Abstract
Methods are presented for incorporation of parent-of-origin effects into linkage analysis of quantitative traits. The estimated proportion of marker alleles shared identical by descent is first partitioned into a component derived from the mother and a component derived from the father. These parent-specific estimates of allele sharing are used in variance-components or Haseman-Elston methods of linkage analysis so that the effect of the quantitative-trait locus carried on the maternally derived chromosome is potentially different from the effect of the locus on the paternally derived chromosome. Statistics for linkage between trait and marker loci derived from either or both parents are then calculated, as are statistics for testing whether the effect of the maternally derived locus is equal to that of the paternally derived locus. Analyses of data simulated for 956 siblings from 263 nuclear families who had participated in a linkage study revealed that type I error rates for these statistics were generally similar to nominal values. Power to detect an imprinted locus was substantially increased when analyzed with a model allowing for parent-of-origin effects, compared with analyses that assumed equal effects; for example, for an imprinted locus accounting for 30% of the phenotypic variance, the expected LOD score was 4.5 when parent-of-origin effects were incorporated into the analysis, compared with 3.1 when these effects were ignored. The ability to include parent-of-origin effects within linkage analysis of quantitative traits will facilitate genetic dissection of complex traits.
Introduction
For some chromosomal regions, the genomic segment inherited from one parent is inactivated, so that expression of an allele in one of these regions is dependent on the sex of the parent from whom it was inherited. This phenomenon, known as “genomic imprinting,” plays an important role in several genetic diseases in humans, notably Prader-Willi syndrome, most cases of which are caused by a chromosome 15q deletion carried on the paternally derived chromosome (MIM 176270). Similarly, Angelman syndrome is caused by a chromosome 15q deletion carried on the maternally derived chromosome (MIM 105830). Imprinting may play a role in the inheritance of complex diseases, and a potential role for imprinting has been suggested for diseases such as bipolar affective disorder (Grigoroiu-Serbanescu et al. 1995), atopy (Ruiz et al. 1992; Sandford et al. 1993), and both type 1 and type 2 diabetes mellitus (Margaritte-Jeannin et al. 1995; Huxtable et al. 2000; Lindsay et al. 2000a). To facilitate detection of imprinted loci influencing susceptibility to disease, a number of methods have been developed to incorporate parent-of-origin effects into linkage analysis. These methods have generally employed parametric models (Heutink et al. 1992; Stine et al. 1995; Strauch et al. 2000) or have assessed allele sharing between pairs of affected siblings (Gershon et al. 1996; Paterson et al. 1999). Methods for incorporation of parent-of-origin effects into the linkage analysis of quantitative traits have received little attention. In the present article, allele-sharing methods for linkage analysis of quantitative traits are extended to incorporate parent-of-origin effects, and the statistical properties of the resulting methods are assessed via simulation.
Methods
Parent-Specific Allele Sharing in Siblings
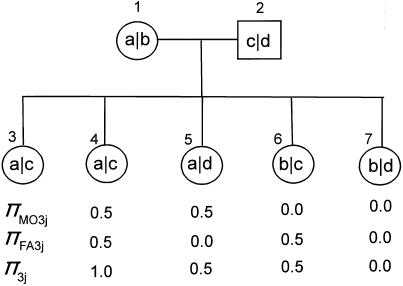
The most widely used nonparametric methods for linkage analysis of quantitative traits are the variance-components method (Amos 1994) and the regression method of Haseman and Elston (1972). Both methods are based on the principle that, if a marker is closely linked to a locus influencing a trait, pairs of relatives who share a larger proportion of marker alleles identical by descent (IBD) will tend to have more-similar trait values than are seen in pairs of relatives who share fewer alleles IBD. Consequently, a critical part of these methods is estimation of the proportion of marker alleles shared IBD between pairs of relatives (denoted as “πij,” where i and j denote the two individuals constituting the pair). A pair of siblings, for example, may share 0, 1, or 2 alleles IBD, corresponding to πij=.0, .5, and 1.0, respectively, and, on average, πij=.5 for sibling pairs. To assess parent-of-origin effects, it is necessary to partition πij into a component representing the proportion of alleles shared IBD derived from the mother (a component denoted as “πMOij”) and a component derived from the father (a component denoted as “πFAij”; see fig. 1). For sibling pairs, both πMOij and πFAij can be either .0 or .5 and will average .25. Note that, as presently defined, πMOij+πFAij=πij.
Figure 1.
Calculation of parent-specific IBD for a hypothetical family containing five siblings. Individual identification numbers are shown above the symbols, and alleles (designated “a,” “b,” “ c,” or “d”) at a hypothetical marker locus are shown within the symbols. For each of the siblings numbered “4”–“7,” calculation of allele sharing with individual 3 is shown for maternally derived alleles (πMO3j), paternally derived alleles (πFA3j), and all alleles (π3j).
If markers are not perfectly informative, πij must be estimated. A number of algorithms have been developed to accomplish this; in the present analyses, a modification of the method described by Curtis and Sham (1994) was employed. This method makes use of the risk-calculation facility of the LINKAGE program (Lathrop and Lalouel 1984; Lathrop et al. 1984). One member of a pair of relatives is considered affected by a hypothetical rare recessive disease, and risk calculation is performed for the other member of the pair, under the assumptions that there is no recombination between the marker and disease loci and that the affection status of all other members of the family is unknown. In these conditions, πij can be derived from the probability that the individual is a homozygous carrier (PHOM) and the probability that the individual is a heterozygous carrier (PHET), according to the equation πij=PHOM+(PHET/2). For siblings (and half-siblings), it is straightforward to extend this method to estimate parent-specific IBD, by employing separate recombination fractions for females (θMO) and males (θFA). To estimate the maternal contribution to allele sharing, the calculation is performed under the assumption that θMO=.0 and θFA=.5. Since, with θFA=.5, transmission of the hypothetical disease allele from the father is independent of the marker, the paternal contribution to the resulting estimate of πij will equal the expected value of πFAij: E(πFAij)=.25 for siblings. Thus, the maternal contribution to allele sharing can be calculated from the extent to which total allele sharing deviates from this expectation: πMOij=PHOM+(PHET/2)-E(πFAij). Similarly, πFAij can be derived by setting θMO=.5 and θFA=.0 and subtracting E(πMOij) from the estimated allele sharing.
Variance-Components Linkage Analysis
The variance-components method is a widely used and powerful tool for linkage analysis of quantitative traits. It involves fitting a linear “mixed” model, which estimates the trait mean (μ) and partitions the variance into a number of components (Amos 1994). Three components of variance are generally included: σ2Q, a monogenic component that reflects the influence of a quantitative-trait locus linked to the region of interest; σ2G, a “polygenic” component that reflects the effects of unlinked genes or other familial influences, including environmental factors shared by families; and σ2E, an “environmental” component that reflects effects unique to the individual. Under the assumptions of no recombination between trait and marker loci and multivariate normality, the phenotypic variance-covariance matrix (Ω) between individuals in a pedigree is
where Π is a matrix of the proportion of marker alleles shared IBD (i.e., the elements are the individual estimates of πij), Φ is a matrix of the expected proportion of alleles shared IBD (.5 for sibling pairs), and I is an identity matrix.
The logarithm of the likelihood (lnL) for a pedigree with n individuals is
where (xi-μ) represents a vector of the differences between the individual trait values xi and the mean and where C is a constant whose value depends on n (Lange et al. 1976). The parameters of these models are estimated by maximizing the sum of lnL over all pedigrees. Evidence for linkage is typically assessed by the likelihood-ratio test comparing the value of twice the negative of lnL from a model in which σ2Q is estimated versus that from a model in which σ2Q is constrained to equal 0. Conventionally, variance components are constrained to be ⩾0, in order to preserve a biologically meaningful interpretation. With this constraint, the distribution of the likelihood-ratio test under the null hypothesis (σ2Q=0) is not the usual χ2 with 1 df but, instead, is a mixture of distributions that is one-half-part χ2 with 1 df and one-half-part point mass at 0 (Hopper and Mathews 1982; Self and Liang 1987). To account for the one-sided nature of the test, the P value for the likelihood-ratio test is typically calculated by dividing the corresponding P value for a 1 df χ2 by 2. Such P values are asymptotically valid, provided that the assumption of multivariate normality is not violated (Allison et al. 1999). The likelihood-ratio test can also be converted to a LOD score, by division by 2×loge(10).
To accommodate parent-of-origin effects, the monogenic component of variance can be partitioned into (a) a component that reflects the influence of the quantitative-trait locus carried on the maternally derived chromosome (σ2QMO) and (b) a component that reflects the influence of the locus carried on the paternally derived chromosome (σ2QFA). The phenotypic variance-covariance matrix then becomes
where ΠMO is a matrix of the proportion of marker alleles shared IBD that are derived from the mother, ΠFA is a matrix of the proportion of alleles shared IBD that are derived from the father, and Φ and I are defined as above.
A number of statistical tests for linkage are possible under this model. In a fashion analogous to that described for the model without parent-of-origin effects, the likelihood-ratio test can be used to test for a trait-influencing locus on either the maternally derived chromosome (LODMO) or the paternally derived chromosome (LODFA). If both tests are conducted and the maximum of either LODMO or LODFA (MAXLODMO,FA) is taken as the evidence for linkage, then one would expect inflation of the type I error rate unless a correction for the fact that two statistical tests have been conducted is applied. This can be accomplished with a Bonferroni correction, by which the corrected P value is calculated as 1-(1-Pmax)2 , where Pmax is the nominal P value associated with the maximum of either LODMO or LODFA (Ott 1991). Alternatively, a global test for linkage with alleles derived from either or both parents can be conducted by comparing the model allowing for imprinting (eq. [2]) versus a model in which both monogenic components are constrained to equal 0 (LODIMP). In this case, the distribution of the likelihood-ratio test under the null hypothesis will be a mixture of distributions that is one-fourth-part χ2 with 2 df, one-half-part χ2 with 1 df and one-fourth-part point mass at 0 (Hopper and Mathews 1982; Self and Liang 1987). To account for the constraints on the variance components, the P value associated with LODIMP can be calculated by dividing the P value associated with the 2 df χ2 distribution by 2, in a fashion analogous to the approach used for one component of variance. Since ΠMO+ΠFA=Π, the model in equation (1) is a special case of that in equation (2)—a special case in which σ2QMO and σ2QFA are constrained to be equal. Therefore, the usual test for linkage, assessed in the absence of parent-of-origin effects (eq. [1]), can also be seen as a test of the null hypothesis that both monogenic variance components are 0 versus the alternative that both are >0 and equal to one another (HA: σ2QMO=σ2QFA>0). For the purposes of the present article, this conventional linkage test will therefore be termed “LODEQ.” Finally, statistical significance of parent-of-origin effects can be assessed by comparison of the full model allowing for such effects (eq. [2]) versus the model in which the monogenic components are constrained to be equal (eq. [1]). The P value for this test of whether the monogenic components are different (Pdiff) can be calculated from a χ2 distribution with 1 df.
Haseman-Elston Regression
The Haseman-Elston regression method is another technique that is widely used for linkage analysis of quantitative traits, especially for sibling pairs (Haseman and Elston 1972). In this method, the squared difference between the trait values for a pair of relatives, (xi-xj)2, is regressed against the proportion of marker alleles shared IBD, πij):
A negative value of β reflects a tendency for siblings to be more similar with respect to the trait as they share a greater proportion of marker alleles IBD and, thus, implies linkage between the trait and marker loci. Therefore, a test of the null hypothesis, that β=0, against the alternative, that β<0, provides statistical assessment of the evidence for linkage—analogous to that provided by LODEQ in the variance-components method. The model parameters can be estimated by ordinary least-squares regression, but the P value is calculated from a t-statistic with a reduced number of degrees of freedom, to account for any nonindependence introduced by analysis of multiple pairs within sibships (Wilson and Elston 1993).
The Haseman-Elston method can be readily extended to accommodate parent-of-origin effects, by estimation of separate β coefficients, according to the source of allele sharing:
Linkage with the maternally or paternally derived chromosome can be assessed by testing whether βMO<0 (i.e., LODMO) or βFA<0 (i.e., LODFA), and the statistic with the strongest evidence for linkage (MAXLODMO,FA) can be taken, by employing a Bonferroni correction, as for the variance-components analysis. The global test for linkage (i.e., LODIMP) can be conducted with an F-test (with 2 df in the numerator) to test the null hypothesis that βMO and βFA are both 0. Since the linear-regression method does not constrain βMO and βFA to be <0, adjustment of the P value to produce an equivalent to a one-sided test is appropriate only if both βMO and βFA are <0; if this is not the case, then linkage should be declared only if strong evidence for linkage is seen with at least one of the parental chromosomes. Determining the significance of the parent-of-origin effect requires an assessment of whether βMO=βFA, and this can be accomplished by dividing the difference between βMO and βFA by its standard error, determined from the variances of βMO and βFA and from the covariance between the β coefficients (Kleinbaum et al. 1982):
 |
The two-tailed P value for the resulting statistic (Pdiff) can be calculated from a t-distribution, with calculation, as described above, of the number of degrees of freedom. Null and alternate hypotheses associated with the various statistics are shown in table 1, for both variance-components and Haseman-Elston methods.
Table 1.
Null and Alternate Hypotheses for Statistics Used in Variance-Components and Haseman-Elston Analyses Allowing for Parent-of-Origin Effects[Note]
|
Variance-Components Analyses |
Haseman-Elston Analyses |
|||
| Statistic | Null Hypothesis | Alternate Hypothesis | Null Hypothesis | Alternate Hypothesis |
| LODMO | σ2QMO=0 | σ2QMO>0 | βMO=0 | βMO<0 |
| LODFA | σ2QFA=0 | σ2QFA>0 | βFA=0 | βFA<0 |
| MAXLODMO,FA | σ2QMO=σ2QFA=0 | σ2QMO>0 or σ2QFA> 0 | βMO=βFA=0 | βMO<0 or βFA< 0 |
| LODIMP | σ2QMO=σ2QFA=0 | σ2QMO>0 or σ2QFA> 0 | βMO=βFA=0 | βMO<0 or βFA< 0 |
| LODEQ | σ2QMO=σ2QFA=0 (σ2Q=0) | σ2QMO=σ2QFA>0 (σ2Q>0) | βMO=βFA=0 (β=0) | βMO=βFA<0 (β<0) |
| Pdiff | σ2QMO=σ2QFA | σ2QMO≠σ2QFA | βMO=βFA | βMO≠βFA |
Note.— Hypotheses are defined in terms of equation (2), for the variance-components method, and in terms of equation (4), for the Haseman-Elston method; in parentheses are LODEQ hypotheses, which are defined in terms of equation (1), for the variance-components method, and in terms of equation (3), for the Haseman-Elston method.
Simulations
A simulation study was conducted to assess the performance of these methods. Genotypic and phenotypic data were simulated under a number of genetic models for a set of nuclear families that had participated in a genomewide linkage study of loci influencing susceptibility to type 2 diabetes mellitus and related traits (Hanson et al. 1998). The ability to partition, into maternal and paternal components, alleles shared IBD requires genotypic data on at least one parent (either measured directly or inferred through extended relationships), and the present sample was selected to approximate the subset of families, from the linkage study, for which this information was available and to which we first applied this method (Lindsay et al. 2000b). The data set consisted of 263 nuclear families containing 956 siblings; the median sibship size was 3, with a range of 2–11. Genotypic data were considered to be available from both parents in 32% of the families, from the mother only in 48%, and from the father only in 20%, as was the case in the families with at least one parent genotyped that had been reported in our previous study (Hanson et al. 1998).
Data were simulated by SLINK (Ott 1989; Weeks et al. 1990) and by programs written by the authors of the present article. Two unlinked biallelic loci with equally frequent alleles were initially generated, and a marker locus with four equally frequent alleles that was tightly linked (θ=0) to one of the biallelic loci was also simulated. A total of 500 replicates of genotypic data were thus created. Quantitative-trait data were generated by allowing the two biallelic loci to influence the trait, with sampling of values from a mixture of normal distributions. To assess type I error, the trait locus linked to the marker was assumed to have no influence on the trait, whereas the unlinked trait locus was assumed to have an effect that resulted in a heritability (h2) of .5. To assess type I error under a variety of conditions, data were generated for a model in which only the maternally derived allele at the unlinked trait locus was expressed and for a model with no imprinting; in addition, data were generated for a model in which the trait was influenced by a factor shared among all siblings, as might occur, for example, in mitochondrial inheritance or with an environmental factor affecting an entire sibship. To assess power, data were generated for models in which the biallelic locus linked to the marker influenced the quantitative trait, with varying locus-specific heritability. To determine power for assessment of parent-of-origin effects, data were simulated such that only the maternally derived allele at the biallelic locus linked to the marker influenced the trait. The biallelic locus not linked to the marker also influenced the trait, but without an imprinting effect, such that the total heritability was .5; this allows power to be examined for cases in which there is oligogenic inheritance. To determine type I error in detection of parent-of-origin effects (Pdiff) and to compare power to detect imprinted loci versus that to detect nonimprinted loci, data were also generated under models in which the trait locus linked to the marker was not imprinted. In these simulations, the maternally derived allele at the unlinked trait locus was also assumed to influence the trait, such that the total heritability was .5; this allows type I error for Pdiff to be determined for the case in which a locus linked to the marker is not imprinted but in which an imprinted locus occurs elsewhere in the genome. Models employed in the simulations are summarized in table 2.
Table 2.
Summary of Models Used in Simulation Studies
|
Trait Locus Linked to Marker |
Trait Locus Unlinked to Marker |
||||
| Mechanism of Action | h2 | Mechanism of Action | h2 | Total h2 | |
| Models Used to Assess Type I Error of Linkage Statistics |
|||||
| Maternal expression | … | .00 | Maternally expressed | .50 | .50 |
| Nonimprinted | … | .00 | Nonimprinted | .50 | .50 |
| Common sibling | … | .00 | … | .00 | .50a |
| Models Used to Assess Power to Detect Nonimprinted Loci |
|||||
| Locus-specific h2: | |||||
| .10 | Nonimprinted | .10 | Maternally expressed | .40 | .50 |
| .20 | Nonimprinted | .20 | Maternally expressed | .30 | .50 |
| .30 | Nonimprinted | .30 | Maternally expressed | .20 | .50 |
| .40 | Nonimprinted | .40 | Maternally expressed | .10 | .50 |
| .50 | Nonimprinted | .50 | … | .00 | .50 |
| Models Used to Assess Power to Detect Imprinted Loci |
|||||
| .10 | Maternally expressed | .10 | Nonimprinted | .40 | .50 |
| .20 | Maternally expressed | .20 | Nonimprinted | .30 | .50 |
| .30 | Maternally expressed | .30 | Nonimprinted | .20 | .50 |
| .40 | Maternally expressed | .40 | Nonimprinted | .10 | .50 |
| .50 | Maternally expressed | .50 | … | .00 | .50 |
Neither genetic locus influenced the trait, but a familial factor influenced the trait to give a total h2 of .50.
Analyses of linkage between the simulated markers and quantitative traits were conducted by use of the procedures outlined above. The FASTLINK (Cottingham et al. 1993; Schäffer et al. 1994) modification of the LINKAGE program (Lathrop and Lalouel 1984; Lathrop et al. 1984) was used to estimate parent-specific IBD for pairs of siblings, at each simulated marker. Variance components were estimated by the scoring algorithm (Lange et al. 1976), implemented in PROC IML of SAS. Haseman-Elston analyses were conducted in SAS, by use of standard regression methods.
Results
Type I error rates for the various linkage statistics, determined by analysis of replicates generated under models in which there was no linkage between the trait and marker loci, are shown in table 3. For both variance-components and Haseman-Elston methods, type I error rates were similar to the nominal values, for all statistics. When the evidence for linkage was maximized over both maternal and paternal chromosomes (MAXLODMO,FA), correction for multiple testing was necessary; in the absence of the Bonferroni correction, type I error rates for MAXLODMO,FA were approximately double the values reported in table 3.
Table 3.
Type I Error Rates (with 95% Confidence Intervals) for Linkage Statistics from Models Incorporating Parent-of-Origin Effects
|
Type I Error Rate (95% Confidence Interval) |
||||
| Variance-Components Analysis |
Haseman-Elston Analysisa |
|||
| Generating Model and Statistic | P<.05 | P<.01 | P<.05 | P<.01 |
| Maternal expression:b | ||||
| LODEQ | .070 (.050–.097) | .010 (.004–.025) | .050 (.033–.074) | .010 (.004–.025) |
| LODMO | .062 (.043–.088) | .008 (.003–.022) | .050 (.033–.074) | .012 (.005–.027) |
| LODFA | .038 (.024–.060) | .004 (.001–.016) | .046 (.030–.069) | .008 (.003–.022) |
| MAXLODMO,FAc | .046 (.030–.069) | .008 (.003–.022) | .046 (.030–.069) | .012 (.005–.027) |
| LODIMP | .040 (.025–.062) | .006 (.002–.019) | .044 (.028–.067) | .012 (.005–.027) |
| Nonimprinted:b | ||||
| LODEQ | .038 (.024–.060) | .006 (.002–.019) | .034 (.021–.055) | .008 (.003–.022) |
| LODMO | .054 (.037–.079) | .010 (.004–.025) | .044 (.028–.067) | .010 (.004–.025) |
| LODFA | .042 (.027–.065) | .008 (.003–.022) | .054 (.037–.079) | .010 (.004–.025) |
| MAXLODMO,FAc | .044 (.028–.067) | .004 (.001–.016) | .054 (.037–.079) | .012 (.005–.027) |
| LODIMP | .042 (.027–.065) | .002 (.0001–.013) | .046 (.030–.069) | .010 (.004–.025) |
| Common sibling:b | ||||
| LODEQ | .038 (.024–.060) | .010 (.004–.025) | .046 (.030–.069) | .010 (.004–.025) |
| LODMO | .040 (.025–.062) | .002 (.0001–.013) | .048 (.032–.072) | .004 (.001–.016) |
| LODFA | .032 (.019–.053) | .010 (.004–.025) | .034 (.021–.055) | .012 (.005–.027) |
| MAXLODMO,FAc | .032 (.019–.053) | .006 (.002–.019) | .042 (.027–.065) | .006 (.002–.019) |
| LODIMPc | .022 (.012–.040) | .006 (.002–.019) | .036 (.022–.057) | .008 (.003–.022) |
| Combined:b | ||||
| LODEQ | .049 (.039–.061) | .009 (.005–.015) | .043 (.034–.055) | .009 (.005–.016) |
| LODMO | .052 (.042–.065) | .007 (.003–.013) | .047 (.037–.060) | .009 (.005–.015) |
| LODFA | .037 (.029–.049) | .007 (.004–.014) | .045 (.035–.057) | .010 (.006–.017) |
| MAXLODMO,FAc | .041 (.031–.052) | .006 (.003–.012) | .047 (.037–.060) | .010 (.006–.017) |
| LODIMP | .035 (.026–.046) | .005 (.002–.010) | .042 (.033–.054) | .010 (.006–.017) |
For Haseman-Elston analyses, results are considered positive only if both βMO and βFA are <0 or if linkage, at the relevant P value, is seen with at least one of the parental chromosomes.
Results are based on analysis of 500 replicates for each model; thus, “Combined” represents 1,500 replicates. Confidence intervals were calculated as described by Fleiss (1981).
A Bonferroni correction is applied to the nominal P value from the maximum of LODMO and LODFA.
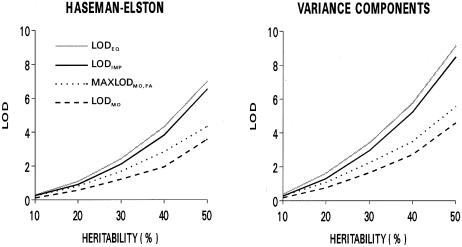
Median LOD scores for replicates generated under models in which a nonimprinted trait locus was linked to the marker are shown in figure 2. The analysis model that did not incorporate parent-of-origin effects (LODEQ) was the most powerful, as expected, since its alternative hypothesis corresponds most closely to the generating model. The general test allowing for parent-of-origin effects (i.e., LODIMP) was modestly less powerful, a consequence of the additional degree of freedom required for estimation of the additional parameter. Tests of linkage that were specific to alleles shared from a single parent (i.e., LODMO and LODFA) were substantially less powerful, and this loss of power was only modestly overcome by use of the maximum LOD score for either parent. Variance-components analyses were somewhat more powerful than Haseman-Elston analyses, for all statistics. Table 4 shows type I error rates for tests of parent-of-origin effects derived from analyses of these replicates. Type I error rates for the variance-components analyses were close to the nominal values, but those for the Haseman-Elston methods were slightly higher than the nominal values. To examine the possibility that the inflated type I error rates reflect the failure of the Haseman-Elston method to adequately account for nonindependence of pairs in sibships with more than two siblings, data were analyzed for one sibling pair selected at random from each family. In this situation, the type I error rates for the Haseman-Elston method were close to the nominal values (although the expected LOD scores were much lower).
Figure 2.
Median LOD scores, in analysis of 500 replicates generated for a nonimprinted quantitative-trait locus, for each of the tests of linkage, by the proportion of phenotypic variance attributed to the locus (heritability). Values for LODFA were virtually identical to those for LODMO and, thus, are not shown. For comparability, P values for LODIMP and MAXLODMO,FA have been converted to equivalent LOD scores.
Table 4.
Type I Error Rates (with 95% Confidence Intervals) at Pdiff<.05, for Tests of Parent-of-Origin Effects, by h2
|
Type I Error Rate (95% Confidence Interval)a |
|||
| Haseman-Elston Analysis |
|||
| h2 | Variance-Components Analysis | All Pairs | One Pair per Sibship |
| .10 | .034 (.021–.055) | .072 (.052–.099) | .034 (.021–.055) |
| .20 | .032 (.019–.053) | .086 (.064–.115) | .056 (.038–.081) |
| .30 | .054 (.037–.079) | .102 (.078–.133) | .048 (.032–.072) |
| .40 | .040 (.025–.062) | .130 (.102–.163) | .032 (.019–.053) |
| .50 | .050 (.033–.074) | .116 (.090–.148) | .048 (.032–.072) |
| Overall | .042 (.035–.051) | .101 (.090–.114) | .044 (.036–.053) |
Based on analysis of 500 replicates for each value of h2; confidence intervals were calculated as described by Fleiss (1981).
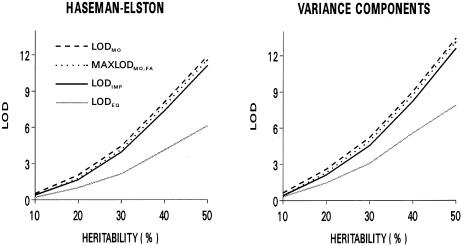
Median LOD scores for replicates generated under a model with an imprinted maternally expressed quantitative-trait locus linked to the marker are shown in figure 3. In this case, the statistic assessing linkage to the maternal chromosome (i.e., LODMO) was the most powerful, which is not surprising, since this statistic provides the most direct test of the generating model; however, taking the maximum of LODMO and LODFA was only slightly less powerful (the power loss being a consequence of the correction for multiple testing), and the general test allowing for parent-of-origin effects (i.e., LODIMP) was only modestly less powerful, than LODMO. Although linkage analysis that did not allow for imprinting (i.e., LODEQ) provided some power for detection of an imprinted locus, it was substantially less powerful than the tests that explicitly allowed for parent-of-origin effects. Again, the variance-components method was more powerful than the Haseman-Elston method. To further examine the potential increase in power for analyses that account for imprinting, the distribution of LODEQ was examined for replicates in which analyses accounting for imprinting gave some evidence for linkage (LODIMP>1.18; P<.01). The results show that, in a significant number of situations for which linkage can be detected by LODIMP, the analysis ignoring imprinting gives little to no evidence for linkage, particularly for loci with more-moderate effects (table 5); for example, for an imprinted locus accounting for 10% of the phenotypic variance, only 60% of replicates with LODIMP>1.18 also had LODEQ>1.18, and 11% had LODEQ⩽0.59 (P⩾.05).
Figure 3.
Median LOD scores, in analysis of 500 replicates generated for a maternally expressed quantitative-trait locus, for each of the tests of linkage, by the proportion of phenotypic variance attributed to the locus (heritability). Values for LODFA were 0 for all values of heritability and, thus, are not shown. For comparability, P values for LODIMP and MAXLODMO,FA have been converted to equivalent LOD scores.
Table 5.
Distribution of LODEQ Scores for Replicates Generated under an Imprinted Model for Which an Analysis Accounting for Imprinting Gave Evidence for Linkage (LODIMP>1.18), by h2[Note]
|
Variance-Components Analysis |
Haseman-Elston Analysis |
|||||||
| Proportion of N |
Proportion of N |
|||||||
| h2 | Na | LODEQ⩽0.59 | 0.59<LODEQ⩽1.18 | LODEQ>1.18 | Na | LODEQ⩽0.59 | 0.59<LODEQ⩽1.18 | LODEQ>1.18 |
| .1 | 93 | .108 | .290 | .602 | 85 | .306 | .200 | .494 |
| .2 | 382 | .055 | .194 | .754 | 331 | .160 | .245 | .595 |
| .3 | 485 | .016 | .072 | .911 | 462 | .052 | .113 | .835 |
| .4 | 499 | .002 | .004 | .994 | 495 | .014 | .020 | .966 |
| .5 | 500 | .000 | .002 | .998 | 499 | .004 | .010 | .986 |
Note.— P values corresponding to LOD scores of 1.18 and .59 are .01 and .05, respectively.
N = number of replicates (of a total of 500) for which LODIMP>1.18.
The power of the tests for assessment of parent-of-origin effects in analyses of data generated under a model with an imprinted maternally expressed locus is shown in table 6. For models with a locus-specific heritability of 10%, the power to detect these effects was low, but it increased rapidly for loci with larger effects. Restriction of the Haseman-Elston analysis to a single sibling pair for each sibship resulted in a substantial loss of power, versus analysis of all pairs.
Table 6.
Proportion (with 95% Confidence Intervals) of Replicates Demonstrating Statistically Significant Parent-of-Origin Effects at Pdiff<.05, for Data Generated under Imprinted Models, by h2
|
Proportion (95% Confidence Interval) of Replicates with Significant Parent-of-Origin Effects at Pdiff<.05a |
|||
| Haseman-Elston Analysis |
|||
| h2 | Variance-Components Analysis | All Pairs | One Pair per Sibship |
| .10 | .196 (.163–.234) | .212 (.178–.251) | .042 (.027–.065) |
| .20 | .622 (.578–.664) | .592 (.547–.635) | .076 (.055–.104) |
| .30 | .904 (.874–.928) | .858 (.824–.887) | .232 (.196–.272) |
| .40 | .994 (.981–.998) | .978 (.960–.988) | .340 (.299–.384) |
| .50 | .998 (.987–.9999) | .994 (.981–.998) | .520 (.475–.564) |
Based on analysis of 500 replicates for each value of h2; 95% confidence intervals were calculated as described by Fleiss (1981).
Discussion
The knowledge that genomic imprinting plays an important role in some human diseases—and the possibility that it might be involved in others—has led several investigators to incorporate parent-of-origin effects into linkage analyses. These efforts have been largely conducted by use of likelihood-based parametric-linkage methods. Parent-of-origin effects can be incorporated into such analyses by classification of families according to the sex of the transmitting parent (Stine et al. 1995; Nöthen et al. 1999); by the use of different penetrances, depending on whether an individual has an affected father or mother (Heutink et al. 1992; Meijers-Heijboer et al. 1992); or by allowance for different penetrances, depending on the parental origin of the disease allele (Strauch et al. 2000). In addition, family-based tests of allelic association can be extended to allow for differential effects, according to the sex of the transmitting parent, by use of either the transmission/disequilibrium test or log-linear models (Rice et al. 1995; Weinberg 1999). Affected-sib-pair methods of linkage analysis can also be stratified by the sex of the transmitting parent (Gershon et al. 1996) and can be modified to assess allele sharing according to parent of origin, in a fashion similar to that employed in the present analyses (Paterson et al. 1999); however, methods for incorporation of parent-of-origin effects into linkage analysis of quantitative traits have received little attention. The present work shows that standard allele-sharing methods for linkage analysis of quantitative traits, such as the variance-components and Haseman-Elston methods, can be extended to incorporate parent-of-origin effects and that this approach can result in substantially increased power to detect imprinted loci, versus analyses that ignore parent-of-origin effects. Statistical tests can also be conducted to determine whether the effects of a locus differ according to parent of origin, thus allowing one to directly assess the hypothesis of imprinting.
Analyses of simulated data showed that type I error rates were close to the nominal values for statistics assessing linkage between the trait and marker loci (statistics such as LODEQ, LODMO, LODFA, MAXLODMO,FA, and LODIMP). The extent to which results of analyses of simulated data can be generalized beyond the family structures and genetic models for which the data were simulated is uncertain, but the present analyses were conducted for families that had participated in a linkage study, and they incorporated a number of features likely to be encountered in practice; these features include variable sibship size, less than perfectly informative markers, and missing genotypic data on some parents. Thus, the present analyses suggest that these factors do not produce substantial inflation of type I error rates. Similar results have been found, in a variety of situations, for both variance-components and Haseman-Elston analyses that do not include parent-of-origin effects (Amos et al. 1996; Pratt et al. 2000). The present simulations were not designed to produce the severe departures from multivariate normality (e.g., kurtosis) that, at least for variance-components analyses, can produce inflation of type I error rates (Allison et al. 1999). Application of the present methods to linkage analysis of traits with severely kurtotic distributions may require either the use of a normalizing transformation or the calculation of empirical P values by simulation. Tests of statistical significance of parent-of-origin effects (Pdiff) had type I error rates for the variance-components method that were near the nominal values but had type I error rates for the Haseman-Elston method that were somewhat inflated. The inflation of type I error rates for the Haseman-Elston method appears to reflect the failure of this method to account adequately for the nonindependence of sibling pairs derived from sibships with more than two siblings. Although restriction of the Haseman-Elston analysis to a single pair from each sibship results in appropriate type I error rates, this strategy can result in a substantial loss of power to detect parent-of-origin effects.
The power of linkage analysis depends on the quantity and structure of the available family material, as well as on the effect of the locus in question. Simulations for nonimprinted loci have suggested that, for sample sizes typical of linkage studies, quantitative-trait loci need to have at least moderate effects (accounting for 20%–30% of the phenotypic variance) in order to have reasonable power to detect strong (LOD >3) linkage (Amos et al. 1996; Almasy and Blangero 1998). The present analyses are consistent with this and show that power to detect imprinted loci is comparable to that for nonimprinted loci. Obviously, power is greatest for those analyses that best capture the underlying genetic architecture—for example, LODMO for maternally expressed loci and LODEQ for nonimprinted loci. If linkage is assessed for both parental chromosomes separately, the LOD scores need to be adjusted accordingly, in the form of either a Bonferroni correction for MAXLODMO, FA or an extra degree of freedom for LODIMP, in order to reduce type I error. This will result in some loss of power. The variance-components method was more powerful than the Haseman-Elston method, for detection of both linkage and parent-of-origin effects. This finding has also been seen in other analyses of simulated data on nonimprinted loci (Pugh et al. 1997; Pratt et al. 2000); it is probably due to additional information derived from the covariance between siblings, information that is not contained in the squared trait difference (Drigalenko 1998).
The present analyses were restricted to sibships, and additional work is needed to devise analytic strategies for detection of imprinted quantitative-trait loci in extended pedigrees. For many collections of family data, however, it is likely that sibships provide most of the information for assessment of parent-of-origin effects. The variance-components method can readily accommodate more-distant relatives (Almasy and Blangero 1998), but, to account for additional sources of familial resemblance, analyses of such data may require models more complex than those employed here. In analyses including both siblings and half-siblings, for example, it is probably necessary to partition the polygenic variance (i.e., σ2G) into components representing maternal and paternal contributions; these separate polygenic components are not identifiable in analyses of sibships.
Recombination rates are sex specific; for most chromosomal regions, θMO>θFA, and, on average, the genetic distance between markers is 1.6 times greater for females than for males (Broman et al. 1998). Significant parent-of-origin effects detected with the present methods may thus reflect greater recombination between trait and marker loci in females versus that in males, or vice versa, rather than genomic imprinting. To examine the sensitivity of the present analyses to sex-specific differences in recombination, additional data were simulated under a model that included a nonimprinted quantitative-trait locus (h2=.5) linked to a marker with different θFA and θMO values. Results of analyses of these data (table 7) suggest that, even with a fairly strong effect of the locus, the method is not very sensitive to modest sex differences in recombination (i.e., a ratio of sex-specific genetic distances <10). It is difficult to distinguish, statistically, between imprinting and a sex difference in recombination, since imprinting will result in recombination rates that are apparently different between sexes (Smalley 1993). However, it may be possible to determine the plausibility of each mechanism by examination of both the female:male ratio of genetic distance and the pattern of parent-specific linkage results in the region of interest.
Table 7.
Proportion (with 95% Confidence Intervals) of Replicates with Pdiff<.05 in Simulations of a Nonimprinted Locus Influencing the Quantitative Trait (h2=.5) with Variable Differences between θFA and θMO [Note]
|
Proportion (95% Confidence Interval) of Replicates with Pdiff<.05 |
|||
| θFA | θMO | Variance-Components Analysis | Haseman-Elston Analysis |
| .01 | .02 | .04 (.01–.11) | .08 (.04–.16) |
| .01 | .05 | .09 (.04–.17) | .14 (.08–.23) |
| .01 | .10 | .08 (.04–.16) | .14 (.08–.23) |
| .01 | .15 | .15 (.09–.24) | .21 (.14–.31) |
| .01 | .20 | .34 (.25–.44) | .31 (.22–.41) |
Note— Results are based on analysis of 100 replicates for each combination of θFA and θMO. Confidence intervals were calculated as described by Fleiss (1981).
These methods recently have been applied to data from a genomic scan to detect loci influencing obesity and type 2 diabetes in Pima Indians (Hanson et al. 1998; Lindsay et al. 2000b). Analyses that allowed for imprinting detected tentative evidence for linkage in additional regions that were not detected in the initial analyses, which did not take into account the parent of origin. For body-mass index, for example, evidence was obtained for a maternally expressed locus on chromosome 5p (LODMO=1.7; LODFA=0; LODIMP=1.2; LODEQ=0.1; Pdiff=.01) and for a paternally expressed locus on chromosome 10p (LODMO=0; LODFA=1.7; LODIMP=1.1; LODEQ=0.7; Pdiff=.04). The strongest evidence for linkage with body-mass index was seen in a chromosome 11 region that was detected in the initial analyses; this locus did not appear to be imprinted (LODIMP=2.1; LODEQ=2.7; Pdiff=.60). Assessment of parent-of-origin effects requires that genotypic data be available from at least one parent, and this resulted in exclusion of some families that could be used in the analyses that did not take into account the parent of origin. The results of the initial analyses of a larger sample, therefore, gave stronger evidence for linkage on chromosome 11 (LODEQ=3.6) than was seen in analyses of families informative for parent-of-origin effects (LODEQ=2.7).
The optimal strategy for conducting linkage analyses with parent-of-origin effects depends on the nature of the available families and on the characteristics of the trait being analyzed. To maximize power to detect nonimprinted loci and to minimize false-positive signals, it may be reasonable to conduct initial analyses that do not allow for imprinting (i.e., LODEQ) and to test for parent-of-origin effects in regions that show suggestive linkage. This approach, however, may result in failure to detect imprinted loci that could be detected if a genomewide linkage analysis accounting for imprinting was conducted. It therefore may be useful to assess a priori whether imprinted loci are likely—by use of epidemiologic data, for example. Of course, if imprinting is strongly suspected, then the optimal approach would be to assess linkage to the chromosome of the parent whose alleles are putatively expressed. The extent to which the human genome is imprinted is currently unknown. It is likely, on the basis of homology with the mouse genome, that several large blocks of imprinted genes exist but that most of the genome is probably not imprinted (Shire 1989; Bartolomei and Tilghman 1997). Many of the known imprinted genes influence fetal development (Bartolomei and Tilghman 1997), and, therefore, incorporation of parent-of-origin effects into linkage analyses may be particularly useful for study of development-related traits, such as birth weight. Regardless of the strategy by which linkage is detected initially, knowledge regarding parent-of-origin effects may be useful for positional cloning efforts. The ability to conduct quantitative-trait linkage analyses incorporating parent-of-origin effects thus provides a useful tool for genetic dissection of complex traits.
Acknowledgments
The authors thank Dr. Laura Almasy, of the Southwest Foundation for Biomedical Research, for reviewing the manuscript. These data were presented, in part, at the 50th annual meeting of The American Society of Human Genetics, in Philadelphia, on October 3–7, 2000.
Electronic-Database Information
Accession numbers and the URL for data in this article are as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for Prader-Willi syndrome [MIM 176270] and Angelman syndrome [MIM 105830])
References
- Allison DB, Neale MC, Zannolli R, Schork NJ, Amos CI, Blangero J (1999) Testing the robustness of the likelihood-ratio test in a variance-component quantitative-trait loci–mapping procedure. Am J Hum Genet 65:531–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almasy L, Blangero J (1998) Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet 62:1198–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amos CI (1994) Robust variance-components approach for assessing genetic linkage in pedigrees. Am J Hum Genet 54:535–543 [PMC free article] [PubMed] [Google Scholar]
- Amos CI, Zhu DK, Boerwinkle E (1996) Assessing genetic linkage and association with robust components of variance approaches. Ann Hum Genet 60:143–160 [DOI] [PubMed] [Google Scholar]
- Bartolomei MS, Tilghman SM (1997) Genomic imprinting in mammals. Annu Rev Genet 31:493–525 [DOI] [PubMed] [Google Scholar]
- Broman KW, Murray JC, Sheffield VC, White RL, Weber JL (1998) Comprehensive human genetic maps: individual and sex-specific variation in recombination. Am J Hum Genet 63:861–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottingham RW, Idury RM, Schäffer AA (1993) Faster sequential genetic linkage computations. Am J Hum Genet 53:252–263 [PMC free article] [PubMed] [Google Scholar]
- Curtis D, Sham PC (1994) Using risk calculation to implement an extended relative pair analysis. Ann Hum Genet 58:151–162 [DOI] [PubMed] [Google Scholar]
- Drigalenko E (1998) How sib pairs reveal linkage. Am J Hum Genet 63:1242–1245 [PMC free article] [PubMed] [Google Scholar]
- Fleiss JL (1981) An introduction to applied probability. In: Statistical methods for rates and proportions. Wiley & Sons, New York, pp 1–18 [Google Scholar]
- Gershon ES, Badner JA, Detera-Wadleigh SD, Ferraro TN, Berrettini WH (1996) Maternal inheritance and chromosome 18 allele sharing in unilineal bipolar illness pedigrees. Am J Med Genet 67:202–207 [DOI] [PubMed] [Google Scholar]
- Grigoroiu-Serbanescu M, Nöthen M, Propping P, Poutska F, Magureanu S, Vasilescu R, Marinescu E, Ardelean V (1995) Clinical evidence for genomic imprinting in bipolar I disorder. Acta Pyschiatr Scand 92:365–370 [DOI] [PubMed] [Google Scholar]
- Hanson RL, Ehm MG, Pettitt DJ, Prochazka M, Thompson DB, Timberlake D, Foroud T, Kobes S, Baier L, Burns DK, Almasy L, Blangero J, Garvey WT, Bennett PH, Knowler WC (1998) An autosomal genomic scan for loci linked to type II diabetes and body-mass index in Pima Indians. Am J Hum Genet 63:1130–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseman JK, Elston RC (1972) The investigation of linkage between a quantitative trait and a marker locus. Behav Genet 2:3–19 [DOI] [PubMed] [Google Scholar]
- Heutink P, van der Mey AGL, Sankuijl LA, van Gils APG, Bardoel A, Breedveld GJ, van Vliet M, van Ommen GJB, Cornelisse CJ, Oostra BA, Weber JL, Devilee P (1992) A gene subject to genomic imprinting and responsible for hereditary paragangliomas maps to chromosome 11q23-qter. Hum Mol Genet 1:7–10 [DOI] [PubMed] [Google Scholar]
- Huxtable S, Saker PJ, Haddad L, Walker M, Frayling TW, Levy JC, Hitman GA, O’Rahilly S, Hattersley AT, McCarthy MI (2000) Analysis of parent-offspring trios provides evidence for linkage and association between the insulin gene and type 2 diabetes mediated exclusively through paternally transmitted class III variable number tandem repeat alleles. Diabetes 49:126–130 [DOI] [PubMed] [Google Scholar]
- Hopper JL, Mathews JD (1982) Extensions to multivariate normal models for pedigree analysis. Ann Hum Genet 46:373–383 [DOI] [PubMed] [Google Scholar]
- Kleinbaum DG, Kupper LL, Morgenstern H (1982) Modeling: theoretical considerations. In: Epidemiologic research: principles and quantitative methods. Van Nostrand Reinhold, New York, pp 419–446 [Google Scholar]
- Lange K, Westlake J, Spence MA (1976) Extensions to pedigree analysis III: variance components by the scoring method. Ann Hum Genet 39:485–491 [DOI] [PubMed] [Google Scholar]
- Lathrop GM, Lalouel JM (1984) Easy calculations of lod scores and genetic risks on small computers. Am J Hum Genet 36:460–465 [PMC free article] [PubMed] [Google Scholar]
- Lathrop GM, Lalouel JM, Julier C, Ott J (1984) Strategies for multilocus linkage analysis in humans. Proc Natl Acad Sci USA 81:3443–3446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay RS, Dabelea D, Roumain J, Hanson RL, Bennett PH, Knowler WC (2000a) Type 2 diabetes and low birth weight: the role of paternal inheritance in the association of low birth weight and diabetes. Diabetes 49:445–449 [DOI] [PubMed] [Google Scholar]
- Lindsay RS, Kobes S, Knowler WC, Hanson RL (2000b) A variance components method of linkage analysis to assess parent-of-origin effects on body mass index. Am J Hum Genet 67 Suppl 2:218 [Google Scholar]
- Margaritte-Jeannin P, Clerget-Darpoux F, Hors J, Deschamps I (1995) Testing parental imprinting in insulin-dependent diabetes mellitus by the marker-association-segregation–χ2 method. Am J Hum Genet 56:1080–1087 [PMC free article] [PubMed] [Google Scholar]
- Meiers-Heijboer EJ, Sandkuijl LA, Brunner HG, Smeets HJM, Hoogeboom AJM, Deelen WH, van Hemal JO, Nelen MR, Smeets DFCM, Niermeijer MF, Halley DJJ (1992) Linkage analysis with chromosome 15q11-13 markers shows genomic imprinting in familial Angelman syndrome. J Med Genet 29:853–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nöthen MM, Cichon S, Rohleder H, Hemmer S, Franzek E, Fritze J, Fritze J, Albus M, Borrmann-Hassenbach M, Kreiner R, Weigelt B, Minges J, Lichtermann D, Maier W, Craddock N, Fimmers R, Höller T, Baur MP, Rietschel M, Propping P (1999) Evaluation of linkage of bipolar affective disorder to chromosome 18 in a sample of 57 German families. Mol Psychiatry 4:76–84 [DOI] [PubMed] [Google Scholar]
- Ott J (1989) Computer-simulation methods in human linkage analysis. Proc Natl Acad Sci USA 86:4175–4178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ——— (1991) Methods of linkage analysis. In: Analysis of human genetic linkage. Johns Hopkins University Press, Baltimore, pp 54–81 [Google Scholar]
- Paterson AD, Naimark DMJ, Petronis A (1999) The analysis of parental origin of alleles may detect susceptibility loci for complex disorders. Hum Hered 49:197–204 [DOI] [PubMed] [Google Scholar]
- Pratt SC, Daly MK, Kruglyak L (2000) Exact multipoint quantitative-trait linkage analysis in pedigrees by variance components. Am J Hum Genet 66:1153–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugh EW, Jaquish CE, Sorant AJM, Doetsch JP, Bailey-Wilson JE, Wilson AF (1997) Comparison of sib-pair and variance-components methods of genomic screening. Genet Epidemiol 14:867–872 [DOI] [PubMed] [Google Scholar]
- Rice JP, Neuman RJ, Hoshaw SL, Daw EW, Gu C (1995) TDT with covariates and genomic screens with mod scores: their behavior on simulated data. Genet Epidemiol 12:659–664 [DOI] [PubMed] [Google Scholar]
- Ruiz RGG, Kemeny DM, Price JF (1992) Higher risk of infantile atopic dermatitis from maternal atopy than from paternal atopy. Clin Exp Allergy 22:762–766 [DOI] [PubMed] [Google Scholar]
- Sandford AJ, Shirakawa T, Moffatt MF, Daniels SE, Ra C, Faux JA, Young RP, Nakamura Y, Lathrop GM, Cookson WOCM, Hopkin JM (1993) Localisation of atopy and β subunit of high-affinity IgE receptor (FceRI) on chromosome 11q. Lancet 341:332–334 [DOI] [PubMed] [Google Scholar]
- Schäffer AA, Gupta SK, Shriram K, Cottingham RW (1994) Avoiding recomputation in linkage analysis. Hum Hered 44:225–237 [DOI] [PubMed] [Google Scholar]
- Self SG, Liang KY (1987) Asymptotic properties of maximum likelihood estimators and likelihood ratio tests under nonstandard conditions. J Am Stat Assoc 82:605–610 [Google Scholar]
- Shire JG (1989) Unequal parental contributions: genomic imprinting in mammals. New Biol 1:115–120 [PubMed] [Google Scholar]
- Smalley SL (1993) Sex-specific recombination frequencies: a consequence of imprinting? Am J Hum Genet 52:210–212 [PMC free article] [PubMed] [Google Scholar]
- Stine OC, Xu J, Koskela R, McMahon FJ, Gschwend M, Friddle C, Clark CD, McInnis MG, Simpson SG, Breschel TS, Vishio E, Riskin K, Feilotter H, Chen E, Shen S, Folstein S, Meyers DA, Botstein D, Marr TG, DePaulo JR (1995) Evidence for linkage of bipolar disorder to chromosome 18 with a parent-of-origin effect. Am J Hum Genet 57:1384–1394 [PMC free article] [PubMed] [Google Scholar]
- Strauch K, Fimmers R, Kurz T, Deichmann KA, Wienker TF, Baur MP (2000) Parametric and nonparametric multipoint linkage analysis with imprinting and two-locus-trait models: application to mite sensitization. Am J Hum Genet 66:1945–1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks DE, Ott J, Lathrop M (1990) SLINK: a general simulation program for linkage analysis. Am J Hum Genet Suppl 47:A204 [Google Scholar]
- Weinberg CR (1999) Methods for detection of parent-of-origin effects in genetic studies of case-parents triads. Am J Hum Genet 65:229–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson AF, Elston RC (1993) Statistical validity of the Haseman-Elston sib-pair test in small samples. Genet Epidemiol 10:593–598 [DOI] [PubMed] [Google Scholar]