Abstract
The cause of Parkinson disease (PD) is still unknown, but genetic factors have recently been implicated in the etiology of the disease. So far, four loci responsible for autosomal dominant PD have been identified. Autosomal recessive juvenile parkinsonism (ARJP) is a clinically and genetically distinct entity; typical PD features are associated with early onset, sustained response to levodopa, and early occurrence of levodopa-induced dyskinesias, which are often severe. To date, only one ARJP gene, Parkin, has been identified, and multiple mutations have been detected both in families with autosomal recessive parkinsonism and in sporadic cases. The Parkin-associated phenotype is broad, and some cases are indistinguishable from idiopathic PD. In ⩾50% of families with ARJP that have been analyzed, no mutations could be detected in the Parkin gene. We identified a large Sicilian family with four definitely affected members (the Marsala kindred). The phenotype was characterized by early-onset (range 32–48 years) parkinsonism, with slow progression and sustained response to levodopa. Linkage of the disease to the Parkin gene was excluded. A genomewide homozygosity screen was performed in the family. Linkage analysis and haplotype construction allowed identification of a single region of homozygosity shared by all the affected members, spanning 12.5 cM on the short arm of chromosome 1. This region contains a novel locus for autosomal recessive early-onset parkinsonism, PARK6. A maximum LOD score 4.01 at recombination fraction .00 was obtained for marker D1S199.
Introduction
Parkinson disease (PD [MIM 168600]) is a common neurodegenerative disorder that usually occurs later in life, presenting with bradykinesia, resting tremor, muscular rigidity, and postural instability, with sustained response to levodopa. The pathological hallmarks are the occurrence of Lewy bodies (cytoplasmic eosinophilic inclusions) and neuronal loss, which affects the substantia nigra pars compacta (SNpc) extensively and other regions of the brain less specifically (Fearnley and Lees 1991). A genetic contribution to the etiology of PD is now well established and appears to be particularly important in cases with early onset of the disease (Tanner et al. 1999). The gene α-synuclein (Polymeropoulos et al. 1997) has been proved in families with autosomal dominant PD, and a second gene, that for ubiquitin carboxy-terminal hydrolase L1, has been implicated in disease causation (Leroy et al. 1998). Moreover, two other genetic loci have been mapped: PARK3 on chromosome 2p13 and PARK4 on chromosome 4p14-16.3 (Gasser et al. 1998; Farrer et al. 1999). However, autosomal dominant forms are rare and seem to account for only a small number of families. Autosomal recessive juvenile parkinsonism (ARJP [MIM 600116]) is a distinct clinical and genetic entity within familial PD; it is characterized by typical PD features associated with early age at onset (<40 years), slow progression of the disease, sustained response to levodopa, early levodopa-induced complications (fluctuations and dyskinesias) that are often severe, hyperreflexia, and mild dystonia mainly in the feet (Yamamura et al. 1973; Ishikawa and Tsuji 1996). Pathological studies have demonstrated a highly selective degeneration of dopaminergic neurons in the SNpc and the locus coeruleus and an absence of Lewy bodies (Takahashi et al. 1994). ARJP was initially described in Japanese patients (Yamamura et al. 1973). Recently, Matsumine and coworkers (1997) identified a novel gene, Parkin, located on chromosome 6q25.2-27 (Kitada et al. 1998). A wide variety of mutations have been detected in families with autosomal recessive parkinsonism and in sporadic cases of different ethnic origin (Hattori et al. 1998; Leroy et al. 1998; Abbas et al. 1999; Nisipeanu et al. 1999; Lücking et al. 2000). The inclusion criteria adopted in these studies were age at onset ⩽45 years (in at least one affected member of the family, for familial cases) and unaffected parents. It has been proved that, especially in non-Japanese cases, mutations in the Parkin gene are associated with a wide range in both age at onset (⩽64 years) and clinical signs, and they can result in parkinsonism that is clinically indistinguishable from PD, making genotype-phenotype correlation uncertain (Abbas et al. 1999; Klein et al. 2000). Nevertheless, only ∼50% of cases of autosomal recessive early-onset parkinsonism are related to the Parkin gene, and in most patients, especially sporadic cases, the contribution of genetic factors remains to be elucidated (Lücking et al. 2000). It is likely that a novel, as yet unidentified, gene is responsible for early-onset parkinsonism in some of these cases. We mapped a novel locus, named “PARK6,” to a 12.5-cM region on chromosome 1p35-36, in a large Italian family with autosomal recessive early-onset parkinsonism, confirming the genetic heterogeneity of this disorder.
Subjects and Methods
Subjects
A large Italian family (the Marsala kindred) comprising 122 members was investigated. The methodology and the clinical characterization of the family have been reported elsewhere (Valente et al. 1999). We examined 35 subjects, including 30 family members and 5 spouses. After informed consent was obtained, venous-blood samples were taken from 27 family members and the 5 spouses, for DNA analysis.
DNA and Linkage Analysis
DNA was extracted from leukocytes, by standard techniques. Exclusion of linkage between the disease and PARK2 on chromosome 6q has been reported elsewhere (Valente et al. 1999). A simulation study performed with the program SLINK (Ott 1989) revealed a maximum expected LOD score of 4.2 at recombination fraction (θ) .00. The family was then considered suitable for a genomewide homozygosity mapping (Lander and Botstein 1987). In the first screen, only the four definitely affected individuals were genotyped. We analyzed 400 highly polymorphic fluorescent microsatellite markers spanning the 22 autosomes and having an average distance of 10 cM (Linkage Mapping Set version 2; Applied Biosystems). Microsatellite markers were amplified from genomic DNA by the PCR technique, as specified by the manufacturers, and were electrophoresed on a denaturing acrylamide gel by a 377 XL DNA sequencer (Applied Biosystems). DNA fragment-size analysis was performed semiautomatically by GENESCAN and GENOTYPER software (Applied Biosystems), to determine genotypes.
When all the affected individuals were shown to be homozygous for the same allele at a given marker, the surrounding region was saturated with closely spaced microsatellite markers. All the available family members were then genotyped, and haplotypes were constructed manually. Phase was assigned on the basis of the minimum number of recombinants.
Two-point LOD scores were generated using the FASTLINK version of the MLINK program (Cottingham et al. 1993; Dwarkadas et al. 1994), an assumption of equal recombination rate for the two sexes, autosomal recessive inheritance, reduced penetrance (.90), gene frequency .001, and equal allele frequencies for each marker. Loops were broken by the automated loop-breaking procedure described by Becker and coworkers (1998). Marker order and genetic distances were based on the chromosome 1 genetic map of the Center for Medical Genetics, Marshfield Medical Research Foundation, and on the chromosome 1 physical map of the National Center for Biotechnology Information, available at The Human Genome website.
Results
Clinical Analysis
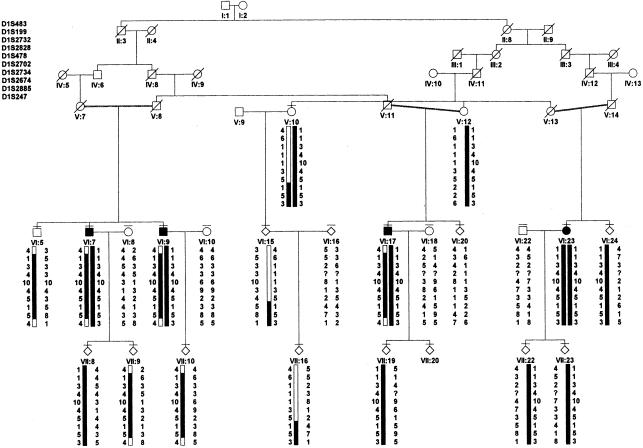
A simplified pedigree of the family is shown in figure 1. The age at onset in affected subjects was 32–48 years. The patients presented with the typical parkinsonian phenotype, including slow progression of the disease, sustained response to levodopa, and occurrence of levodopa-associated dyskinesias of variable severity. Foot dystonia and sleep benefit were not observed. No pathological data are available.
Figure 1.
Simplified pedigree of the family, and haplotypes of marker loci spanning the linked region on chromosome 1p35-36. Black symbols denote definitely affected individuals, and deceased members are denoted by a diagonal bar. A short thin horizontal bar above a symbol indicates a family member who was examined clinically. The black bar denotes the disease-associated haplotype. To protect patient confidentiality, a diamond symbol has been used to mask the sex of some unaffected individuals.
Both the high frequency of consanguineous marriages within the family and the presence of affected members of both sexes in only one generation strongly suggest an autosomal recessive mode of inheritance. Moreover, genealogic records permitted us to trace the origin of the family to a common ancestral couple seven generations back (18th century). A summary of the clinical presentation in the family and of the Hoehn and Yahr (1967) stage (on-state) are given in table 1.
Table 1.
Clinical Phenotype in Four Individuals with PD
|
Status of AffectedFamily Membera |
||||
| Patient Characteristics | VI:7 | VI:9 | VI:17 | VI:23 |
| Sex | M | M | M | F |
| Age at onset (years) | 45 | 48 | 32 | 38 |
| Asymmetry at onset? | Yes | No | Yes | Yes |
| Disease duration (years) | 29 | 20 | 20 | 7 |
| Levodopa-induced dyskinesias | ++ | ++ | +++ | + |
| Phenotype at examination: | ||||
| Resting tremor | 81 | ± | 69 | 68 |
| Bradykinesia | 0 | 77 | 0 | 0 |
| Rigidity | 0 | 0 | 0 | 0 |
| Postural instability | 0 | 0 | 0 | - |
| Hoehn and Yahr (1967) stage (on-state) | II.5 | II.5 | III | II |
| Daily medication: | ||||
| Levodopa (mg) | 300 | 625 | 800 | 500 |
| Pergolide (mg) | 4 | … | 3 | … |
+ = Mild; ++ = moderate; +++ = severe; ± = uncertain.
Homozygosity Mapping and Linkage Analysis
Linkage to PARK2 has been excluded by a previous study (Valente et al. 1999). Given the autosomal recessive pattern of inheritance, we expected homozygosity at the level of the disease locus in affected individuals, owing to identity by descent from a common progenitor (Lander and Botstein 1987). In the four definitely affected family members, 400 microsatellite markers covering all autosomes were analyzed. These individuals were homozygous for the same allele at seven marker loci, located on chromosomes 1, 8, 10, 14, and 19. The regions surrounding these seven loci and all regions surrounding noninformative markers were saturated with closely spaced microsatellite markers. All available family members were genotyped, and haplotypes were constructed. A combination of negative LOD scores and the detection of different haplotypes carried by the affected individuals allowed exclusion of all the autosomes, except for a region on the short arm of chromosome 1. All markers spanning this candidate interval produced positive LOD scores, with a maximum LOD score of 4.01 (θ = .00), between the disease and marker D1S199 (table 2). Calculation of pairwise LOD scores under different penetrance values and under the assumption of affected individuals–only analysis did not result in a significant change (data not shown). All affected individuals in the family shared a region of homozygosity, between D1S199 and D1S2885 (fig. 1), allowing the identification of a 12.5-cM interval containing a novel gene (i.e., PARK6) for autosomal recessive parkinsonism. The upper and lower extents of the region are determined by recombinations occurring between markers D1S483 and D1S199 (telomeric end) and between markers D1S2885 and D1S247 (centromeric end), respectively, which were detected in subjects VI:7, VI:9, and VI:17.
Table 2.
Pairwise LOD Scores for PD and Markers on Chromosome 1p
|
LOD Scores at θ = |
|||||||
| Marker | .0 | .01 | .05 | .1 | .2 | .3 | .4 |
| D1S483 | .18 | .53 | .83 | .83 | .59 | .31 | .10 |
| D1S199 | 4.01 | 3.92 | 3.55 | 3.08 | 2.14 | 1.24 | .48 |
| D1S2732 | 2.65 | 2.60 | 2.39 | 2.10 | 1.46 | .83 | .32 |
| D1S2828 | 3.49 | 3.42 | 3.12 | 2.72 | 1.89 | 1.08 | .41 |
| D1S478 | 3.45 | 3.36 | 3.03 | 2.61 | 1.78 | 1.00 | .38 |
| D1S2702 | 3.36 | 3.16 | 2.84 | 2.44 | 1.65 | .93 | .36 |
| D1S2734 | 3.62 | 3.55 | 3.26 | 2.87 | 2.03 | 1.19 | .46 |
| D1S2674 | 3.29 | 3.21 | 2.87 | 2.44 | 1.60 | .87 | .32 |
| D1S2885 | 3.94 | 3.85 | 3.49 | 3.03 | 2.11 | 1.23 | .49 |
| D1S247 | 1.01 | 1.35 | 1.59 | 1.48 | 1.03 | .58 | .19 |
Discussion
We have identified, by homozygosity mapping, a second locus for autosomal recessive early-onset parkinsonism, a locus named “PARK6,” on the short arm of chromosome 1 in a family from Sicily. The phenotype is characterized by early-onset parkinsonism with slow progression, sustained response to levodopa, and frequent levodopa-induced fluctuations and dyskinesias. Other signs often reported in ARJP, such as dystonia at onset and sleep benefit, are absent. The clinical picture in this family overlaps with that described in Parkin-positive families of non-Japanese origin (Abbas et al. 1999). Lücking and coworkers screened 73 families with onset age ⩽45 years in at least one affected member and 100 sporadic cases with onset age ⩽45 years (Lücking 2000). They found Parkin mutations in approximately half of the families, but in only 18/100 sporadic cases, most (14/18) with onset age ⩽20 years. The typical ARJP features—such as dystonia, hyperreflexia, sustained response to levodopa, and early levodopa-induced dyskinesias—were more frequent in the Parkin-positive patients than in the Parkin-negative group. However, these signs were less frequent than in previous reports, and none could be used to specifically distinguish between the two groups. The age at onset in the Parkin-positive group was, on average, lower than that in the Parkin-negative group, but mutations were identified in patients with onset age ⩾45 years (⩽64 years) and with clinical features difficult to distinguish from those of PD (Klein et al. 2000; Lücking et al. 2000).
The identification of several families that have autosomal recessive parkinsonism but that do not have Parkin mutations and are clinically indistinguishable from the Parkin-positive families strongly suggests the existence of at least one other, possibly similar, gene responsible for autosomal recessive parkinsonism. The identification of a novel locus for autosomal recessive early-onset parkinsonism in a family with a phenotype similar to that reported for Parkin is a crucial step toward the identification of another gene that could account for at least a subset of the Parkin-negative cases. It is noteworthy that the disease in this family is more characteristic of sporadic, Lewy-body PD, since it has both a later onset age than is seen in Parkin-positive patients and fewer atypical features. Still, the absence of autoptic confirmation does not allow us to assign a definite diagnosis of PD. The term “early-onset parkinsonism” is preferred in the case of families, such as those studied in the present article, for which information on the occurrence of Lewy bodies and on the pattern of neuronal loss is missing.
The role of this novel locus needs to be tested in other autosomal recessive Parkin-negative families and in cases of early-onset sporadic parkinsonism, since a sporadic presentation may underlie an autosomal recessive mode of inheritance. Until the PARK6 gene has been identified, a screen will be possible only by a search of regions of homozygosity within the candidate interval. Homozygosity mapping is a powerful technique to map genes responsible for autosomal recessive diseases in either consanguineous or nonconsanguineous pedigrees (Wang et al. 1997). However, this method is not completely accurate in determining the frequency of a gene.
It is worth noting that individual V:10 shares with the affected individuals a smaller region of homozygosity, from D1S2674 to D1S247. At the latest examination, this individual was 84 years old and did not show any signs of PD. It is likely that the gene is located outside the region of homozygosity shared by the unaffected person. In this case, the linked interval would be reduced to a 9-cM region between D1S483 and D1S2674. Alternatively, the identification of a definitely unaffected family member partially sharing the region of homozygosity can be explained if a reduced penetrance of the underlying gene is assumed. This would be in agreement with the reduced (.9) penetrance of the Parkin gene, and, in fact, we used this penetrance value for linkage calculations in this family. This issue will be clarified by detection of recombinations in affected individuals from other families.
The Parkin protein appears to be involved in protein degradation, as a ubiquitin-protein ligase interacting with the ubiquitin-conjugating enzyme UbcH7 (Shimura et al. 2000). The protein is abundant in the melanin-containing neurons of SNpc in normal individuals and in cases of sporadic parkinsonism, where it is also detected in association with α-synuclein within Lewy bodies. Conversely, mutations in the Parkin gene completely abolish the expression of the Parkin protein, which is not detectable in any region of patients' brains (Shimura et al. 1999). The mechanism of selective neural cell death remains to be clarified; however, it is likely that the absence of Parkin protein results in accumulation of still-unidentified proteins leading to selective neuronal degeneration.
The work of the human genome–sequencing project has resulted in availability of a draft sequence of much of the genome, including the 12.5-cM region that we have identified on chromosome 1. This region, which is equivalent to ∼10.5 Mb on physical maps of the region, contains a large number of genes, both predicted and already known. Included in this group are a number of homeobox genes and the genes for endothelin-converting enzyme 1 and a G-protein–coupled receptor. However, none of them either presents similarities with Parkin or represents an obvious candidate for PD. The identification of other families with linkage to chromosome 1p will help in refinement of the locus position on the genetic map, which is an essential step toward the identification of the gene and its function.
Acknowledgments
The authors are grateful to all family members who kindly participated in this study; to Dr. Giulio Cesare De Santis, who substantially contributed to an understanding of the family genealogy; and to Dr. Alejandro Schäffer, who offered helpful suggestions for linkage analysis. P.H.D. and N.W.W. are supported by MRC program grant G9706148, The Parkinson’s Disease Society (U.K.), and the Brain Research Trust. This project was supported in part by Telethon grants C38 (to A.R.B.) and E1165 (to A.A.).
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Center for Medical Genetics, Marshfield Medical Research Foundation, http://research.marshfieldclinic.org/genetics/ (for the chromosome 1 genetic map)
- Human Genome, The, http://www.ncbi.nlm.nih.gov/genome/guide/human/ (for the chromosome 1 physical map)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for PD [MIM 168600] and ARJP [MIM 600116])
References
- Abbas N, Lücking CB, Ricard S, Dürr A, Bonifati V, De Michele G, Bouley S, Vaughan JR, Gasser T, Marconi R, Broussolle E, Brefel-Courbon C, Harhangi BS, Oostra BA, Fabrizio E, Bohme GA, Pradier L, Wood NW, Filla A, Meco G, Denefle P, Agid Y, Brice A (1999) A wide variety of mutations in the parkin gene are responsible for autosomal recessive parkinsonism in Europe. Hum Mol Genet 8:567–574 [DOI] [PubMed] [Google Scholar]
- Becker A, Geiger D, Schäffer AA (1998) Automatic selection of loop breakers for genetic linkage analysis. Hum Hered 48:49–60 [DOI] [PubMed] [Google Scholar]
- Cottingham RW Jr, Idury RM, Schäffer AA (1993) Faster sequential genetic linkage computations. Am J Hum Genet 53:252–263 [PMC free article] [PubMed] [Google Scholar]
- Dwarkadas S, Schäffer AA, Cottingham RW Jr, Cox AL, Keleher P, Zwaenepoel W (1994) Parallelization of general linkage analysis problems. Hum Hered 44:127–141 [DOI] [PubMed] [Google Scholar]
- Farrer M, Gwinn-Hardy K, Muenter M, DE Vrieze FW, Crook R, Perez-Tur J, Lincoln S, Maraganore D, Adler C, Newman S, MacElwee K, McCarthy P, Miller C, Waters C, Hardy J (1999) A chromosome 4p haplotype segregating with Parkinson’s disease and postural tremor. Hum Mol Genet 8:81–85 [DOI] [PubMed] [Google Scholar]
- Fearnley JM, Lees AJ (1991) Ageing and Parkinson’s disease: substantia nigra regional selectivity. Brain 114:2283–2301 [DOI] [PubMed] [Google Scholar]
- Gasser T, Müller-Myhsok B, Wszolek ZK, Oehlmann R, Calne DB, Bonifati V, Bereznai B, Fabrizio E, Vieregge P, Horstmann RD (1998) A susceptibility locus for Parkinson's disease maps to chromosome 2p13. Nat Genet 18:262–265 [DOI] [PubMed] [Google Scholar]
- Hattori N, Kitada T, Matsumine H, Asakawa S, Yamamura Y, Yoshino H, Kobayashi T, Yokochi M, Wang M, Yoritaka A, Kondo T, Kuzuhara S, Nakamura S, Shimizu N, Mizuno Y (1998) Molecular genetic analysis of a novel Parkin gene in Japanese families with autosomal recessive juvenile parkinsonism: evidence for variable homozygous deletions in the Parkin gene in affected individuals. Ann Neurol 44:935–941 [DOI] [PubMed] [Google Scholar]
- Hoehn MM, Yahr MD (1967) Parkinsonism: onset, progression, and mortality. Neurology 17:427–442 [DOI] [PubMed] [Google Scholar]
- Ishikawa A, Tsuji S (1996) Clinical analysis of 17 patients in 12 Japanese families with autosomal recessive type juvenile parkinsonism. Neurology 47:160–166 [DOI] [PubMed] [Google Scholar]
- Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, Yokochi M, Mizuno Y, Shimizu N (1998) Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature 392:605–608 [DOI] [PubMed] [Google Scholar]
- Klein C, Pramstaller PP, Kis B, Page CC, Kann M, Leung J, Woodward H, Castellan CC, Scherer M, Vieregge P, Breakefield XO, Kramer PL, Ozelius LJ (2000) Parkin deletions in a family with adult-onset, tremor-dominant parkinsonism: expanding the phenotype. Ann Neurol 48:65–71 [PubMed] [Google Scholar]
- Lander ES, Botstein D (1987) Homozygosity mapping: a way to map human recessive traits with the DNA of inbred children. Science 236:1567–1570 [DOI] [PubMed] [Google Scholar]
- Leroy E, Anastasopoulos D, Konitsiotis S, Lavedan C, Polymeropoulos MH (1998) Deletions in the Parkin gene and genetic heterogeneity in a Greek family with early onset Parkinson’s disease. Hum Genet 103:424–427 [DOI] [PubMed] [Google Scholar]
- Lücking CB, Dürr A, Bonifati V, Vaughan J, De Michele G, Gasser T, Harhangi BS, Meco G, Denefle P, Wood NW, Agid Y, Brice A (2000) Association between early-onset Parkinson’s disease and mutations in the parkin gene. N Engl J Med 342:1560–1567 [DOI] [PubMed] [Google Scholar]
- Matsumine H, Saito M, Shimoda-Matsubayashi S, Tanaka H, Ishikawa A, Nakagawa-Hattori Y, Yokochi M, Kobayashi T, Igarashi S, Takano H, Sanpei K, Koike R, Mori H, Kondo T, Mizutani Y, Schaffer AA, Yamamura Y, Nakamura S, Kuzuhara S, Tsuji S, Mizuno Y (1997) Localization of a gene for an autosomal recessive form of juvenile parkinsonism to chromosome 6q25.2-27. Am J Hum Genet 60:588–596 [PMC free article] [PubMed] [Google Scholar]
- Nisipeanu P, Inzelberg R, Blumen SC, Carasso RL, Hattori N, Matsumine H, Mizuno Y (1999) Autosomal recessive juvenile parkinsonism in a Jewish Yemenite kindred: mutation of the Parkin gene. Neurology 53:1602–1604 [DOI] [PubMed] [Google Scholar]
- Ott J (1989) Computer-simulation methods in human linkage analysis. Proc Natl Acad Sci USA 86:4175–4178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polymeropoulos MH, Ladevan C, Leroy C, Ide SE, Dehejia A, Dutra A, Pike B, Root H, Rubenstein J, Boyer R, Stenroos ES, Chandrasekharappa S, Athanassiadou A, Papapetropoulos T, Johnson WG, Lazzarini AM, Duvoisin RC, Di Iorio G, Golbe LI, Nussbaum RL (1997) Mutation in the a-synuclein gene identified in families with Parkinson’s disease. Science 276:2045–2047 [DOI] [PubMed] [Google Scholar]
- Shimura H, Hattori N, Kubo S, Mizuno Y, Asakawa S, Minoshima S, Shimizu N, Iwai K, Chiba T, Tanaka K, Suzuki T (2000) Familial Parkinson disease gene product, parkin, is a ubiquitin-protein ligase. Nat Genet 25:302–305 [DOI] [PubMed] [Google Scholar]
- Shimura H, Hattori N, Kubo S, Yoshikawa M, Kitada T, Matsumine H, Asakawa S, Minoshima S, Yamamura Y, Shimizu N, Mizuno Y (1999) Immunohistochemical and subcellular localization of Parkin protein: absence of protein in autosomal recessive juvenile parkinsonism patients. Ann Neurol 45:668–672 [DOI] [PubMed] [Google Scholar]
- Takahashi H, Ohama E, Suzuki S, Horikawa Y, Ishikawa A, Morita T, Tsuji S, Ikuta F (1994) Familial juvenile parkinsonism: clinical and pathological study in a family. Neurology 44:437–441 [DOI] [PubMed] [Google Scholar]
- Tanner CM, Ottmann R, Goldman SM, Ellenberg J, Chan P, Mayeux R, Langston JW (1999) Parkinson’s disease in twins: an etiologic study. JAMA 281:341–346 [DOI] [PubMed] [Google Scholar]
- Valente EM, Bentivoglio AR, Ialongo T, Ferraris A, Frontali M, Wood NW, Albanese A (1999) A new family with autosomal recessive juvenile parkinsonism not linked to the Parkin gene. J Neurol 246 Suppl 1:166 [Google Scholar]
- Wang CY, Hawkins-Lee B, Ochoa B, Walker DR, She JX (1997) Homozygosity and linkage-disequilibrium mapping of the urofacial (Ochoa) syndrome gene to a 1-cM interval on chromosome 10q23-q24. Am J Hum Genet 60:1461–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamura Y, Sobue I, Ando K, Iida M, Yanagi T (1973) Paralysis agitans of early onset with marked diurnal fluctuation of symptoms. Neurology 23:239–244 [DOI] [PubMed] [Google Scholar]