Abstract
Rhizobium leguminosarum bv. viciae UPM791 induces hydrogenase activity in pea (Pisum sativum L.) bacteroids but not in free-living cells. The symbiotic induction of hydrogenase structural genes (hupSL) is mediated by NifA, the general regulator of the nitrogen fixation process. So far, no culture conditions have been found to induce NifA-dependent promoters in vegetative cells of this bacterium. This hampers the study of the R. leguminosarum hydrogenase system. We have replaced the native NifA-dependent hupSL promoter with the FnrN-dependent fixN promoter, generating strain SPF25, which expresses the hup system in microaerobic free-living cells. SPF25 reaches levels of hydrogenase activity in microaerobiosis similar to those induced in UPM791 bacteroids. A sixfold increase in hydrogenase activity was detected in merodiploid strain SPF25(pALPF1). A time course induction of hydrogenase activity in microaerobic free-living cells of SPF25(pALPF1) shows that hydrogenase activity is detected after 3 h of microaerobic incubation. Maximal hydrogen uptake activity was observed after 10 h of microaerobiosis. Immunoblot analysis of microaerobically induced SPF25(pALPF1) cell fractions indicated that the HupL active form is located in the membrane, whereas the unprocessed protein remains in the soluble fraction. Symbiotic hydrogenase activity of strain SPF25 was not impaired by the promoter replacement. Moreover, bacteroids from pea plants grown in low-nickel concentrations induced higher levels of hydrogenase activity than the wild-type strain and were able to recycle all hydrogen evolved by nodules. This constitutes a new strategy to improve hydrogenase activity in symbiosis.
In the nitrogen fixation process in legume root nodules, a large amount of hydrogen is released from nodules as an obligate by-product of the nitrogenase reaction. This hydrogen production is one of the major factors that affect the efficiency of symbiotic nitrogen fixation (39). In certain strains of Bradyrhizobium japonicum and Rhizobium leguminosarum, a hydrogen uptake (Hup) system oxidizes the hydrogen produced by the nitrogenase. This H2 recycling reduces energy losses and has been shown to enhance legume productivity (11, 25).
The hydrogenase systems from B. japonicum and R. leguminosarum bv. viciae have been extensively characterized (for a review, see reference 34). In both cases, a membrane-bound heterodimeric [NiFe] hydrogenase is responsible for hydrogen uptake. The genetic determinants for hydrogen oxidation are clustered in large DNA regions spanning ca. 20 kb (21, 22); on the basis of sequence analysis, these regions have similar hup gene organizations. These hydrogenase clusters are composed by 17 common genes, hupSLCDFGHIJK and hypABFCDEX, which display a high sequence similarity (32 to 69% identity [34]). Most of these genes are also found in hydrogenase systems from other H2-oxidizing bacteria, suggesting that they encode proteins with essential functions in hydrogenase biosynthesis. Among them, the hupS and hupL genes code for the small and large hydrogenase structural polypeptides, respectively (16, 37). These polypeptides are synthesized as precursors in a soluble, immature form; after insertion of nickel, iron, and other metal groups, the polypeptides are processed into the active subunits and translocated into the membrane (for a review see reference 9). As has been shown for the hydrogenase-3 of Escherichia coli, the processing step implies the proteolytic cleavage of both an N-terminal signal peptide in HupS and a C-terminal peptide in HupL by a specific protease (32, 43). The recruitment and incorporation of metallic groups into the hydrogenase active site require the contribution of the remaining Hup and Hyp proteins in a complex and not completely elucidated process.
Despite the similarities, B. japonicum and R. leguminosarum hydrogenase systems show important differences, mainly in terms of gene composition and hydrogenase regulation (35). The B. japonicum hup gene cluster lacks the hupE gene present in R. leguminosarum but contains the hupNOP, hupUV, and hoxAT operons. It has been shown that HupNOP are involved in nickel metabolism (12) and that HoxA is a transcriptional regulator required for hydrogenase activation in microaerobic free-living cultures in the presence of hydrogen and nickel (10, 18, 44, 46). Functions of HupUV and HupT are not well established, but it has been proposed that HupUV might constitute a pseudohydrogenase that would act as a sensor for the adequate environmental conditions required for hydrogenase activity (4), while HupT could transduce the signal sensed by HupUV to HoxA by modifying its transcription-inducing activity via phosphorylation (45). In contrast, R. leguminosarum UPM791 contains a truncated, functionally inactive HoxA, and no genes homologous to either hupNOP, hupUV, or hupT (7, 8). The absence of these genes may explain why hydrogenase expression in R. leguminosarum is observed only in symbiosis, while B. japonicum is able to induce hydrogenase activity in free-living as well as in symbiotic conditions. The analysis of hupSL promoter (PhupSL) expression showed that hup gene transcription is activated by NifA, the key regulator of the nitrogen fixation process (8). The fact that NifA-dependent promoters, such as nif promoters, are not induced in free-living conditions confines R. leguminosarum hup gene expression to the nodule environment.
The requirement for symbiotic hydrogenase expression has hampered the characterization of the hydrogenase system in R. leguminosarum bv. viciae. Experiments related to hydrogenase purification, the analysis of its regulation, and the elucidation of specific functions for the hup and hyp gene products have been delayed due to this fact. In an attempt to overcome this drawback, we have engineered R. leguminosarum strain UPM791 to obtain hydrogenase activity in microaerobic free-living cells. Interestingly, the modified strain also displays higher levels of hydrogenase activity in symbiotic conditions.
MATERIALS AND METHODS
Bacterial strains, plasmids and growth conditions.
Bacterial strains and plasmids used in this work are listed in Table 1. R. leguminosarum strain UPM791 (24) was routinely grown in tryptone-yeast extract (2) or yeast-mannitol (YMB) (47) media at 28°C. E. coli strains were grown in Luria-Bertani medium. Antibiotics were added at the following concentrations: tetracycline, 5 μg·ml−1; kanamycin, 50 μg·ml−1; ampicillin, 100 μg·ml−1; spectinomycin, 50 μg·ml−1. Cosmid pAL618 is a pLAFR1 derivative carrying the hydrogenase gene cluster from UPM791 in a 20-kb DNA fragment (23). Strain SPF25 is a UPM791 derivative in which the NifA-dependent PhupSL promoter has been replaced by the microaerobically expressed PfixN promoter (see below). Cosmid pALPF1 is a pAL618 derivative carrying the PfixN::hupSL construct. Plasmids were introduced into R. leguminosarum by conjugation, and transconjugants were selected in Rhizobium minimal medium (Rm) (28) supplemented with the corresponding antibiotic. For stoppered-tube hydrogenase induction assays with free-living microaerobic cells, cultures were aerobically grown in Rm or YMB medium to an optical density at 600 nm (OD600) of 0.2 in a 200-ml flask, which was then tightly capped, evacuated, and flushed several times with 1% O2 and incubated for 16 h at 28°C. For hydrogenase induction in the fermentor (BIOFLO C30; New Brunswick Scientific, Edison, N.J.), R. leguminosarum cultures were aerobically grown to an OD600 of 0.35 in a flask and then incubated in the fermentor at 28°C, 400-rpm agitation, and continuous flow of 0.8% O2.
TABLE 1.
Bacterial strains and plasmids used in this work
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| R. leguminosarum | ||
| UPM791 | 128C53 Strr | 24 |
| SPF25 | UPM791 with PfixN::hupSL | This work |
| DG2 | UPM791 fnrN1 fnrN2 | 13 |
| E. coli | ||
| S17.1 | thi pro hsdR hsdM+recA RP4 2-Tc::Mu-Km::Tn7 | 41 |
| C2110 | polA Nalr | G. Ditta |
| Plasmids | ||
| pAL618 | Cosmid containing the UPM791 hup cluster; Tcr | 23 |
| pK18mobsac | pK18 derivative sacB; Kmr | 38 |
| pSPV4 | pMP220 derivative; Tcr | 29 |
| PCR2.1-TOPO | PCR product cloning vector; Apr | Invitrogen |
| pALPF1 | PfixN::hupSL in pAL618 | This work |
| pSPF1 | PfixN::hupSL in pSPV4 | This work |
| pKPF1 | PfixN::hupSL in pK18mobsac | This work |
| pPF1 | PfixN in PCR2.1-TOPO | This work |
| pPF2 | PfixN::hupSL in PCR2.1-TOPO | This work |
| pDH1 | hupSL in PCR2.1-TOPO | This work |
| pIM1 | ′orf3orf4 in PCR2.1-TOPO | This work |
DNA manipulation techniques.
Plasmid DNA preparations, restriction enzyme digestions, agarose gel electrophoresis, DNA cloning, and transformation of DNA into E. coli cells were carried out by standard protocols (36). Genomic DNA of R. leguminosarum was extracted as previously described (24). For Southern hybridizations, DNA probes were labeled with digoxigenin and the hybridizing bands were visualized with a chemiluminescent DIG detection kit as described by manufacturer (Roche Molecular Biochemicals, Mannheim, Germany). DNA sequencing was carried out using the rhodamine terminator cycle sequencing ready reaction kit and an ABI377 automatic sequencer (PE Biosystems, Foster City, Calif.)
Construction of the PfixN::hupSL fusion.
A 320-bp DNA fragment containing the PfixN promoter of R. leguminosarum was amplified by PCR using primers PF1 (5′-GGATCCTGCTCACACATAAACCTG-3′) and PF2 (5′-CATATGGTCGTCCTCAATGCGCCG-3′). The 1,550-bp DNA region upstream of the hupSL promoter (PhupSL) and corresponding to ′orf3orf4 (8) was amplified with primers M7D (5′-AAGGTGGATAGCAAAAT-3′) and IM2 (5′-CGGGATCCAGCCCTCCGGTCACCAAG-3′), whereas a 1,756-bp DNA fragment carrying the hupSL′ genes was obtained with primers DH1 (5′-CATATGGCAACTGCCGAGAC-3′) and DH2 (5′-TCTAGACTTGACGATCTCCTTCTG-3′). These DNA fragments were independently cloned in the PCR2.1-TOPO vector (Invitrogen BV, Groningen, The Netherlands), resulting in constructs pPF1, pIM1, and pDH1, respectively. Correct DNA amplification was confirmed by sequencing. The DNA region containing the hupSL′ genes (from plasmid pDH1) was digested at the ATG start site of hupS and cloned in frame with the fixN promoter in pPF1, resulting in plasmid pPF2. For this cloning, we took advantage of the NdeI sites generated with primers PF2 and DH1. The ′orf3orf4 region from pIM1 was inserted into the EcoRI/BamHI sites upstream of the PfixN promoter in plasmid pPF2. The resulting 3,626-bp insert was cloned in pK18 mobsac (38), producing plasmid pKPF1, which was introduced in R. leguminosarum UPM791. Transconjugants where PhupSL was replaced by the fixN promoter by double crossover were identified by Southern blotting experiments using a 320-bp DNA probe from PfixN and genomic DNA from candidate strains. To generate cosmid pALPF1, plasmid pKPF1 was transformed into E. coli C2110(pAL618) and the double crossover was selected by using the sacB system. Cosmids carrying the PfixN promoter were identified by Southern hybridizations. An EcoRI/XbaI DNA fragment carrying the PfixN::hupSL fusion was also cloned in plasmid pSPV4 (29) in front of the promoterless lacZ gene, resulting in plasmid pSPF1.
Plant tests and enzyme assays.
Inoculation of pea (Pisum sativum L. cv. Frisson) seedlings with R. leguminosarum strains and plant growth under bacteriologically controlled conditions were carried out as previously described (24, 33). The nitrogen-free plant nutrient solution was supplemented with 20 μM NiCl2 from day 10 after seedling inoculation. Pea bacteroids from 21-day-old nodulated plants were prepared as described by Leyva et al. (24). Hydrogenase activity in bacteroid suspensions and free-living cells was measured by an amperometric method with oxygen as the terminal electron acceptor (33). Hydrogen evolution in intact nodules was determined by gas chromatography (5). The protein contents of bacteroids and free-living cells were measured by the bicinchoninic acid method (42) with the modifications described by Brito et al. (6).
Cell fractionation of R. leguminosarum cultures.
A 400-ml overnight microaerobic culture was collected by centrifugation, washed, and resuspended in 4 ml of 20 mM Tris-HCl, pH 8.0, buffer containing a protease inhibitor mixture (complete-Mini; Roche Molecular Biochemicals). Cells were broken by three passages through a French pressure cell at 12,000 lb/in2, and the cell lysate was centrifuged for 20 min at 13,000 × g. The resultant supernatant (S10) was removed and centrifuged in a TL-100 ultracentrifuge (Beckman Inc., Palo Alto, Calif.) at 135,000 × g for 1 h at 4°C. The supernatant (S100) was transferred to another tube, and the membrane fraction (MB) was resuspended in the same volume of Tris buffer. Both fractions were cleared by an additional centrifugation at 135,000 × g for 1 h at 4°C. All fractions were aliquoted and stored at −40°C.
Immunological detection of Hup and Hyp proteins.
Immunological detection of HupL and HypB in free-living cells, bacteroids, and cell fractions was carried out by immunoblotting as described previously (6) using antibodies raised against B. japonicum HupL and R. leguminosarum HypB. Both antisera were used at a 1:2,000 dilution.
RESULTS
Construction of a hup gene system expressing hydrogenase activity in microaerobic free-living cells of R. leguminosarum bv. viciae UPM791.
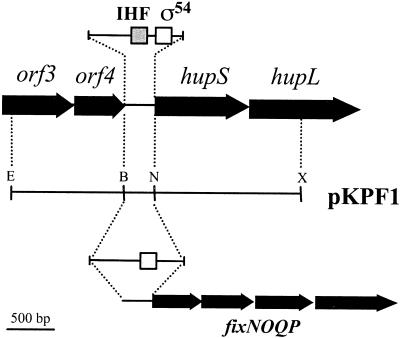
To obtain hydrogenase activity in free-living cells of R. leguminosarum UPM791, we replaced the NifA-dependent hupSL promoter (PhupSL) by a promoter known to be induced in free-living cells of R. leguminosarum UPM791. Since it was known that the hypBFCDEX operon of the UPM791 hydrogenase cluster is induced in microaerobiosis by FnrN (13, 14), we chose an FnrN-regulated promoter. The fixNOQP operon from R. leguminosarum UPM791 had previously been cloned in our laboratory and had been shown to be controlled by FnrN (13). A 320-bp DNA region containing the fixN promoter (PfixN) was amplified by PCR and cloned in front of the start site of the hupS gene (Fig. 1; see Materials and Methods). A DNA region upstream of PhupSL covering ′orf3orf4 was also amplified by PCR and cloned upstream of the PfixN promoter in plasmid pKPF1, a derivative of pK18 mobsac. This construction was transferred by conjugation into R. leguminosarum strain UPM791, and replacement of PhupSL by PfixN was selected by using sucrose sensitivity, mediated by the sacB system. Correct promoter insertion in the resulting strain, SPF25, was confirmed by Southern blotting experiments and by sequencing the promoter fusion. In parallel, and by a similar approach, pKPF1 was used to replace PhupSL with PfixN in cosmid pAL618, which carries the entire hup gene cluster, resulting in cosmid pALPF1.
FIG. 1.
Construction of a microaerobically expressed hydrogenase system in R. leguminosarum bv. viciae UPM791. The σ54-dependent promoter of hupSL expressed in symbiosis (top) was replaced by the FnrN-dependent PfixN promoter from R. leguminosarum UPM791. This construct was introduced into the pK18 mobsac plasmid, producing pKPF1. This plasmid was used to exchange the PhupSL promoter in strain UPM791 and in cosmid pAL618, obtaining strain SPF25 and cosmid pALPF1, respectively. Arrows, genes flanking the PhupSL promoter and the fixNOQP genes; white and grey boxes (top), σ54 and integration host factor binding sequences in PhupSL; bottom box, FnrN consensus binding site (anaerobox). B, BamHI; E, EcoRI; N, NdeI; X, XbaI.
Induction of hydrogenase activity in aerobic and microaerobic cultures of UPM791, SPF25, and merodiploid strains UPM791(pAL618) and SPF25(pALPF1) was measured (Table 2). Strain SPF25 expressed hydrogenase activity in microaerobic free-living cells at a level similar to that observed in pea bacteroids from wild-type strain UPM791 (23). A sixfold increase in hydrogenase activity in microaerobic cells of SPF25(pALPF1), compared with that in SPF25 cells, was observed (Table 2). No hydrogen uptake in microaerobic UPM791 or UPM791(pAL618) cells or in aerobic cultures of any strain was detected. The presence of HupL and HypB proteins in these cultures was analyzed by immunoblotting experiments (data not shown). In microaerobic SPF25 cells, the HupL antibody cross-reacted with a 65-kDa band corresponding to the form of processed and active HupL, and this band was more intense in microaerobic cells of merodiploid strain SPF25(pALPF1). In addition, a prominent band corresponding to the unprocessed form was detected in this culture. No HupL immunoreactive bands were observed in microaerobic or aerobic cells of UPM791 and UPM791(pAL618). As expected, the HypB antiserum specifically recognized a 39-kDa protein in microaerobic cells of all strains (data not shown). The FnrN dependence of microaerobic hydrogenase induction was tested in strain DG2 (mutant fnrN1 and fnrN2); no hydrogenase activity was observed in this strain (data not shown). The effect of nickel addition to the culture medium on microaerobic hydrogenase activity was also measured. Supplementation of the culture medium with 1 μM NiCl2 did not significantly increase the levels of hydrogenase activity in our experimental conditions. In fact, addition of 10 μM NiCl2 had an adverse effect on hydrogenase activity, whereas a final concentration of 20 μM NiCl2 in the culture medium completely abolished hydrogen uptake (data not shown). Addition of hydrogen (5%) to the induction atmosphere was also tested and did not affect hydrogenase activity levels (data not shown).
TABLE 2.
Hydrogenase activity in R. leguminosarum free-living cells cultured under different oxygen tensions
| Strain | Hydrogenase activitya at O2 tension (%) of:
|
|
|---|---|---|
| 20 | 1 | |
| UPM791 | <10 | <10 |
| SPF25 | <10 | 930 ± 190 |
| UPM791 (pAL618) | <10 | <10 |
| SPF25(pALPF1) | <10 | 6,300 ± 1,290 |
Values of hydrogenase activity are expressed as nanomoles of hydrogen per hour per milligram of protein and are the averages of three assays ± standard deviations.
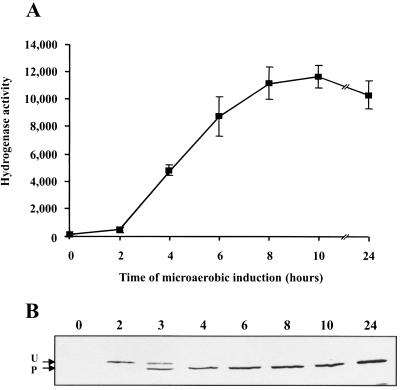
To better characterize hydrogenase induction in microaerobic free-living cells, SPF25(pALPF1) hydrogenase activity was monitored for 24 h in cultures continuously sparged with 0.8% O2 (Fig. 2A). Hydrogen uptake was clearly detected after 4 h and reached a maximum at 10 h. No increase was observed after 24 h of microaerobiosis. This is twice the activity observed in stoppered cultures, probably because in the latter condition the oxygen supply limits rhizobial metabolism. Transcription of hydrogenase genes during the time course of induction was analyzed by using a PfixN::hupSL-lacZ transcriptional fusion. Expression of the PfixN::hupSL promoter was detected after 2 h of microaerobic incubation (data not shown). Also, the status of the HupL subunit during the experiment was studied by immunoblot assays (Fig. 2B). The immunoblot revealed that the inactive, unprocessed HupL form also appeared after 2 h of microaerobiosis. One hour later, both the unprocessed and processed HupL forms were observed; their appearance correlates with the increase in the level of hydrogenase activity. Only the processed HupL form was observed after 4 h of incubation.
FIG. 2.
Time course of microaerobic hydrogenase induction in R. leguminosarum. (A) SPF25(pALPF1) cells were induced in a fermentor and amperometrically analyzed for hydrogenase activity with oxygen as the electron acceptor. Values are given as nanomoles of hydrogen per hour per milligram of protein. (B) Immunodetection of HupL in SPF25(pALPF1) cell extracts at 0, 2, 3, 4, 6, 8, 10, and 24 h of microaerobic incubation. Arrows, unprocessed (U) and processed (P) forms of HupL. Bars, standard deviations.
Subcellular localization of hydrogenase in free-living microaerobic cells of R. leguminosarum.
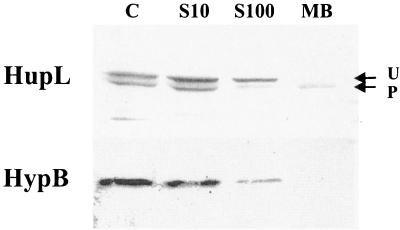
As a further characterization of the microaerobically induced hydrogenase activity we attempted the identification and localization of the active- and inactive-hydrogenase forms in the cell. Cellular fractionation of R. leguminosarum pea bacteroids has been attempted in the past, but methodological problems, due to the fragility of bacteroids and the shortage of biological material, made this assay a tedious and complicated task that led only to contradictory results. Microaerobic free-living cells of strain SPF25(pALPF1) were fractionated as described in Materials and Methods. Fractions corresponding to the crude extract (S10), the soluble fraction (S100), and membrane fraction (MB) were analyzed for the presence of the processed and unprocessed HupL forms in immunoblot assays (Fig. 3). As expected, two HupL immunoreactive bands were observed in whole cells, as well as in the S10 crude extracts. The slow-migrating band, corresponding to the unprocessed HupL form, was mostly observed in the soluble S100 fraction, whereas the fast-moving, processed subunit was only found in the membrane fraction. This result indicates that the active hydrogenase of R. leguminosarum is inserted in the membrane, while the inactive form remains soluble in the cytoplasm. As a control for contamination of the cellular fractions, we confirmed by immunoblot assays with the HypB antiserum that HypB is found only in the soluble fraction, as it is expected for a cytoplasmic protein (Fig. 3).
FIG. 3.
Subcellular localization of HupL and HypB in microaerobic, free-living R. leguminosarum cells. SPF25(pALPF1) whole cells (C), crude extracts (S10), the soluble fraction (S100), and membranes (MB) were analyzed for the presence of HupL and HypB proteins by immunoblotting. Arrows, HupL unprocessed (U) and processed (P) forms.
Increased symbiotic hydrogenase activity of the FnrN-dependent hup system.
To ascertain that the PhupSL replacement had not impaired the hydrogen uptake ability in symbiosis, we measured the hydrogenase activity of SPF25 pea bacteroids. To do this, pea plants were inoculated with cultures of strains UPM791 and SPF25 and grown in a standard plant nutrient solution without nickel supplementation or supplemented with 20 μM NiCl2. Hydrogenase activity in 21-day-old bacteroids, with oxygen as the terminal electron acceptor, was measured (Table 3). SPF25 bacteroids displayed high levels of hydrogenase activity, indicating that the promoter replacement had not affected hydrogen uptake in symbiosis. In fact, SPF25 bacteroids showed a twofold increase in hydrogenase activity compared to the wild-type strain, UPM791, regardless the nickel supplementation. This increment in hydrogenase activity was correlated with an increase of the processed-HupL band as revealed by immunoassays carried out with HupL antiserum (data not shown). Hydrogen evolution assays with intact nodules showed that SPF25 bacteroids, even those from plants not exposed to nickel addition, were able to recycle virtually all hydrogen produced by the nitrogenase, whereas nodules from UPM791 grown in the same condition still evolved significant amounts of hydrogen (Table 3). This result suggests that our construction may improve symbiotic hydrogenase activity under the nickel-limiting conditions normally found in agricultural soils.
TABLE 3.
Hydrogen metabolism in bacteroids and nodules of R. leguminosarum bv. viciae strains
| Strain | Bacteroid hydrogenase activitya
|
Nodule H2 evolutionb
|
||
|---|---|---|---|---|
| −Ni | +Ni | −Ni | +Ni | |
| UPM791 | 1,080 ± 40 | 2,930 ± 320 | 2.13 ± 0.31 | <0.25 |
| SPF25 | 2,210 ± 190 | 5,060 ± 350 | 0.34 ± 0.09 | <0.25 |
Values are the averages of three assays ± standard deviations and are expressed as nanomoles of hydrogen per hour per milligram of protein. +Ni, addition of 20 μM NiCl2 to the standard plant nutrient solution; −Ni, no nickel addition.
Data are the means of four independent determinations ± standard deviations and are expressed as micromoles of hydrogen evolved per hour per gram of nodule fresh weight.
DISCUSSION
Hydrogenase systems from B. japonicum and R. leguminosarum bv. viciae have been extensively characterized in the last 10 years. Although the systems are similar in gene composition and organization, they differ markedly in their regulation (35). The B. japonicum hydrogenase is induced in soybean bacteroids and in microaerobic free-living cultures in the presence of hydrogen and nickel, whereas R. leguminosarum expresses hydrogenase activity only in symbiotic conditions. This requirement for symbiosis, likely due to the fact that the PhupSL promoter is controlled by NifA (8), has obviously hampered the study of the R. leguminosarum hydrogenase. In this paper, we develop a strategy to induce hydrogenase activity in free-living cells of R. leguminosarum. Replacement of the NifA-regulated hupSL promoter by the FnrN-dependent promoter of the fixNOQP operon led to hydrogenase activity in microaerobic free-living cells. Synthesis of hydrogenase polypeptides started as early as 2 h after microaerobic incubation, as revealed by the accumulation of the unprocessed HupL form. One hour later, low levels of hydrogenase activity, which were also correlated with the processing of the large subunit, were detected, and maximal hydrogenase activity was obtained after 10 h of microaerobiosis.
The results obtained in these experiments highlight two main points. First, microaerobic hydrogenase activity is not limited by nickel availability in our experimental conditions, since nickel traces in the culture medium are enough for maximum levels of hydrogenase activity in microaerobiosis. In contrast, maximum symbiotic hydrogenase activity in pea bacteroids is obtained only when the plant nutrient solution is supplemented with NiCl2 (6). This effect is correlated with a nickel-dependent processing of the hydrogenase subunits and not with a nickel-dependent regulation of PhupSL (6). The different behaviors of microaerobic and symbiotic hydrogenase activity indicate that bacterial nickel transport does not limit hydrogenase activity in free-living cultures of R. leguminosarum and suggest that nickel limitation in symbiosis is probably exerted by the host plant.
Second, our results indicate that the PhupSL substitution places the expression of the entire hup gene cluster under the control of FnrN, given that microaerobiosis is the only requirement for expression of hydrogenase activity in free-living cells and that this activity is abolished in a R. leguminosarum fnrN double mutant. This is interesting, since two promoters putatively expressed in symbiosis, in addition to PhupSL, have been identified in the hup gene cluster of R. leguminosarum. The first one was found in front of the hupGHIJK operon (22), and the second was found upstream of hypA and within hupK (15). No regulators for these two putative promoters have been identified, but the involvement of FnrN can be ruled out because they are expressed only in symbiosis. In addition, the hupGHJIK and hypA gene products are essential for hydrogenase activity in symbiosis (15, 30), as well as in microaerobic free-living cells (B. Brito, unpublished data). This implies that the PfixN::hupS promoter should transcribe the hupSLCDEF, hupGHIJK, and hypA operons in microaerobiosis without any transcriptional stop. This is not surprising since previous evidence indicates that an unusually large transcript of 17 kb from the promoter of the hoxKG genes of Ralstonia eutropha (homologous to hupSL of R. leguminosarum) spans the whole hydrogenase cluster (40). Our results suggest that the hupGHIJK and hypA promoters are somehow dispensable and only required to reinforce transcription from the main promoter, PhupSL. The relevance of the hyp promoter in R. leguminosarum is still an open question that is currently being elucidated by generating point mutations that specifically inactivate the two overlapping promoters expressing the hyp operon (15).
The availability of strain SPF25 provides a fast and easy system to generate hydrogenase-induced R. leguminosarum cells and opens new perspectives for the characterization of this enzyme and the molecular functions of Hup and Hyp products. As a first step in the R. leguminosarum hydrogenase characterization, we carried out the localization of the hydrogenase in the cell. Although this type of information has been obtained for hydrogenases from other microorganisms, it remained elusive in R. leguminosarum due to the constrictions imposed by the symbiosis. We now have been able to localize the processed and unprocessed hydrogenase forms in the cell compartments. Cellular fractionation of microaerobic free-living cells of SPF25(pALPF1) showed that the unprocessed HupL protein is found in the soluble fraction corresponding to the cytoplasm, whereas the processed HupL subunit is mostly associated with the membrane. This result is in agreement with observations reported for Ralstonia eutropha (19), Azotobacter vinelandii (26), and E. coli hydrogenase-1 (27) and fits with the current model of hydrogenase maturation (9). In this model, incorporation of metal groups leads to the processing of the HupL precursor, which is targeted to the membrane as a complex with the HupS subunit by the Tat (twin-arginine translocation) system (31).
Several strategies to improve and extend hydrogenase activity in different rhizobial strains have been followed in our and other laboratories (1, 3, 17, 20, 21, 25). Recently a novel hup minitransposon has been reported to generate new hydrogen-recycling strains of Rhizobiaceae (1). This minitransposon has been successfully applied to introduce the Hup trait in R. leguminosarum, Rhizobium etli, Sinorhizobium meliloti, and Mesorhizobium loti Hup− strains. Different levels of hydrogenase activity were obtained depending on the rhizobial background and the legume host partner. Previous studies established that hupSL activation by NifA and nickel availability to the bacteroid could be major factors limiting hydrogenase activity in certain rhizobial strains (5). As soils with high nickel content are rare in nature and since addition of nickel to the soil is not an acceptable agricultural practice, the selection of Rhizobium-legume partners with optimal hydrogenase activity in poor-nickel conditions has been proposed as a way to improve nitrogen fixation through the activity of an efficient hydrogenase enzyme (5). Here, we propose an alternative strategy based on the replacement of the NifA-dependent promoter of hupSL. We demonstrate that SPF25 expressing the hup genes from an FnrN-dependent promoter shows an increased hydrogenase activity in symbiosis. This results in the recycling of all hydrogen produced by nitrogenase even in plants grown without nickel addition. This increment in hydrogen uptake might be due to an enhanced transcription of the hup and, probably, the hyp operons by PfixN. In this situation, a higher production of Hup and Hyp proteins would lead to improved nickel incorporation in the hydrogenase. This effect has already been shown in bacteroids that carry extra copies of the hup and hyp genes and that are exposed to low nickel concentrations (6). It will be interesting to know whether the promoter substitution causes an increase in hydrogenase activity in other rhizobial strains that have been shown to be limited in their hydrogen uptake ability.
Acknowledgments
We are grateful to R. J. Maier for the antisera against B. japonicum hydrogenase large subunit.
This work was supported by grants PB98-0723 (DGES) (Spain) to J.P. and BIO095-0232 from DGICYT (Spain), CT960027 (IMPACT2) from the EU Biotech Programme, and Programa de Grupos Estratégicos (III PRICIT) of Comunidad Autónoma de Madrid to T.R.A. B.B. is the recipient of a Contrato de Incorporación de Doctores y Tecnólogos from the Ministerio de Educación y Cultura (Spain).
REFERENCES
- 1.Báscones, E., J. Imperial, T. Ruiz-Argüeso, and J. M. Palacios. 2000. Generation of new hydrogen-recycling Rhizobiaceae strains by introduction of a novel hup minitransposon. Appl. Environ. Microbiol. 66:4292-4299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beringer, J. 1974. R factor transfer in Rhizobium leguminosarum. J. Gen. Microbiol. 84:188-198. [DOI] [PubMed] [Google Scholar]
- 3.Bhanu, N., S. Khanuja, and M. Lhoda. 1994. Integration of hup genes into the genome of chickpea-Rhizobium through site-specific recombination. J. Plant Biochem. Biotechnol. 3:19-24. [Google Scholar]
- 4.Black, L. K., C. Fu, and R. J. Maier. 1994. Sequence and characterization of hupU and hupV genes of Bradyrhizobium japonicum encoding a possible nickel-sensing complex involved in hydrogenase expression. J. Bacteriol. 176:7102-7106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brito, B., J. Monza, J. Imperial, T. Ruiz-Argüeso, and J. M. Palacios. 2000. Nickel availability and hupSL activation by heterologous regulators limit symbiotic expression of the Rhizobium leguminosarum bv. viciae hydrogenase system in Hup− rhizobia. Appl. Environ. Microbiol. 66:937-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brito, B., J. M. Palacios, E. Hidalgo, J. Imperial, and T. Ruiz-Argüeso. 1994. Nickel availability to pea (Pisum sativum L.) plants limits hydrogenase activity of Rhizobium leguminosarum bv. viciae bacteroids by affecting the processing of the hydrogenase structural subunits. J. Bacteriol. 176:5297-5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brito, B., J. M. Palacios, T. Ruiz-Argüeso, and J. Imperial. 1996. Identification of a gene for a chemoreceptor of the methyl-accepting type in the symbiotic plasmid of Rhizobium leguminosarum bv. viciae UPM791. Biochim. Biophys. Acta 1308:7-11. [DOI] [PubMed] [Google Scholar]
- 8.Brito, B., M. Martínez, D. Fernández, L. Rey, E. Cabrera, J. M. Palacios, J. Imperial, and T. Ruiz-Argüeso. 1997. Hydrogenase genes from Rhizobium leguminosarum bv. viciae are controlled by the nitrogen fixation regulatory protein NifA. Proc. Natl. Acad. Sci. USA 94:6019-6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casalot, L., and M. Rousset. 2001. Maturation of the [NiFe] hydrogenases. Trends Microbiol. 9:228-237. [DOI] [PubMed] [Google Scholar]
- 10.Durmowicz, M. C., and R. J. Maier. 1997. Roles of HoxX and HoxA in biosynthesis of hydrogenase in Bradyrhizobium japonicum. J. Bacteriol. 179:3676-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Evans, H. J., S. A. Russell, F. J. Hanus, and T. Ruiz-Argüeso. 1988. The importance of hydrogen recycling in nitrogen fixation by legumes, p. 777-791. In R. J. Summerfield (ed.), World crops: cool season food legumes. Kluwer Academic Publishers, Boston, Mass.
- 12.Fu, C., S. Javedan, F. Moshiri, and R. J. Maier. 1994. Bacterial genes involved in incorporation of nickel into a hydrogenase enzyme. Proc. Natl. Acad. Sci. USA 91:5099-5103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gutiérrez, D., Y. Hernando, J. M. Palacios, J. Imperial, and T. Ruiz-Argüeso. 1997. FnrN controls symbiotic nitrogen fixation and hydrogenase activities in Rhizobium leguminosarum bv. viciae UPM791. J. Bacteriol. 179:5264-5270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hernando, Y., J. M. Palacios, J. Imperial, and T. Ruiz-Argüeso. 1995. The hypBFCDE operon from Rhizobium leguminosarum bv. viciae is expressed from an Fnr-type promoter that escapes mutagenesis of the fnrN gene. J. Bacteriol. 177:5661-5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hernando, Y., J. M. Palacios, J. Imperial, and T. Ruiz-Argüeso. 1998. Rhizobium leguminosarum bv. viciae hypA is expressed in pea (Pisum sativum) bacteroids and is required for hydrogenase activity and processing. FEMS Microbiol. Lett. 169:295-302. [DOI] [PubMed] [Google Scholar]
- 16.Hidalgo, E., A. Leyva, and T. Ruiz-Argüeso. 1990. Nucleotide sequence of the hydrogenase structural genes from Rhizobium leguminosarum. Plant Mol. Biol. 15:367-370. [DOI] [PubMed] [Google Scholar]
- 17.Kent, A. D., M. L. Wojtasiak, E. A. Robleto, and E. W. Triplett. 1998. A transposable partitioning locus used to stabilize plasmid-borne hydrogen oxidation and trifolitoxin production genes in a Sinorhizobium strain. Appl. Environ. Microbiol. 64:1657-1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim, H., C. Yu, and R. J. Maier. 1991. Common cis-acting region responsible for transcriptional regulation of Bradyrhizobium japonicum hydrogenase by nickel, oxygen, and hydrogen. J. Bacteriol. 173:3993-3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kortlüke, C., and B. Friedrich. 1992. Maturation of membrane-bound hydrogenase of Alcaligenes eutrophus H16. J. Bacteriol. 174:6290-6293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kunnimalaiyaan, M., and M. L. Lodha. 1992. Expression of chromosome-integrated hydrogen-uptake genes in Cicer-Rhizobium. J. Plant Biochem. Biotechnol. 1:19-21. [Google Scholar]
- 21.Lambert, G. R., A. R. Harker, M. A. Cantrell, F. J. Hanus, S. A. Russell, R. A. Haugland, and H. J. Evans. 1987. Symbiotic expression of cosmid-borne Bradyrhizobium japonicum hydrogenase genes. Appl. Environ. Microbiol. 53:422-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leyva, A., J. M. Palacios, J. Murillo, and T. Ruiz-Argüeso. 1990. Genetic organization of the hydrogen uptake (hup) cluster from Rhizobium leguminosarum. J. Bacteriol. 172:1647-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leyva, A., J. M. Palacios, T. Mozo, and T. Ruiz-Argüeso. 1987. Cloning and characterization of hydrogen uptake genes from Rhizobium leguminosarum. J. Bacteriol. 169:4929-4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leyva, A., J. M. Palacios, and T. Ruiz-Argüeso. 1987. Conserved plasmid hydrogen-uptake (hup)-specific sequences within Hup+ Rhizobium leguminosarum strains. Appl. Environ. Microbiol. 53:2539-2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maier, R. J., and E. W. Triplett. 1996. Towards more productive, efficient and competitive nitrogen-fixing symbiotic bacteria. Crit. Rev. Plant Sci. 15:191-234. [Google Scholar]
- 26.Menon, A. L., and R. L. Robson. 1994. In vivo and in vitro nickel-dependent processing of the [NiFe] hydrogenase in Azotobacter vinelandii. J. Bacteriol. 176:291-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Menon, N. K., J. Robbins, J. C. Wendt, K. T. Shanmugam, and A. E. Przybyla. 1991. Mutational analysis and characterization of the Escherichia coli hya operon, which encodes [NiFe] hydrogenase 1. J. Bacteriol. 173:4851-4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Gara, F., and K. T. Shanmugam. 1976. Regulation of nitrogen fixation by rhizobia: export of fixed nitrogen as NH4+. Biochim. Biophys. Acta 437:313-321. [DOI] [PubMed] [Google Scholar]
- 29.Parry, S. K., S. B. Sharma, and E. A. Terzaghi. 1994. Construction of a bidirectional promoter probe vector and its use in analysing nod gene expression in Rhizobium loti. Gene 150:105-109. [DOI] [PubMed] [Google Scholar]
- 30.Rey, L., E. Hidalgo, J. Palacios, and T. Ruiz-Argüeso. 1992. Nucleotide sequence and organization of an H2-uptake gene cluster from Rhizobium leguminosarum bv. viciae containing a rubredoxin-like gene and four additional open reading frames. J. Mol. Biol. 228:998-1002. [DOI] [PubMed] [Google Scholar]
- 31.Rodrigue, A., A. Chanal, K. Beck, M. Müller, and L.-F. Wu. 1999. Co-translocation of a periplasmic enzyme complex by a hitchhiker mechanism through the bacterial Tat pathway. J. Biol. Chem. 274:13223-13228. [DOI] [PubMed] [Google Scholar]
- 32.Rossmann, R., T. Maier, F. Lottspeich, and A. Böck. 1995. Characterization of a protease from Escherichia coli involved in hydrogenase maturation. Eur. J. Biochem. 227:545-550. [DOI] [PubMed] [Google Scholar]
- 33.Ruiz-Argüeso, T., F. J. Hanus, and H. J. Evans. 1978. Hydrogen production and uptake by pea nodules as affected by strains of Rhizobium leguminosarum. Arch. Microbiol. 116:113-118. [Google Scholar]
- 34.Ruiz-Argüeso, T., J. M. Palacios, and J. Imperial. 2000. Uptake hydrogenases in root nodule bacteria, p. 489-507. In E. W. Triplett (ed.), Prokaryotic nitrogen fixation. A model system for the analysis of a biological process. Horizon Scientific Press, Norfolk, England.
- 35.Ruiz-Argüeso, T., J. M. Palacios, and J. Imperial. 2001. Regulation of the hydrogenase system in Rhizobium leguminosarum. Plant Soil 230:49-57. [Google Scholar]
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 37.Sayavedra-Soto, L. A., G. K. Powell, H. J. Evans, and R. O. Morris. 1988. Nucleotide sequence of the genetic loci encoding subunits of Bradyrhizobium japonicum uptake hydrogenase. Proc. Natl. Acad. Sci. USA 85:8395-8399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schäfer, A., A. Tauch, W. Jäger, J. Kalinowski, G. Thierbach, and A. Pühler. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 39.Schubert, K. R., and H. J. Evans. 1976. Hydrogen evolution: a major factor affecting the efficiency of nitrogen fixation in nodulated symbionts. Proc. Natl. Acad. Sci. USA 73:1207-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schwartz, E., T. Buhrke, U. Gerischer, and B. Friedrich. 1999. Positive transcriptional feedback controls hydrogenase expression in Alcaligenes eutrophus H16. J. Bacteriol. 181:5684-5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simon, R., U. Priefer, and A. Pühler. 1983. Vector plasmids for in-vivo and in-vitro manipulations of gram-negative bacteria, p. 98-106. In A. Pühler (ed.), Molecular genetics of the bacteria-plant interactions. Springer-Verlag KG, Berlin, Germany.
- 42.Smith, P. K., R. I. Krohn, G. T. Hermanson, A. K. Mallia, F. H. Gartner, M. D. Provenzano, E. K. Fujimoto, N. M. Goeke, B. J. Olson, and D. C. Klenk. 1985. Measurement of protein using bicinchoninic acid. Anal. Biochem. 150:76-85. [DOI] [PubMed] [Google Scholar]
- 43.Theodoratou, E., A. Paschos, A. Magalon, E. Fritsche, R. Huber, and A. Bock. 2000. Nickel serves as a substrate recognition motif for the endopeptidase involved in hydrogenase maturation. Eur. J. Biochem. 267:1995-1999. [DOI] [PubMed] [Google Scholar]
- 44.Van Soom, C., C. Verreth, M. J. Sampaio, and J. Vanderleyden. 1993. Identification of a potential transcriptional regulator of hydrogenase activity in free-living Bradyrhizobium japonicum strains. Mol. Gen. Genet. 239:235-240. [DOI] [PubMed] [Google Scholar]
- 45.Van Soom, C., I. Lerouge, J. Vanderleyden, T. Ruiz-Argüeso, and J. M. Palacios. 1999. Identification and characterization of hupT, a gene involved in negative regulation of hydrogen oxidation in Bradyrhizobium japonicum. J. Bacteriol. 181:5085-5089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Van Soom, C., P. de Wilde, and J. Vanderleyden. 1997. HoxA is a transcriptional regulator for expression of the hup structural genes in free-living Bradyrhizobium japonicum. Mol. Microbiol. 23:967-977. [DOI] [PubMed] [Google Scholar]
- 47.Vincent, J. M. 1970. A manual for the practical study of root-nodule bacteria. Blackwell Scientific Publications, Ltd., Oxford, United Kingdom.