaneous squamous cell carcinomas and basal cell carcinomas, together commonly called “nonmelanoma skin cancer,” are among the most common cancers in white populations (Preston and Stern 1992). Basal cell carcinomas can be subdivided into nodular and superficial multifocal basal cell carcinoma, for both of which the distribution varies according to age, sex, and site (Bastiaens et al. 1998). Ultraviolet (UV) radiation is the major environmental risk factor of nonmelanoma skin cancer (Kricker et al. 1994), whereas fair skin and red hair are considered to be the most important genetically determined risk factors (Kricker et al. 1991).
Melanocortin-1 receptor (MC1R) gene variants (MC1R [MIM 155555]), particularly the Arg151Cys, Arg160Trp, and Asp294His variants, are associated with fair skin and red hair (Valaverde 1995; Box et al. 1997). Fair skin and red hair are also associated with an increased risk of cutaneous malignant melanoma (Kricker et al. 1991; Bliss et al. 1995). Increased risks of malignant melanoma have been shown in subjects carrying different MC1R gene variant alleles (Valverde et al. 1996; Healy et al. 1999; Palmer et al. 2000). Carriers of the Asp84Glu variant allele were found to have an increased risk of malignant melanoma in a study reported by Valverde and coworkers (1996), but later studies by the same research group (Healy et al. 1999) and by other research groups (Ichii-Jones et al. 1999; Palmer et al. 2000) were not able to confirm this association. More recently, the Arg151Cys, Arg160Trp, and Asp294His variant alleles appeared to be associated with an increased risk of malignant melanoma (Palmer et al. 2000). The association between MC1R variants and malignant melanoma suggests that the MC1R gene is a susceptibility gene for this skin malignancy (Rees and Healy 1997). The role of MC1R gene variants in modulating the risk of nonmelanoma skin cancer, however, is largely unknown. In a small group of British patients with nonmelanoma skin cancer, an overrepresentation of the Asp294His variant allele was found (Smith et al. 1998). Such an overrepresentation, however, could not be confirmed in a study of patients with basal cell carcinoma (Ichii-Jones et al. 1999).
Melanin pigmentation plays an important role in protecting the skin against the damaging effects of UV radiation. In humans, α-melanocyte–stimulating hormone (α-MSH) and other pro-opiomelanocortin (POMC) peptides, such as ACTH, modulate pigment metabolism via MC1R (Suzuki et al. 1996; Thody and Graham 1998). The enhanced pigmentation of the skin after UVB exposure is probably also mediated through α-MSH and MSH receptors (Abdel-Malek et al. 1999). Two pigments have been identified in human skin: the black eumelanin and the red pheomelanin (Thody et al. 1991). Melanocytes, which are stimulated by α-MSH acting through MC1R, synthesize the black eumelanin pigment instead of red pheomelanin (Hunt et al. 1995). Eumelanin is photoprotective, but pheomelanin may contribute to skin carcinogenesis by producing free radicals in response to UV radiation (Ranadive et al. 1986). Individuals with red hair and fair skin predominantly synthesize pheomelanin, which may explain their sun sensitivity and inability to tan (Thody et al. 1991). Apart from their function in pigment metabolism, α-MSH and other POMC peptides also have immunomodulatory and anti-inflammatory activity (Wintzen and Gilchrest 1996; Luger et al. 1998). In addition, α-MSH affects proliferation and differentiation of melanocytes (Lunec et al. 1990, 1992; De Luca et al. 1993; Suzuki et al. 1996) and keratinocytes (Slominski et al. 1991).
The MC1R is a seven-pass transmembrane G-protein–coupled receptor of 317 amino acids, which is expressed in several cell types, including melanocytes and, possibly, keratinocytes (Mountjoy et al. 1992; Chakraborty and Pawelek 1993; Chakraborty et al. 1999b). Suzuki et al. (1996), however, could not confirm by northern blot analyses that human epidermal keratinocytes express MC1R mRNA. The human MC1R gene has been cloned and localized to chromosome 16q24.3 (Gantz et al. 1993; Magenis et al. 1994). In the MC1R gene, common polymorphisms have been identified (Valverde et al. 1996; Box et al. 1997; Koppula et al. 1997). In an Irish population, >75% of individuals tested had one or more MC1R gene variants (Smith et al. 1998), several of which are able to alter the function of the receptor. In mice, MC1R gene variants leading to loss of function of the receptor result in an overproduction of pheomelanin and consequently yellow hair (Cone et al. 1996), whereas dominant variants extend the amount of black pigment and diminish the amount of yellow (Robbins et al. 1993). In humans, loss-of-function variants have been identified (Val60Leu, Arg142His, Arg151Cys, Arg160Trp, and Asp294His), which were unable to stimulate cAMP production as strongly as the wild-type receptor in response to α-MSH stimulation (Frändberg et al. 1994, 1998; Schiöth et al. 1999). Some of these variant alleles have also been found to be associated with red hair and to be overrepresented in individuals with fair skin (Box et al. 1997; Smith et al. 1998; Flanagan et al. 2000; Palmer et al. 2000).
The purpose of the present study was to compare individuals who had nonmelanoma skin cancer with control subjects who had no history of skin cancer and to focus on the presence of MC1R gene variants. We examined the association of MC1R gene variants with fair skin and red hair. We then investigated whether subjects with any MC1R variant alleles were at increased risk of developing cutaneous squamous cell carcinoma, nodular basal cell carcinoma, or superficial multifocal basal cell carcinoma and, if so, whether this increased risk was mediated by fair skin and red hair. Finally, we assessed the relative risks of nonmelanoma skin cancer for carriers of the individual MC1R-variant alleles.
Subjects and Methods
Study Population
The Leiden Skin Cancer Study (LSS) is an extensive effort begun in 1997 to study environmental and genetic risk factors for various skin cancers in the Dutch population. The medical ethics committees of Leiden University Medical Center (LUMC) and the other medical centers involved approved the protocol, and all participants gave informed consent.
Patients who were 30–80 years old and had histologically proved squamous cell carcinoma or nodular or superficial multifocal basal cell carcinoma of the skin were included in the study; tumors were newly diagnosed between January 1985 and December 1997 at the LUMC Department of Dermatology or in three hospitals in the same region as the LUMC (St. Franciscus Hospital in Rotterdam, the Rijnland Hospital in Leiderdorp, and the Westeinde Hospital in The Hague).
Control subjects in the same age range as the patients were selected from the ophthalmology outpatient clinic of the LUMC. They had no history of skin cancer, and individuals with a history of intraocular melanoma were excluded. Organ transplant recipients and patients with xeroderma pigmentosum or nevoid basal cell carcinoma (Gorlin's syndrome) were excluded, because these persons are at greatly increased risk of skin cancer. For this part of the study we also excluded all individuals with a history of cutaneous malignant melanoma. Only patients and control subjects with skin types I through IV, as defined by Fitzpatrick (1988), were included.
Interview and Physical Examination
Patients eligible for the study were asked by mail to participate, and the participation of control subjects was requested in a letter that was handed to them during a visit to the ophthalmology outpatient clinic. All subjects were contacted by telephone to arrange an appointment at the dermatology outpatient clinic for the interview, physical examination, and collection of a blood sample.
Trained interviewers performed a standardized personal interview. The invitation letter instructed participants not to mention during the interview and physical examination whether they were treated for skin cancer. Skin type was assessed as follows: always burn, never tan (type I), always burn, then tan (type II), always tan, sometimes burn (type III), and always tan, never burn (type IV). Hair was assessed for its color at the age of 20 years and was classified into four categories: red, light blond, dark blond, and brown or black.
The physical examination was performed by dermatologists, using a standard protocol. Eye color was categorized as brown, hazel/gray, green, or blue. If nonmelanoma skin cancer was found in control subjects, they were grouped with the patients in all subsequent analyses.
Detection of MC1R Gene Variants
Genomic DNA was isolated from peripheral blood leukocytes, using routine methods (Miller et al. 1988). The MC1R gene coding sequence (GenBank accession number X65634) was amplified by PCR in the following reaction: 200 ng genomic template DNA; 60 mM Tris HCl (pH 10.0); 2.0 mM MgCl2; 15 mM (NH4)2SO4; 100 μM each dGTP, dTTP, dATP, and dCTP; 1 μl α-[32P] dCTP (3,000 Ci mM−1); 500 ng of each PCR primer; 2 U AmpliTaq (Applied Biosystems); and 10% DMSO in a total volume of 100 μl. Samples were covered with mineral oil, denatured for 4 min at 92°C, and passed through 33 cycles of amplification, consisting of 50 s of denaturation at 92°C, 50 s of primer annealing at 58°C, and 2 min of elongation at 72°C. The amplifications were performed in 0.5-ml tubes (Applied Biosystems). The DNA sequences of the primers were as follows: forward 5′-CAACGACTCCTTCCTGCTTC-3′ and reverse 5′-TGCCCAGCACACTTAAAGC-3′. The resulting 1,018-bp PCR fragments were digested by 2 U of either RsaI or MspI and were screened for mutations by SSCP analysis (Orita et al. 1989) on a 6% polyacrylamide gel with 10% glycerol. The gel analyses were performed at room temperature for 6 h at 28 W or for 16 h at 20 W for MspI and RsaI digests, respectively.
Sequence Analysis
DNA samples for sequencing were obtained by PCR, as described above, with the following M13-tailed MC1R gene primers: M13MC1R-F (5′-TGTAAAACGACGGCCAGTCAACGACTCCTTCCTGCTTC-3′) and M13MC1R-IR (5′-CAGGAAACAGCTATGACCATGAGTCACGATGCTGTGGTAGC-3′), resulting in a 542-bp fragment, and the primers M13MC1R-IF (5′-GACGTTGTAAACGACGGCCAGTACCTGCAGCTCCATGCTGTC-3′) and M13MC1R-R (5′-CAGGAAACAGCTATGACCATGATGCCCAGCACACTTAAAGC-3′), resulting in a 661-bp fragment.
Sequence analysis was performed on an ABI 377 automated DNA sequencer, using Big-Dye Terminator cycle sequencing kits (Applied Biosystems) according to the manufacturer's protocol.
Statistical Analyses and Strategy of Analyses
Logistic regression analysis was used to compare MC1R gene variants independently of skin type and hair color in patients and control subjects. Exposure odds ratios (ORs) with 95% confidence intervals (CIs) were calculated to estimate the relative risk for the presence and the number of the nonmelanoma skin cancer types dependent on the MC1R gene variants. Further stratification was performed on the basis of the following hypotheses.
Hypothesis 1
The first hypothesis was that the MC1R-gene status determines skin type and red hair, which determine the risk of skin cancer. In this line of reasoning, skin type and hair color are considered confounding factors, and the association between MC1R gene variants and nonmelanoma skin cancer is expected to disappear in strata of skin type (type I/II and III/IV) and red hair (red and non-red). A Mantel-Haenszel weighted OR was calculated to adjust ORs for the MC1R gene variant according to skin type and hair color.
Hypothesis 2
The second hypothesis was that fair skin and red hair are only indicators of the presence of MC1R gene variants, and it is the gene variant that is the true risk factor for skin cancer. If this hypothesis were true, the association between skin type and nonmelanoma skin cancer and that between red hair and nonmelanoma skin cancer would disappear within strata of MC1R gene variants.
We decided to first analyze the association of any MC1R variant allele (i.e., all variant alleles taken together) with nonmelanoma skin cancer. At a later stage we analyzed the relative contribution of the individual MC1R variant alleles to the nonmelanoma skin cancer risk.
Hypothesis 3
The third hypothesis was that fair skin, red hair, and MC1R gene variants are independent risk factors for nonmelanoma skin cancer. If the hypothesis were correct an association would persist between MC1R gene variants and nonmelanoma skin cancer and between fair skin, red hair, and nonmelanoma skin cancer in strata mentioned above.
Results
Composition of Study Population
A total of 889 participants were interviewed and examined at the dermatology outpatient clinic of the LUMC. Of the subjects who agreed to participate in the study, 51 were excluded because they did not fulfill the inclusion criteria. Among these, 26 individuals had a history solely of basal cell carcinoma subtypes other than nodular and superficial types, and in 4 individuals MC1R genotyping was not successful, because no DNA or PCR product could be obtained.
The final series for analyses comprised 838 subjects: 153 subjects with squamous cell carcinoma, 284 with nodular basal cell carcinoma, and 142 with superficial multifocal basal cell carcinoma; 385 control subjects were studied. Of the 453 patients with skin cancer, 103 had only squamous cell carcinoma, 182 only nodular basal cell carcinoma, and 57 only superficial multifocal basal cell carcinoma. A total of 111 subjects had more than one type of skin cancer: 26 had squamous cell carcinoma in combination with nodular basal cell carcinoma, 9 had squamous cell carcinoma in combination with superficial multifocal basal cell carcinoma, 15 had squamous cell carcinoma in combination with both types of basal cell carcinoma, and 61 had nodular in combination with superficial multifocal basal cell carcinoma. In the analyses, patient groups consisted of subjects with a certain skin cancer regardless of the history of any other skin cancer; when patient groups consisting of patients with only one type of skin cancer were analyzed, similar relative risks were obtained.
Table 1 shows the characteristics of the study population. As was expected, squamous cell carcinoma was significantly associated with skin type (P=.0002) and hair color (P<.0001). Nodular basal cell carcinoma was significantly associated with skin type (P=.008), and the P value approached a level that would indicate an association with hair color (P=.08). Superficial multifocal basal cell carcinoma was significantly associated with both skin type (P=.007) and hair color (P=.04).
Table 1.
Characteristics of the Study Population
| Characteristic | Controls (n = 385) | Any Type of Skin Cancera(n = 453) | Squamous Cell Carcinoma (n = 153) | Nodular Basal Cell Carcinoma (n = 284) | Superficial Multifocal Basal Cell Carcinoma (n = 142) |
| Sex (no. [%]) | |||||
| Male | 163 [42.3] | 254 [56.1] | 99 [64.7] | 162 [57.0] | 74 [52.1] |
| Female | 222 [57.7] | 199 [43.9] | 54 [35.3] | 122 [43.0] | 68 [47.9] |
| Age at interview (years): | |||||
| Mean | 58.5 | 63.0 | 66.2 | 63.3 | 60.4 |
| Range | 28.6–79.9 | 31.6–78.2 | 36.9–78.2 | 31.6–78.2 | 31.7–78.2 |
| Skin type (no. [%]):b | |||||
| IV: Tan, never burn | 24 [6.2] | 24 [5.3] | 7 [4.6] | 16 [5.6] | 9 [6.3] |
| III: Tan, sometimes burn | 181 [47.0] | 167 [36.9] | 46 [30.1] | 107 [37.7] | 50 [35.2] |
| II: Burn, then tan | 155 [40.3] | 202 [44.6] | 76 [49.6] | 123 [43.3] | 61 [43.0] |
| I: Burn, never tan | 25 [6.5] | 60 [13.2] | 24 [15.7] | 38 [13.4] | 22 [15.5] |
| Hair color at age 20 years (no. [%]):c | |||||
| Black | 27 [7.1] | 22 [4.8] | 5 [3.3] | 16 [5.6] | 7 [4.9] |
| Brown | 97 [25.3] | 79 [17.4] | 13 [8.5] | 58 [20.4] | 28 [19.7] |
| Dark blond | 143 [37.3] | 162 [35.8] | 55 [35.9] | 94 [33.1] | 46 [32.4] |
| Light blond | 96 [25.1] | 148 [32.7] | 58 [37.9] | 93 [32.8] | 44 [31.0] |
| Red | 20 [5.2] | 42 [9.3] | 22 [14.4] | 23 [8.1] | 17 [12.0] |
More than one type of skin cancer was present in 111 subjects (see text).
Distribution compared with that in control subjects (P <.05).
Not assessed in two control subjects.
MC1R Gene Variants and the Association with Skin Type and Hair Color
A total of 27 MC1R gene variants were found (table 2). Eighteen variants showed a frequency <.5% of total alleles. Of the 1,676 alleles, only 8 showed two variants within the same allele, a result observed in eight individuals. Of these eight individuals, seven also showed a variant in the other allele, and in one individual the other allele was the wild type. A polymorphism resulting from an A→G synonymous substitution at nucleotide 942 was found in 178 (10.6%) alleles. Because this substitution did not alter the amino acid, A942G was not used as a MC1R gene variant in the analyses. The Val92Met variant was always observed together with the A942G polymorphism.
Table 2.
Allele Frequencies of MC1R Gene Variants in the Various Skin Types[Note]
| Variant (Nucleotide Change) | Skin Types I–IV (n= 1,676) | Skin Type IV (n = 96) | Skin Type III (n = 696) | Skin Type II (n = 714) | Skin Type I (n = 170) | Pa |
| No. (%) of Patients with Skin Typea |
||||||
| Val60Leu (178G→T) | 156 | 9 (9.4) | 59 (8.5) | 72 (10.1) | 16 (9.4) | .04 |
| Asp84Glu (252C→A) | 24 | 1 (1.0) | 6 (.9) | 14 (2.0) | 3 (1.8) | .10 |
| Val92Met (284G→A) | 146 | 6 (6.3) | 58 (8.3) | 62 (8.7) | 20 (11.8) | .004 |
| Arg142His (425G→A) | 13 | 0 (.0) | 4 (.6) | 7 (1.0) | 2 (1.2) | .41 |
| Arg151Cys (451C→T) | 133 | 1 (1.0) | 27 (3.9) | 60 (8.4) | 25 (14.7) | <.0001 |
| Arg160Trp (478C→T) | 189 | 7 (7.3) | 53 (7.6) | 97 (13.6) | 32 (18.8) | <.0001 |
| Arg163Gln (488G→A) | 75 | 4 (4.2) | 29 (4.2) | 34 (4.8) | 8 (4.7) | .22 |
| His260Pro (779A→C) | 13 | 1 (1.0) | 5 (.7) | 5 (.7) | 2 (1.2) | .61 |
| Asp294His (880G→C) | 19 | 0 (.0) | 6 (.9) | 8 (1.1) | 5 (2.9) | .02 |
| Othersb | 67 | 5 (5.2) |
27 (3.9) |
31 (4.3) |
4 (2.4) |
.58 |
| Frequency(%) |
||||||
| All | 35.5 | 39.2 | 54.7 | 68.8 | ||
Note.— n = no. of alleles.
Calculated in a logistic regression model with the wild-type allele as the reference.
Other variants, constituting <0.5% of total alleles, were as follows: Pro18Ala (52C→G), Ala81Pro (241G→C), Thr95Met (284C→T), Gly104Ser (310G→A), Arg151Arg (453C→G), Ile155Thr (925T→C), Val173del, Val174Ile (981G→A), Pro230Leu (689C→T), Pro230Pro (690G→A), Gln233Gln (699G→A), Ile264Ile (792C→T), Lys278Glu (832A→G), Asn279Ser (835A→G), Asn279Lys (836C→A), 86InsA, and 537InsC.
Of the 838 participants, 244 (29.1%) had no MC1R gene variants, 379 (45.2%) had one MC1R gene variant, 208 (24.8%) had two MC1R gene variants, and 7 (0.9%) had three MC1R gene variants. The group with two variant alleles comprised 29 homozygotes and 179 compound heterozygotes. In the subjects with two variant alleles, the frequencies of the combinations of the MC1R gene variants did not materially differ from the expected frequencies that were calculated from the allele frequencies of the variants (data not shown). Therefore, the MC1R gene variants found in the study can be considered independent variants and, consequently, were analyzed as such.
The Arg151Cys and Arg160Trp variant alleles were strongly associated with fair skin (P<.0001). The variants Val60Leu, Val92Met, and Asp294His were weakly associated with skin type (table 2). Arg142His, Arg151Cys, Arg160Trp, and Asp294His were strongly associated with red hair (P<.0001), whereas Asp84Glu and His260Pro were less strongly associated with red hair (P=.004 and P=.008, respectively). No associations were found between any MC1R gene variants and eye color (data not shown).
Association between All MC1R Gene Variants Combined and Nonmelanoma Skin Cancer
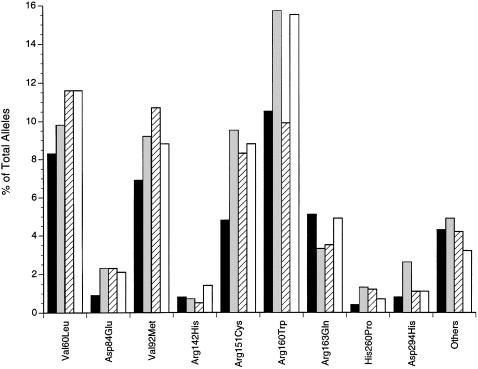
Figure 1 shows the distribution of the MC1R gene variants in the three nonmelanoma skin cancer groups and the control subjects. Most of the variants were overrepresented in the skin cancer groups. Therefore, our first analyses of MC1R gene variants and the nonmelanoma skin cancer groups were performed with all variants combined.
Figure 1.
Distribution of the MC1R gene variants in control subjects and the three nonmelanoma-skin-cancer groups. Blackened bars indicate controls subjects; shaded bars indicate patients with squamous cell carcinoma; bars with slanted lines indicate patients with nodular basal cell carcinoma; and unfilled bars indicate patients with superficial multifocal basal cell carcinoma.
Exposure ORs were calculated to explore whether the presence of one or two MC1R gene variants is associated with an elevated risk for nonmelanoma skin cancer. Table 3 shows that MC1R gene variants were associated with squamous cell carcinoma and with nodular and superficial multifocal basal cell carcinoma.
Table 3.
Risk of Nonmelanoma Skin Cancer Dependent on MC1R Gene Variants Stratified According to Skin Type
| Cancer and Genotype or Comparisona | Skin Types I–IV | Skin Types I and II | Skin Types III and IV | Pooled OR (95% CI) b | |||
| No. of Individuals |
|||||||
| Controls |
Patients |
Controls |
Patients |
Controls |
Patients |
||
| Squamous cell carcinoma: | |||||||
| WT/WT | 137 | 26 | 50 | 15 | 87 | 11 | |
| WT/Var | 174 | 74 | 79 | 44 | 95 | 30 | |
| Var/Var | 74 |
53 |
51 |
41 |
23 |
12 |
|
| OR (95% CI) |
|||||||
| WT/Var vs. WT/WT | 2.24 (1.32–3.81) | 1.86 (.89–3.91) | 2.50 (1.12–5.67) | 2.13 (1.25–3.68) | |||
| Var/Var vs. WT/WT | 3.77 (2.11–6.78) |
2.68 (1.25–5.80) |
4.13 (1.47–11.70) |
2.91 (1.62–5.58) | |||
| No. of Individuals |
|||||||
| Controls |
Patients |
Controls |
Patients |
Controls |
Patients |
||
| Nodular basal cell carcinoma: | |||||||
| WT/WT | 137 | 72 | 50 | 30 | 87 | 42 | |
| WT/Var | 174 | 124 | 79 | 65 | 95 | 59 | |
| Var/Var | 74 |
88 |
51 |
66 |
23 |
22 |
|
| OR (95% CI) |
|||||||
| WT/Var vs. WT/WT | 1.36 (.92–1.99) | 1.37 (.76–2.49) | 1.29 (.76–2.17) | 1.32 (.89–1.95) | |||
| Var/Var vs. WT/WT | 2.26 (1.45–3.52) |
2.16 (1.16–4.03) |
1.98 (.94–4.19) |
1.98 (1.23–3.19) | |||
| No. of Individuals |
|||||||
| Controls |
Patients |
Controls |
Patients |
Controls |
Patients |
||
| Superficial multifocal basal cell carcinoma: | |||||||
| WT/WT | 137 | 27 | 50 | 10 | 87 | 17 | |
| WT/Var | 174 | 65 | 79 | 31 | 95 | 34 | |
| Var/Var | 74 |
50 |
51 |
42 |
23 |
8 |
|
| OR (95% CI) |
|||||||
| WT/Var vs. WT/WT | 1.90 (1.12–3.23) | 1.96 (.83–4.71) | 1.83 (.91–3.70) | 1.87 (1.09–3.21) | |||
| Var/Var vs. WT/WT | 3.43 (1.92–6.15) | 4.12 (1.76–9.86) | 1.78 (.61–5.09) | 2.82 (1.45–5.22) | |||
WT = wild-type allele; Var = variant allele.
Adjusted for skin type, using Mantel-Haenszel weighted OR.
Testing Hypothesis 1
Stratified analyses were performed in subjects with fair and dark skin and in subjects with and without red hair to test the hypothesis that the association between MC1R gene variants and nonmelanoma skin cancer is mediated via fair skin and red hair. The positive association between MC1R gene variants and nonmelanoma skin cancer generally remained in both fair and dark skin strata (table 3) and also in the strata with and without red hair (data not shown), a result indicating that the presence of MC1R gene variants is a risk factor for nonmelanoma skin cancer, independently of fair skin and red hair. The risk of superficial multifocal basal cell carcinoma in carriers of two MC1R gene variants, however, appeared to be lower among subjects with darker skin compared with lighter skin (i.e., 1.78 vs. 4.12, respectively) (table 3). Unfortunately, in this subgroup analysis, the wide CIs indicate that the number of individuals is too small to conclude that the association between MC1R gene variants and superficial multifocal basal cell carcinoma was different among subjects with fair and dark skin. It was also not possible to conclude that the behavior of basal cell carcinoma differs from that of squamous cell carcinoma. It is conceivable that among subjects with darker skin the lower relative risk of superficial multifocal basal cell carcinoma in carriers of two MC1R gene variants was caused by chance. The pooled ORs adjusted for skin type by use of the Mantel-Haenszel test were only slightly lower than the crude OR (table 3), which also strongly supports the conclusion that the association between MC1R gene variants and nonmelanoma skin cancer is independent of skin type.
Testing Hypothesis 2
The risk of squamous cell carcinoma in subjects with fair skin (skin types I and II) compared with dark skin (skin types III and IV) was 2.15 (95% CI 1.43–3.23). For nodular basal cell carcinoma and superficial multifocal basal cell carcinoma, these figures were 1.49 (1.08–2.05) and 1.60 (1.07–2.41), respectively. In all three MC1R-gene strata, fair skin remained a risk factor for the nonmelanoma skin cancers, indicating that fair skin is a risk factor for nonmelanoma skin cancer independently of MC1R gene variants. Analyses performed in subjects with red hair revealed similar results (data not shown).
Testing Hypothesis 3
From the results of the analyses investigating an association between MC1R gene variants and the different types of nonmelanoma skin cancer, and between fair skin, red hair, and nonmelanoma skin cancer, we conclude that MC1R gene variants, fair skin, and red hair were independent risk factors for nonmelanoma skin cancer.
Association between Different MC1R Gene Variants and Nonmelanoma Skin Cancer
Table 4 shows separate analyses with the individual MC1R-variant alleles. The risks of skin cancer for heterozygotes and compound heterozygotes and homozygotes compared with that of subjects who are homozygous for the wild-type allele are listed. Generally, all MC1R gene variants were associated with an increased risk of nonmelanoma skin cancer although statistical significance was not reached for all comparisons, especially when the less-prevalent alleles were tested. Overall, the highest relative risks of nonmelanoma skin cancer were observed in carriers of the Asp84Glu, His260Pro, and Asp294His variant alleles. Slightly lower risks were observed in carriers of the Val60Leu, Val92Met, Arg142His, Arg151Cys, and Arg160Trp variant alleles. Carriers of the Arg163Gln variant allele showed the lowest relative risk, which did not reach statistical significance in any of the comparisons.
Table 4.
Relative Risks of Squamous Cell Carcinoma and Nodular and Superficial Multifocal Basal Cell Carcinoma in Heterozygotes and Compound Heterozygotes and Homozygotes for MC1R-Variant Alleles Compared with Homozygotes for the Wild-Type Allele[Note]
|
Risk (95% CI) in Individuals with |
||||||
| Squamous Cell Carcinoma |
Nodular Basal Cell Carcinoma |
Superficial Multifocal Basal Cell Carcinoma |
||||
| Alleles | Heterozygotes | Cpd Heterozygotes and Homozygotes | Heterozygotes | Cpd Heterozygotes and Homozygotes | Heterozygotes | Cpd Heterozygotes and Homozygotes |
| WT homozygotes | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| Val60Leu | 1.3 (.52–3.0) | 5.0 (2.2–11.4) | 1.1 (.61–2.1) | 3.4 (1.8–6.7) | 1.6 (.70–3.5) | 4.3 (1.9–10.0) |
| Asp84Glu | 7.0 (1.2–42.6) | 7.9 (1.0–71.9) | 4.4 (.99–22.4) | 5.7 (1.0–42.1) | 3.4 (.37–26.6) | 10.2 (1.5–84.7) |
| Val92Met | 2.4 (1.0–5.4) | 3.1 (1.3–7.5) | 1.4 (.70–2.6) | 2.8 (1.4–5.4) | 1.5 (.58–3.7) | 3.5 (1.5–8.1) |
| Arg142His | 0 (0–12.6) | 3.5 (.39–27.7) | 0 (0–4.4) | 1.9 (.30–12.2) | 0 (0–12.1) | 6.8 (1.2–41.0) |
| Arg151Cys | 2.2 (.79–6.1) | 5.6 (2.4–12.9) | 1.8 (.84–3.9) | 2.8 (1.4–5.6) | 2.4 (.89–6.4) | 4.5 (1.9–10.7) |
| Arg160Trp | 2.2 (1.1–4.6) | 4.4 (2.1–9.1) | 1.2 (.68–2.2) | 1.5 (.77–2.8) | 2.3 (1.1–4.6) | 3.6 (1.7–7.6) |
| Arg163Gln | 1.8 (.56–5.3) | 1.2(.31–4.1) | 0.95 (.37–3.4) | 1.2 (.48–2.8) | 2.0 (.67–5.6) | 2.0 (.67–5.6) |
| His260Pro | 7.9 (1.0–71.9) | 5.2 (.0–200.4) | 2.9 (.38–25.0) | 7.6 (.78–182.2) | 2.5 (.0–37.7) | 5.1 (.0–192.7) |
| Asp294His | 4.0 (.65–22.7) | 26.4 (2.8–621.5) | 0.95 (.12–6.2) | 7.6 (.78–182.2) | 1.3 (.20–7.2) | 10.2 (.69–294.0) |
Note— Cpd = compound; WT = wild type.
Only the Arg151Cys and Arg160Trp variant alleles were strongly associated with fair skin, whereas the Val60Leu, Asp84Glu, Val92Met, Arg142His, His260Pro, and Asp294His variant alleles were weakly associated or not associated with fair skin (table 2). Nevertheless, all these variant alleles were associated with skin cancer (table 4), which is an additional strong argument that skin type and nonmelanoma skin cancer risk are independent outcomes of the presence of MC1R gene variants. A similar line of argument holds for red hair.
Association between MC1R Gene Variants and Multiple Skin Tumors
To investigate whether carriers of MC1R gene variants are at an increased risk of developing multiple nonmelanoma skin cancers, all patients were grouped together irrespective of the tumor type and were stratified, according to the number of tumors, into three groups: those with one tumor, those with two to four tumors, and those with five or more tumors. The presence of MC1R gene variants was significantly and independently associated with higher numbers of skin tumors (table 5). Subsequent analyses of each type of skin cancer separately showed that this effect was mostly caused by the occurrence of squamous cell carcinoma and superficial multifocal basal cell carcinoma (data not shown).
Table 5.
Risk of One or Multiple Nonmelanoma Skin Cancers on the Basis of MC1R Gene Variants
| One Skin Tumor | Two to Four Skin Tumors | Five or More Skin Tumors | ||||
|
No. of Individuals with |
||||||
| Genotype or Comparison | Controls | Patients | Controls | Patients | Controls | Patients |
| WT/WT | 137 | 69 | 137 | 30 | 137 | 8 |
| WT/Var | 174 | 112 | 174 | 73 | 174 | 20 |
| Var/Var | 74 |
69 |
74 |
48 |
74 |
24 |
| OR (95% CI) |
||||||
| WT/Var vs. WT/WT | 1.28 (.86–1.89) | 1.92 (1.15–3.19) | 1.97 (.79–5.03) | |||
| Var/Var vs. WT/WT | 1.85 (1.17–2.94) | 2.96 (1.68–5.25) | 5.55 (2.24–14.22) | |||
Wt = wild-type allele; Var = variant allele.
Discussion
Until now, fair skin and red hair have been considered to be the most important genetically determined indicators of increased risk of nonmelanoma skin cancer and malignant melanoma (Kricker et al. 1991; Bliss et al. 1995). In the present study, however, we demonstrate that the presence of MC1R gene variants are also important in modulating an individual's risk of developing any of the three nonmelanoma skin cancers studied. Although the strong association of MC1R gene variants with fair skin and red hair could be confirmed convincingly, our analyses also showed that these factors did not materially influence the association between MC1R gene variants and nonmelanoma skin cancer, a result that undermines hypothesis 1: that the increased risk of nonmelanoma skin cancer is mediated via the pigmentation phenotype.
On the other hand, the association of fair skin and red hair with nonmelanoma skin cancer was not influenced by the presence of MC1R gene variants. This rules out hypothesis 2, that fair skin and red hair are merely indicators of the presence of MC1R gene variants and that the latter factor is the true risk factor for skin cancer.
Therefore, we conclude that MC1R gene variants, fair skin, and red hair are independent risk factors for nonmelanoma skin cancer (thereby verifying hypothesis 3). This conclusion is in agreement with a recent study in which the MC1R gene variants Arg151Cys, Arg160Trp, and Asp294His were found to be associated with cutaneous malignant melanoma in an Australian population and in which the association persisted in individuals with medium or olive/dark complexions (Palmer et al. 2000).
The present data suggest that the MC1R plays an important role in the pathogenesis not only of cutaneous malignant melanoma but also of cutaneous squamous cell carcinoma and both types of basal cell carcinoma studied. Carriers of two MC1R gene variants among subjects with darker skin (skin types III and IV) showed a lower risk of superficial multifocal basal cell carcinoma compared with carriers with lighter skin (skin types I and II). Unfortunately, the number of individuals in this subgroup analysis was too small to determine whether the observed difference between the two skin types represented a true difference or had occurred as a result of chance.
Separate analyses of the individual MC1R variant alleles showed that all of the more common variant alleles were associated with an increased risk of nonmelanoma skin cancer. The highest relative risks of nonmelanoma skin cancer were found in carriers of the Asp84Glu, His260Pro, and Asp294His variant alleles, and the risk was only slightly lower for carriers of the Val60Leu, Val92Met, Arg142His, Arg151Cys, and Arg160Trp variant alleles. With the exception of the Arg151Cys and Arg160Trp variant alleles, these alleles were weakly associated or not associated with fair skin, which further illustrates the finding that fair skin and nonmelanoma skin cancer are independent outcomes of MC1R gene variants. A similar line of argument applies to red hair.
Some MC1R gene variants are associated with fair skin and red hair, and individuals with this pigmentation phenotype predominantly synthesize pheomelanin instead of eumelanin in their melanocytes after stimulation by α-MSH (Thody et al. 1991). Eumelanin is photoprotective, but pheomelanin may contribute to skin carcinogenesis by producing free radicals in response to UV radiation (Ranadive et al. 1986). Free radicals and other reactive oxygen species have been found to cause many types of DNA damage, including single- and double-stranded DNA breaks, which have been implicated in pathological processes such as cancer (Shackelford et al. 2000). Our study, however, indicated that the increased risk of nonmelanoma skin cancer probably is not mediated via pigmentation phenotype. Therefore, apart from pigment metabolism, other mechanisms must be involved in the relationship between MC1R gene variants and the susceptibility of nonmelanoma skin cancer.
It is well documented that α-MSH and other POMC peptides have immunomodulatory and anti-inflammatory functions (Wintzen and Gilchrest, 1996; Luger et al. 1997). Within a mouse model, α-MSH was able to inhibit the induction of a contact hypersensitivity reaction and to induce hapten-specific tolerance (Luger et al. 1999). Also, α-MSH has been shown to play a role in modulating the transcription factor NFκB, which regulates the expression of gene products, such as cytokines and their receptors, that are involved in immune and inflammatory responses (Kalden et al. 1999). UV light induces the release of α-MSH and other POMC peptides and up-regulates the expression of MC1R in the epidermis (Chakraborty et al. 1995 and 1996) indicating that α-MSH may have a role in regulating responses to UV radiation and inflammatory stimuli and that α-MSH may contribute to UV-mediated immunosuppression (Luger et al. 1999).
It has been shown that α-MSH and other POMC peptides influence growth and development of melanocytes and melanoma cells (Lunec et al. 1990; Lunec et al. 1992; De Luca et al. 1993; Suzuki et al. 1996) as well as human keratinocytes (Slominski et al. 1991). Differentiation-driven heat shock protein 70 in keratinocytes was found to be down-regulated by α-MSH, rendering them more sensitive to oxidative stress (Orel et al. 1997). Recently, it was also found that the expression of POMC peptides and their specific receptors was up-regulated during keratinocyte differentiation, which may indicate that α-MSH is part of a mediator network that not only regulates cutaneous inflammation but also plays a role in hyperproliferative skin diseases (Chakraborty et al. 1999a). In conclusion, we have shown that the presence of MC1R gene variants is an important independent genetic risk factor that contributes to the risk of skin cancer.
Acknowledgments
We thank all patients and control subjects, for voluntarily and enthusiastically participating in this study, and Viña Williams-Snijders, for critically reviewing the manuscript. This work was supported by a grant from Zorg Onderzoek Nederland.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- GenBank Overview, http://www.ncbi.nlm.nih.gov/Web/Genbank/Overview.html (for the MC1R gene coding sequence [accession number X65634])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for MC1R [MIM 155555])
References
- Abdel-Malek Z, Suzuki I, Tada A, Im S, Akcali C (1999) The melanocortin-1 receptor and human pigmentation. Ann NY Acad Sci 885:117–133 [DOI] [PubMed] [Google Scholar]
- Bastiaens MT, Hoefnagel JJ, Bruijn JA, Westendorp RGJ, Vermeer BJ, Bouwes Bavinck JN (1998) Differences in age, site distribution, and sex between nodular and superficial basal cell carcinomas indicate different types of tumors. J Invest Dermatol 110:880–884 [DOI] [PubMed] [Google Scholar]
- Bliss JM, Ford D, Swerdlow AJ, Armstrong BK, Cristofolini M, Elwood JM, Green A, Holly EA, Mack T, MacKie R, Østerlind A, Walter SD, Peto J, Easton DF (1995) Risk of cutaneous melanoma associated with pigmentation characteristics and freckling: systematic overview of 10 case-control studies. Int J Cancer 62:367–376 [DOI] [PubMed] [Google Scholar]
- Box NF, Wyeth JR, O'Gorman LE, Martin NG, Sturm RA (1997) Characterisation of melanocyte stimulating hormone receptor variant alleles in twins with red hair. Hum Mol Genet 6:1891–1897 [DOI] [PubMed] [Google Scholar]
- Chakraborty AK, Funasaka Y, Pawelek JM, Nagahama M, Ito A, Ichihashi M (1999a) Enhanced expression of melanocortin-1 receptor (MC1R) in normal human keratinocytes during differentiation: evidence for increased expression of POMC peptides near suprabasal layer of epidermis. J Invest Dermatol 112:853–860 [DOI] [PubMed] [Google Scholar]
- Chakraborty AK, Funasaka Y, Slominski A, Bolognia J, Sodi S, Ichihashi M, Pawelek JM (1999b) UV light and MSH receptors. Ann NY Acad Sci 855:100–116 [DOI] [PubMed] [Google Scholar]
- Chakraborty A, Funasaka Y, Slominski A, Ermak G, Hwang J, Pawelek J, Ichihashi M (1996) Production and release of proopiomelanocortin (POMC) derived peptides by human melanocytes and keratinocytes in culture: regulation by ultraviolet B. Biochim Biophys Acta 1313:130–138 [DOI] [PubMed] [Google Scholar]
- Chakraborty A, Pawelek J (1993) MSH receptors in immortalized human epidermal keratinocytes: a potential mechanism for coordinate regulation of the epidermal-melanin unit. J Cell Physiol 157:344–350 [DOI] [PubMed] [Google Scholar]
- Chakraborty A, Slominski A, Ermak G, Hwang J, Pawelek J (1995) Ultraviolet B and melanocyte-stimulating hormone (MSH) stimulate mRNA production for α-MSH receptors and proopiomelanocortin-derived peptides in mouse melanoma cells and transformed keratinocytes. J Invest Dermatol 105:655–659 [DOI] [PubMed] [Google Scholar]
- Cone R, Lu D, Chen W, Koppula S, Vage DI, Klungland H, Boston B, Orth DN, Pouton C, Kesterson RA (1996) The melanocortin receptors: agonists, antagonists, and the hormonal control of pigmentation. Rec Prog Horm Res 51:287–318 [PubMed] [Google Scholar]
- De Luca M, Siegrist W, Bondanza S, Mathor M, Cancedda R, Eberle AN (1993) Alpha melanocyte stimulating hormone (α-MSH) stimulates normal human melanocyte growth by binding to high-affinity receptors. J Cell Sci 105:1079–1084 [DOI] [PubMed] [Google Scholar]
- Fitzpatrick TB (1988) The validity and practicality of sun-reactive skin type I through VI. Arch Dermatol 124:869–871 [DOI] [PubMed] [Google Scholar]
- Flanagan N, Healy E, Ray A, Philips S, Todd C, Jackson IJ, Birch-Machin MA, Rees JL (2000) Pleiotropic effects of the melanocortin 1 receptor (MC1R) gene on human pigmentation. Hum Mol Genet 9:2531–2537 [DOI] [PubMed] [Google Scholar]
- Frändberg PA, Muceniece R, Prusis P, Wikberg J, Chhajlani V (1994) Evidence for alternate points of attachment for α-MSH and its stereoisomer [Nle4, Dphe7]-α-MSH at the melanocortin-1 receptor. Biochem Biophys Res Commun 202:1266–1271 [DOI] [PubMed] [Google Scholar]
- Frändberg PA, Doufexis M, Kapas S, Chhajlani V (1998) Human pigmentation phenotype: a point mutation generates nonfunctional MSH receptor. Biochem Biophys Res Commun 245:490–492 [DOI] [PubMed] [Google Scholar]
- Gantz I, Konda Y, Tashiro T, Shimoto Y, Miwa H, Munzert G, Watson SJ, DelValle J, Yamada T (1993) Molecular cloning of a novel melanocortin receptor. J Biol Chem 268:8246–8250 [PubMed] [Google Scholar]
- Healy E, Todd C, Jackson IJ, Birch-Machin M, Rees JL (1999) Skin type, melanoma, and melanocortin 1 receptor variants. J Invest Dermatol 112:512–513 [DOI] [PubMed] [Google Scholar]
- Hunt G, Kyne S, Wakamatsu K, Ito S, Thody AJ (1995) Nle4DPhe7 α-MSH increases the eumelanin:phaeomelanin ratio in cultured human melanocytes. J Invest Dermatol 104:83–85 [DOI] [PubMed] [Google Scholar]
- Ichii-Jones F, Ramachandran S, Lear J, Smith A, Bowers B, Ollier WER, Jones P, Fryer AA, Strange RC (1999) The melanocyte stimulating hormone receptor polymorphism: association of the V92M and A294H alleles with basal cell carcinoma. Clin Chim Acta 282:125–134 [DOI] [PubMed] [Google Scholar]
- Kalden DH, Scholzen T, Brzoska T, Luger TA (1999) Mechanisms of the antiinflammatory effects of α-MSH: role of transcription factor NfκB and adhesion molecule expression. Ann NY Acad Sci 885:254–261 [DOI] [PubMed] [Google Scholar]
- Koppula SV, Robbins LS, Lu DS, Baack E, White CR, Swanson NA, Cone RD (1997) Identification of common polymorphisms in the coding sequence of the human MSH receptor (MC1R) with possible biological effects. Hum Mutat 9:30–36 [DOI] [PubMed] [Google Scholar]
- Kricker A, Armstrong BK, English DR, Heenan PJ (1991) Pigmentary and cutaneous risk factors for non-melanocytic skin cancer: a case-control study. Int J Cancer 48:650–662 [DOI] [PubMed] [Google Scholar]
- Kricker A, Armstrong BK, English DR (1994) Sun exposure and non-melanocytic skin cancer. Cancer Causes Control 5:367–392 [DOI] [PubMed] [Google Scholar]
- Luger TA, Scholzen T, Grabbe S (1997) The role of α-melanocyte-stimulating hormone in cutaneous biology. J Investig Dermatol Symp Proc 2:87–93 [DOI] [PubMed] [Google Scholar]
- Luger TA, Scholzen T, Brzoska T, Becher E, Slominski A, Paus R (1998) Cutaneous immunomodulation and coordination of skin stress responses by α-melanocyte-stimulating hormone. Ann NY Acad Sci 840:381–394 [DOI] [PubMed] [Google Scholar]
- Luger TA, Schwartz T, Kalden H, Scholzen T, Schwartz A, Brzoska T (1999) Role of epidermal cell-derived α-melanocyte-stimulating hormone in ultraviolet light mediated local immunosuppression. Ann NY Acad Sci 885:209–216 [DOI] [PubMed] [Google Scholar]
- Lunec J, Pieron C, Sherbet GV, Thody AJ (1990) Alpha-melanocyte-stimulating hormone immunoreactivity in melanoma cells. Pathobiology 58:193–197 [DOI] [PubMed] [Google Scholar]
- Lunec J, Pieron C, Thody AJ (1992) MSH receptor expression and the relationship to melanogenesis and metastatic activity in B16 melanoma. Melanoma Res 2:5–12 [DOI] [PubMed] [Google Scholar]
- Magenis RE, Smith L, Nadeau JH, Johnson KR, Mountjoy KG, Cone RD (1994) Mapping of the ACTH, MSH, and neural (MC3 and MC4) melanocortin receptors in the mouse and human. Mamm Genome 5:503–508 [DOI] [PubMed] [Google Scholar]
- Miller SA, Dykes DD, Polesky HF (1988) A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 16:1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mountjoy KG, Robbins LS, Mortrud MT, Cone RD (1992) The cloning of a family of genes that encode the melanocortin receptors. Science 257:1248–1251 [DOI] [PubMed] [Google Scholar]
- Orel L, Simon MM, Karsleder J, Bhardwaj R, Trautinger F, Schwarz T, Luger TA (1997) α-melanocyte stimulating hormone downregulates differentiation-driven heat shock protein 70 expression in keratinocytes. J Invest Dermatol 108:401–405 [DOI] [PubMed] [Google Scholar]
- Orita M, Suzuki Y, Sekiya T, Hayashi K (1989) Rapid and sensitive detection of point mutations and DNA polymorphisms using the polymerase chain reaction. Genomics 5:874–879 [DOI] [PubMed] [Google Scholar]
- Palmer JS, Duffy DL, Box NF, Aitken JF, O'Gorman LE, Green AC, Hayward NK, Martin NG, Sturm RA (2000) Melanocortin-1 receptor polymorphisms and risk of melanoma: is the association explained solely by pigmentation phenotype? Am J Hum Genet 66:176–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston DS, Stern RS (1992) Nonmelanoma cancers of the skin. N Engl J Med 327:1649–1662 [DOI] [PubMed] [Google Scholar]
- Ranadive NS, Shirwadkar S, Persad S, Menon IA (1986) Effects of melanin induced free radicals on the isolated rat peritoneal mast cells. J Invest Dermatol 86:303–307 [DOI] [PubMed] [Google Scholar]
- Rees JL, Healy E (1997) Melanocortin receptors, red hair, and skin cancer. J Investig Dermatol Symp Proc 2:94–98 [DOI] [PubMed] [Google Scholar]
- Robbins LS, Nadeau JH, Johnson KR, Kelly MA, Roselli-Rehfuss L, Baack E, Mountjoy KG, Cone RD (1993) Pigmentation phenotypes of variant extension locus alleles result from point mutations that alter MSH receptor function. Cell 72:827–834 [DOI] [PubMed] [Google Scholar]
- Schiöth HB, Phillips SR, Rudzish R, Birch-Machin MA, Wikberg JES, Rees JL (1999) Loss of function mutations of the human melanocortin 1 receptor are common and are associated with red hair. Biochem Biophys Res Commun 260:488–491 [DOI] [PubMed] [Google Scholar]
- Shackelford RE, Kaufmann WK, Paules RS (2000) Oxidative stress and cell cycle checkpoint function. Free Radic Biol Med 28:1387–1404 [DOI] [PubMed] [Google Scholar]
- Slominski A, Paus R, Wortsman J (1991) Can some melanotropins modulate keratinocyte proliferation? J Invest Dermatol 97:747 [DOI] [PubMed] [Google Scholar]
- Smith R, Healy E, Siddiqui S, Flanagan N, Steijlen PM, Rosdahl I, Jacques JP, Rogers S, Turner R, Jackson IJ, Birch-Machin MA, Rees JL (1998) Melanocortin 1 receptor variants in an Irish population. J Invest Dermatol 111:119–122 [DOI] [PubMed] [Google Scholar]
- Suzuki I, Cone RD, Sungbin IM, Nordlund J, Abdel-Malek ZA (1996) Binding of melanotrophic hormones to the melanocortin receptor MC1R on human melanocytes stimulates proliferation and melanogenesis. Endocrinology 137:1627–1633 [DOI] [PubMed] [Google Scholar]
- Thody AJ, Graham A (1998) Does α-MSH have a role in regulating skin pigmentation in humans? Pigment Cell Res 11:265–274 [DOI] [PubMed] [Google Scholar]
- Thody AJ, Higgins EM, Wakamatsu K, Ito S, Burchill SA, Marks JM (1991) Phaeomelanin as well as eumelanin is present in human epidermis. J Invest Dermatol 97:340–344 [DOI] [PubMed] [Google Scholar]
- Valverde P, Healy E, Jackson I, Rees JL, Thody AJ (1995) Variants of the melanocyte-stimulating hormone receptor gene are associated with red hair and fair skin in humans. Nat Genet 11:328–330 [DOI] [PubMed] [Google Scholar]
- Valverde P, Healy E, Sikkink S, Haldane F, Thody AJ, Carothers A, Jackson IJ, Rees JL (1996) The Asp84Glu variant of the melanocortin 1 receptor (MC1R) is associated with melanoma. Hum Mol Genet 5:1663–1666 [DOI] [PubMed] [Google Scholar]
- Wintzen M, Gilchrest BA (1996) Proopiomelanocortin, its derived peptides, and the skin. J Invest Dermatol 106:3–10 [DOI] [PubMed] [Google Scholar]