Abstract
A new approach for modulating gene expression, based on randomization of promoter (spacer) sequences, was developed. The method was applied to chromosomal genes in Lactococcus lactis and shown to generate libraries of clones with broad ranges of expression levels of target genes. In one example, overexpression was achieved by introducing an additional gene copy into a phage attachment site on the chromosome. This resulted in a series of strains with phosphofructokinase activities from 1.4 to 11 times the wild-type activity level. In this example, the pfk gene was cloned upstream of a gusA gene encoding β-glucuronidase, resulting in an operon structure in which both genes are transcribed from a common promoter. We show that there is a linear correlation between the expressions of the two genes, which facilitates screening for mutants with suitable enzyme activities. In a second example, we show that the method can be applied to modulating the expression of native genes on the chromosome. We constructed a series of strains in which the expression of the las operon, containing the genes pfk, pyk, and ldh, was modulated by integrating a truncated copy of the pfk gene. Importantly, the modulation affected the activities of all three enzymes to the same extent, and enzyme activities ranging from 0.5 to 3.5 times the wild-type level were obtained.
Microorganisms are used for numerous purposes in industry, including for bulk production of chemicals and enzymes and for food fermentations. These applications have stimulated a great interest in trying to improve the properties of the production organisms through genetic manipulations. Most often, the strategy has been to overproduce (by many fold) an enzyme presumed to be rate limiting or to eliminate a branching pathway flux by deleting a gene. The outcomes of such attempts have often been disappointing (16, 28, 29), and the interest in modulating or tuning enzyme activities instead and thus to perform metabolic optimization (18) is therefore increasing.
Genetic tools for modulating enzyme activities by changing gene expression have long been available and have been employed for fundamental studies of cell physiology, e.g., with inducible systems like the lac-type promoters (31, 15, 26) or the nisin promoter (6). These methods, however, are less well suited for modulating gene expression on an industrial scale. In addition, according to metabolic control analysis (11, 19), truly rate-limiting steps will rarely be found in a metabolic pathway, and it will therefore frequently be necessary to increase the levels of many enzymes to achieve an increased flux.
We recently reported a new approach for obtaining promoter libraries (17, 18), which is based on randomization of the DNA sequences (spacers) that separate the individual consensus sequences of promoters. The method takes advantage of the fact that actual sequences of bases in these spacer areas are less important for the strength of a particular promoter than the resulting DNA structure. By randomizing many base pairs simultaneously in the vicinity of the consensus sequences, it is possible to change the DNA structure and the binding of transcription factors to the promoter sequences, and the promoter libraries obtained by this approach contain promoters with virtually any activity. The individual promoters in such libraries then allow for accurate, simultaneous, and individual modulation of several enzyme activities in a cell, and this method is therefore well suited for both metabolic optimization and metabolic control analysis (1, 2).
A disadvantage of the above-mentioned method is the time-consuming cloning work that is needed when bringing several individual promoters to express a target gene. In addition, the strength of promoters is often context dependent, so that the ranking of individual promoters may change when they are moved into a new context, which may further increase the number of genetic constructions to be made. The method also has the disadvantage that the 5′ end of the mRNA of the modulated gene is likely to be altered, which may affect gene expression and cause problems, particularly in cases where the expression of several genes in an operon should be modulated proportionally (2).
We report here a modification to the Jensen-Hammer method (17, 18) which is far less laborious and at the same time removes the above-mentioned limitations. The method is illustrated for modulating chromosomal gene expression in Lactococcus lactis, but it should in principle be universally applicable.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The bacterial strain MG1363 (8), a plasmid-free derivative of L. lactis subsp. cremoris strain NCDO712, was used as a model organism for modulating gene expression. LB436 is MG1363 containing plasmid pLB65 (3). pLB65 is a derivative of pCI372 (10), an Escherichia coli-L. lactis shuttle vector harboring the cat gene encoding chloramphenicol resistance and the gene coding for the temperate lactococcal bacteriophage TP901-1 integrase. For cloning purposes E. coli strain ABLE-C (Stratagene) {E. coli C lac (LacZ−) [Kanr McrA− McrCB− McrF− Mrr− HsdR (rK− mK−)] [F′ proAB lacIqZΔM15 Tn10(Tetr)]} or KW1 [metB strA purB(aad-uid-man) hsr hsm+ gusA−] (32) was used. The plasmid pRC1 (20), an E. coli vector derived from pBluescript containing genes for erythromycin resistance, was used for the purpose of integrating synthetic promoters upstream of the las operon. The plasmid pG+host8 (21), a replication-thermosensitive derivative of pWV01, was used to replace the native las promoter with a synthetic promoter on the chromosome. The plasmid pLB85 (3), a pMOSBlue derivative (Ampr Ermr) harboring attP of TP901-1 and a promoterless gusA gene encoding β-glucuronidase, was used to introduce an extra copy of pfk, transcribed from synthetic promoters, into the TP901-1 attP locus on the chromosome of MG1363.
Growth media and growth conditions.
E. coli strains were grown aerobically at 37°C in Luria-Bertani broth (27). L. lactis strains were grown in batch cultures (100-ml flasks) without aeration but with slow stirring in M17 broth (30) or defined SA medium (14). The cultures were supplemented with 1% glucose and incubated at 30°C.
Antibiotics.
Erythromycin was used at 5 μg/ml for L. lactis and 200 μg/ml for E. coli. Tetracycline was used at 5 μg/ml for L. lactis and 8 μg/ml for E. coli.
Enzymes.
All enzymes used in connection with the measurements of lactate dehydrogenase (LDH), pyruvate kinase (PK), and phosphofructokinase (PFK) activities were purchased from Boehringer Mannheim, Kvistgård, Denmark.
DNA techniques.
All manipulations were performed as described by Sambrook et al. (27). Taq polymerase was applied for analytical purposes, while PCR products intended for cloning were generated using Elongase enzyme mix (Life Technologies A/S, Tåstrup, Denmark). Chromosomal DNA from L. lactis was isolated using the method described for E. coli with the modification that cells were treated with 20 μg of lysozyme per ml for 2 h before lysis. Cells of L. lactis were made competent by growth in GM17 medium containing 1% glycine and transformed by electroporation as previously described by Holo and Nes (12). After electroporation, cells were plated on GM17. E. coli cells were transformed using electroporation.
RNA isolation, primer labeling, and primer extension.
RNA from L. lactis was isolated from 10 ml of M17 culture (1% glucose) at an optical density (600 nm) of approximately 0.8. The cells were harvested by centrifugation and resuspended in 500 μl of ice-cold TE buffer (Tris [10 mM] and EDTA [1 mM], pH 7.5). The following were mixed in a 2-ml centrifuge tube: 0.6 g of glass beads (100-μm diameter), 0.17 ml of 2% Macaloid (clay), 0.5 ml of phenol-chloroform (1:1), 0.05 ml of 10% sodium dodecyl sulfate, and the resuspended cells. The suspension was vortex mixed for 5 min at maximum speed and then centrifuged for 15 min at 12,000 × g. The aqueous phase was transferred to a new centrifuge tube and extracted once with an equal volume of phenol-chloroform (1:1). The RNA was precipitated by adding 1/10 volume of 3 M sodium acetate and 3 volumes of 96% ethanol. The pellet was washed with 70% ethanol and dissolved in an appropriate volume of TE buffer.
The primer 5′-CCTCCGAAATTGAGATACACAGC-3′ was labeled using T4 polynucleotide kinase (Life Technologies A/S) and [γ-33P]ATP. Primer extension was performed using SuperScript reverse transcriptase (Life Technologies A/S) with 40 μg of RNA in a 20-μl reaction volume.
Enzyme measurements.
The activities of PFK, PK, and LDH were measured in cell extracts obtained by sonication. Cells were grown in SA medium and harvested at an optical density of approximately 0.7 (450 nm). The cells were washed twice with ice-cold 0.2% KCl and then resuspended in ice-cold sonication buffer. The sonication buffer for LDH and PK activity measurements was 50 mM triethanolamine-10 mM KH2PO4-10 mM EDTA-50% glycerol (pH 4.7); that for PFK activity measurements was 50 mM Tris-HCl-0.1 mM EDTA-50% glycerol-1 mM dithiothreitol (pH 7.5). The cell suspension was sonicated three times for 45 s with an interval of 30 s. The preparation was kept on ice during the sonication. Following sonication, the cell debris and intact cells were removed by centrifugation (10 min at 20,000 × g) in a 4°C centrifuge. As a measure of the degree of cell disruption, the optical density at 280 nm was used. The enzyme activities were determined from the consumption of NADH by using a Zeiss M500 spectrophotometer. PFK was assayed as described by Fordyce et al. (7), except that the final concentrations in the assay mixture were 1 mM ATP, 1 mM fructose 6-phosphate, 0.2 mM NADH, 10 mM MgCl2, 10 mM NH4Cl, 0.3 U of triose phosphate isomerase per ml, 1 U of glycerol 3-phosphate dehydrogenase per ml, and 0.3 U of aldolase. PK was assayed as described by Crow and Pritchard (5), with final concentrations in the assay mixture of 1 mM GDP, 1 mM phosphoenolpyruvate, 1 mM fructose 1,6-bisphosphate, 10 mM MgCl2, 0.2 mM NADH, and 6.3 U of LDH per ml. LDH was measured as described by Crow and Pritchard (4), with final concentrations in the assay mixture of 10 mM pyruvate, 0.2 mM NADH, and 1 mM fructose 1,6-bisphosphate. All measured enzyme activities were related to the optical density at 280 nm of the extract for the purpose of determining relative activities.
β-Glucuronidase activities were determined by the procedure described by Miller (23) and modified by Israelsen et al. (13), except that para-nitro-β-glucuronic acid (Biosynth AG) was used as the substrate (25). The activity is given in Miller units and calculated as described by Miller (23).
Construction of strains with elevated PFK activity.
A PCR fragment was generated using the forward primer 5′-ACGACTAGTGGATCCATNNNNNAGTTTATTCTTGACANNNNNNNNNNNNNNTGRTATAATANNCTAGACAACAAATGAACATGG-3′, which carries SpeI and BamHI sites and has homology to the 5′ end of the mRNA of the las operon, and the reverse primer 5′-GAACTGCAGCTCTGCCCTAATTATGCG-3′, which carries a PstI site and has homology to the intergenic region between the pfk gene and the pyk gene. The resulting PCR product, containing synthetic promoters followed by a full-length pfk gene, was digested with SpeI and PstI and cloned in the vector pLB85 digested with XbaI and PstI. Following ligation, the plasmids were transformed directly into L. lactis LB436, a derivative of strain MG1363 containing plasmid pLB65, which encodes the TP901-1 integrase, and the cells were plated on GM17 plates with selection for erythromycin resistance and with 200 μg of X-Gluc (5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid) per ml of medium. In this strain, pLB85 and other plasmids containing the attB site from TP901-1 will integrate with high frequency into the corresponding attachment site for phage TP901-1 on the L. lactis chromosome (3).
Construction of strains with synthetic promoters in front of the las operon.
A PCR fragment was generated using the promoter primer shown above as the forward primer and the reverse primer 5′-CGGGGTACCGGTCTCCTTTATAACCAGC-3′, which carries a KpnI site and has homology to the 3′ end of the pfk gene. The PCR product was digested with BamHI and KpnI, cloned in the vector pRC1 digested with the same enzymes, and transformed into the E. coli host strain ABLE-C. A plasmid library consisting of approximately 5,000 clones was obtained and used for transformation of L. lactis MG1363 with selection for erythromycin resistance. One hundred twenty colonies appeared after incubation, and subsequently chromosomal DNA was prepared from these strains and used to confirm the integration by PCR with the primers 5′-GTAATACGACTCACTATAGGGC-3′ (homologous to the pRC1 vector) and 5′-TCCCCGCGGTCACGAAGCAATTCAACG-3′ (homologous to the C-terminal part of pfk).
Replacement of the native las promoter with a synthetic promoter from CS8.
A PCR fragment containing the las upstream region was generated using the forward primer 5′-GGTACTCGAGGTGGAATTTTCTTTGAAGGTC-3′ (carrying an XhoI site) and the reverse primer 5′-GGAAGGATCCCTTTACAAACTGTATGAAACG-3′ (carrying a BamHI site). The PCR product was digested with XhoI and BamHI and cloned in the vector pG+host8 digested with the same enzymes, using KW1 as the host strain. Next, a PCR fragment was generated with chromosomal DNA from CS8 as the template and the primers 5′-GTAATACGACTCACTATAGGGC-3′ (homologous to the pRC1 vector) and 5′-CTAGTCTAGAGGTCTCCTTTATAACCTTC-3′ (carrying an XbaI site and homologous to the C-terminal region of the pfk gene). The resulting PCR fragment was digested with BamHI (restriction site originating from promoter primer [see above]) and XbaI and cloned in the vector with the upstream region digested with the same restriction enzymes, again using KW1 as the host. The resulting plasmid was used to replace the wild-type las promoter as further described in Results.
RESULTS
A method for modulating gene expression.
We have designed a new approach to facilitate the modulation of gene expression, which was employed here in two different strategies for modulating cellular enzyme activities: (i) introduction of a new gene expressed from a promoter library and (ii) modulation of the expression of preexisting gene copies or operons.
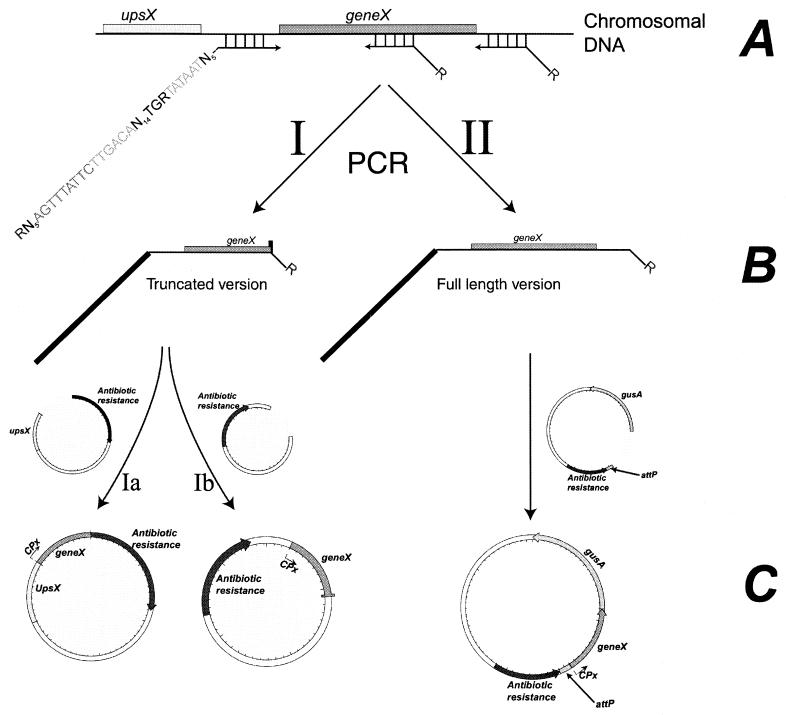
The method requires two PCR primers (Fig. 1). The first primer carries a restriction site, a promoter with randomized spacers, and a sequence with homology to the 5′ end of the mRNA of the target gene. In Fig. 1, N indicates 25% each of A, C, G, and T, while R indicates 50% each of A and G. The second primer also carries a suitable restriction site and a sequence with homology either within the gene in question or downstream of it, allowing for the generation of either a truncated version (strategy I) or a full-length version (strategy II) of the gene behind synthetic promoters. Due to the use of the degenerate promoter primer, a modified PCR protocol is applied. First, a standard PCR is performed. The resulting PCR product is then diluted fourfold and used as a template for a single PCR cycle containing a denaturation step and an annealing step and ending with an elongation step. This last cycle removes annealing between heterologous strands of promoter sequences. The resulting fragment can be cloned after suitable digestion with restriction enzymes into a vector, thereby generating a plasmid library.
FIG. 1.
Overview of the approach used for modulating enzyme activities. (A) Chromosome with an arbitrary gene (geneX) and its upstream region (upsX). The promoter primer (N can be any of the four bases, while R is 50% A and 50% G) and two reverse primers are shown. (B) Two possible PCR products, the first of which contains a truncated geneX and the second of which contains a full-length geneX. (C) The resulting plasmids obtained after cloning of the PCR fragments. If a chromosomal promoter is to be replaced by a synthetic one, then strategy I is used. If the resulting plasmid is to be used in a simple integration event, then strategy Ib is used. If the resulting plasmid is to take part in a double-crossover event, then strategy Ia is used. If the plan is to introduce an additional gene copy into the chromosome, then strategy II is used.
(i) Introduction of a new gene expressed from randomized promoters: modulation of PFK activity above wild-type activity in L. lactis.
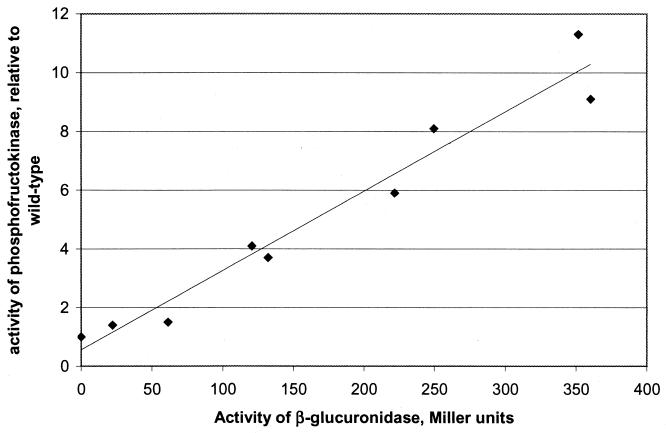
In order to modulate the PFK activity in L. lactis, we generated a PCR fragment containing the entire pfk gene fused to a library of promoters. The fragment was inserted in the multiple cloning site of the vector pLB85 in the same direction as gusA to create a transcriptional fusion between pfk and gusA. The resulting ligation mixture was introduced directly into strain LB436. pLB85 derivatives integrate into the attB site in the chromosome of MG1363 at a high frequency due to the presence of the TP901-1 integrase (3). The transformants were plated on M17-glucose plates containing X-Gluc (a substrate for β-glucuronidase), and the different levels of expression of pfk were indicated by different blue color intensities of the colonies. The PFK activity and the β-glucuronidase activity were determined for nine strains. The PFK activity of the clones varied from 1.4 to 11 times the wild-type activity, and the β-glucuronidase activity varied from 20 to 360 Miller units. Figure 2 shows the measured PFK activity plotted against β-glucuronidase activity, which revealed a fairly good linear correlation between the β-glucuronidase activity and the measured PFK activity. This allowed for an indirect estimate of the PFK activity via the expression of the reporter gene.
FIG. 2.
Correlation between β-glucuronidase activity and relative PFK activity in strains containing an additional gene copy of pfk transcribed from synthetic promoters.
(ii) Simultaneous modulation of PFK, PK, and LDH activities in L. lactis.
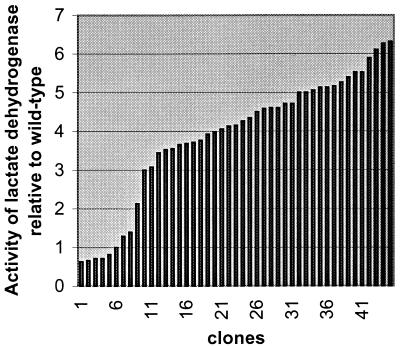
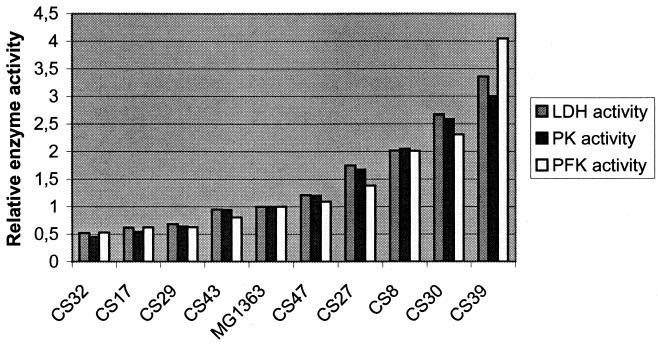
A truncated pfk fragment fused to a library of synthetic promoters was generated by PCR and cloned in the E. coli vector pRC1 (Fig. 1). The resulting plasmid library was subsequently transformed into MG1363, and strains in which the plasmids had crossed into the chromosome were obtained. Chromosomal DNA was isolated from 120 clones and analyzed by PCR using a primer with homology to pRC1 and a primer with homology to pfk beyond the region cloned. In 65 of these clones the truncated pfk fragment was shown to be integrated at the pfk locus. Forty-six of these clones were then tested for LDH activity, and all were found to have altered LDH activity (Fig. 3). Nine clones with different activities were selected and further analyzed for activities of all three las enzymes. Activities both above and below the wild-type enzyme level were obtained, and the activities covered the range from 50 to 350% of the wild-type level (Fig. 4). Importantly, the activities of all three enzymes were perturbed to approximately the same extent.
FIG. 3.
Relative LDH activity measured in strains with synthetic promoters in front of the las operon. Overnight cultures were used for these measurements, and these activities are generally somewhat higher than those measured in extracts from exponentially grown cells.
FIG. 4.
Relative activities of PFK, PK, and LDH measured in extracts from strains with synthetic promoters in front of the las operon during exponential-phase growth. The selected strains cover the entire activity range. MG1363 is the wild-type strain.
Construction of a strain with a synthetic promoter in front of the las operon but without antibiotic resistance and vector.
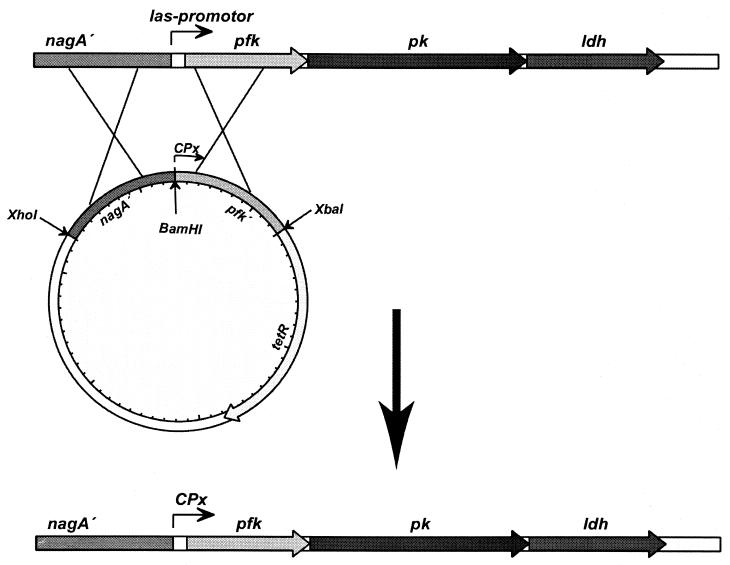
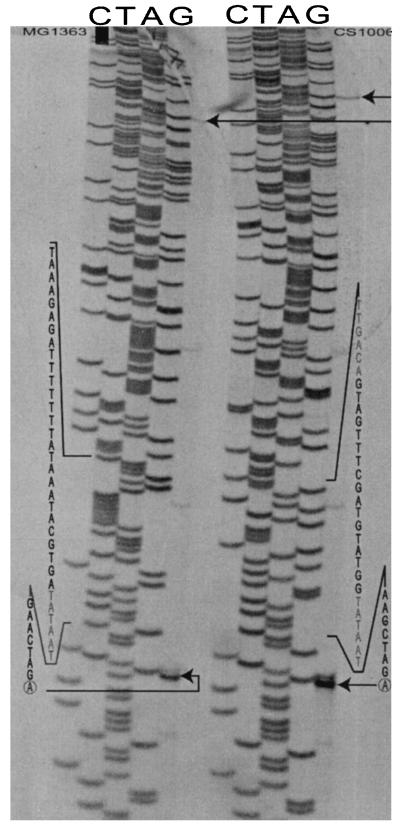
In the example above, we used a vector that does not replicate in L. lactis to achieve integration into the chromosome. Although this is a useful approach for many physiological studies, the method does leave behind an antibiotic marker in the chromosome and also a truncated gene, which can be undesirable for several reasons. Once a strain with proper expression of the target gene has been found, it is possible to construct a strain retaining only the synthetic promoter by gene replacement. In the present case, 1 kb of the DNA sequence upstream of the las promoter, carrying a part of the nagA gene from L. lactis, was cloned into pG+host8, resulting in plasmid pCS564. pG+host8 has a tetracycline marker selectable in L. lactis and a temperature-sensitive origin of replication. Subsequently, the synthetic promoter and the truncated pfk gene were amplified by PCR using chromosomal DNA from strain CS8 and cloned into plasmid pCS564, resulting in plasmid pCS924 (Fig. 5). The general approach is outlined in Fig. 1 as strategy Ia. The resulting plasmid was used to perform two subsequent crossovers to yield a strain in which the native las promoter had been effectively replaced with a synthetic promoter. Plasmid pCS924 was transformed into MG1363 at the permissive temperature (28°C). Subsequently, the resulting strain was plated at 37°C with selection for tetracycline resistance, and clones that were able to grow at 37°C were isolated. PCR performed on chromosomal DNA from these clones revealed a strain in which the plasmid pCS924 had integrated into the pfk locus. When the temperature was lowered to 28°C, there was a strong selection for excision of the plasmid, which can again take place in two different ways to either leave behind the synthetic promoter or restore the wild-type pfk locus (Fig. 5). We isolated a strain, CS1006, in which the synthetic promoter had replaced the wild-type las promoter. Primer extension was performed using RNA from this strain and from the wild-type strain MG1363 to determine the transcription start in the two strains. As seen in Fig. 6, the transcript start is exactly as it is in the wild-type strain. Interestingly, the primer extension indicated the presence of an alternative transcription start in both strains at position −181 that coincided with a putative promoter with the −10 region TGTTAAAAT but without a −35 signal.
FIG. 5.
Strategy used for replacement of the native las promoter with the synthetic promoter obtained from CS8. The plasmid has two regions homologous to the las locus and allowed for the construction of a strain, CS1006, in which the vector has been excised (see text for further details).
FIG. 6.
Primer extension using RNAs from the wild-type strain MG1363 and from strain CS1006, in which a synthetic promoter has replaced the las promoter. Arrows indicate the transcript start. Note the alternative transcript start that was observed in both strains, also indicated by arrows.
DISCUSSION
A new approach for modulating gene expression was developed and tested for modulating the expression of chromosomal genes in L. lactis. One of the advantages of the approach presented here is that the leader 5′ end of the mRNA of a particular gene can be preserved. The sequence of the 5′ end of an mRNA has important implications for the stability of transcripts (9, 24, 22), and changes in this sequence may therefore affect the expression of the downstream gene. In the case of operons containing two or more genes, changes in the mRNA stability may have serious implications for the relative expression of the genes in the operon, as was observed recently for the las operon in L. lactis (2). Here the insertion of synthetic promoter fragments upstream of the las operon resulted in more than twofold-lower expression of the first gene in the operon compared to the other two genes in the operon. This problem was alleviated by the approach presented here, where primer extension showed that the leader was preserved. The introduction of the synthetic promoters resulted in strains in which all of the enzyme activities were modulated to the same extent. The method also allows for improving a leader mRNA sequence or a ribosome binding site by incorporating the relevant sequences into the promoter primer.
We used an extended PCR protocol for introducing the synthetic promoters with randomized DNA stretches in front of the relevant genes. The protocol entailed diluting the PCR product from the standard PCR; adding new primers, buffer, deoxynucleoside triphosphates, and polymerase; and running an extra PCR cycle. This was done in order to avoid possible annealing of heterologous DNA strands in the final rounds of the standard PCR when primers and substrate may be depleted or when the polymerase may no longer be active. However, even with this modified protocol, half of the DNA fragments obtained from the final PCR cycle will still have extensive mismatches in the promoter area, namely, the half of the DNA fragments in which the newly synthesized DNA strand started from a promoter primer. This could in principle also be avoided if a primer with only the constant 5′ DNA sequence of the promoter primer was used in the final PCR cycle (in the present study this primer should be 5′-ACGACTAGTGGATCCAT).
After performing metabolic optimization with the method presented here, it is often desirable to remove plasmids integrated in the chromosome. This can be done in order to improve genetic stability, to allow the reuse of a selectable marker in subsequent steps of optimization, or simply to end up with a strain without antibiotic markers. We have here illustrated how this can be achieved in two steps: first, a promoter of proper strength is identified by integrating a promoter library in front of a target gene and screening for optimal activity of the encoded enzyme, and second, this specific synthetic promoter is amplified along with (part of) the target gene by PCR and cloned together with an upstream DNA fragment in a vector for subsequent gene replacement. The advantage of this approach is that in the first step of the procedure the integration takes place at a high frequency in the correct position, which facilitates the subsequent screening work. However, in some cases it may be an advantage to combine these two steps (which amounts to strategy Ia in Fig. 1), as follows. A library of PCR products with degenerate promoters and target genes is cloned directly into a gene replacement vector which already harbors the relevant upstream DNA region. The resulting plasmid library is then used for consecutive integration and excision events to generate strains with optimized expression levels of the target gene. The screening procedure involved here is somewhat more laborious, but fewer cloning steps are involved.
The method presented in this paper should be applicable to the construction of strains for numerous specific purposes in prokaryotic as well as eukaryotic systems. With respect to metabolic optimization, the method has several advantages over existing inducible expression systems. One advantage is that after the optimal promoter activity has been determined, the strain is in principle ready for use in the industrial fermentation process. Another important feature is the option to simultaneously modulate, to different extents, the expression of many genes or operons located at different positions of the genome in the same strain. With the existing systems now available, one would then quickly run out of expression systems to use.
Acknowledgments
We thank Heidi W. Andersen for donating several primers and for discussions. We also thank Brian Koebmann Jensen for critical comments on the manuscript.
REFERENCES
- 1.Andersen, H. W., M. B. Pedersen, K. Hammer, and P. R. Jensen. 2001. Lactate dehydrogenase has no control on lactate production but has a strong negative control on formate production in Lactococcus lactis. Eur. J. Biochem. 268:6379-6389. [DOI] [PubMed] [Google Scholar]
- 2.Andersen, H. W., C. Solem, K. Hammer, and P. R. Jensen. 2001. Twofold reduction of phosphofructokinase activity in Lactococcus lactis results in strong decreases in growth rate and in glycolytic flux. J. Bacteriol. 183:3458-3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brøndsted, L., and K. Hammer. 1999. Use of the integration elements encoded by the temperate lactococcal bacteriophage TP901-1 to obtain chromosomal single-copy transcriptional fusions in Lactococcus lactis. Appl. Environ. Microbiol. 65:752-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crow, V. L., and C. G. Pritchard. 1977. Fructose 1,6-diphosphate-activated l-lactate dehydrogenase from Streptococcus lactis: kinetic properties and factors affecting activation. J. Bacteriol. 131:82-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crow, V. L., and C. G. Pritchard. 1982. Pyruvate kinase from Streptococcus lactis. Methods Enzymol. 90:165-170. [DOI] [PubMed] [Google Scholar]
- 6.de Ruyter, P. G. G. A., O. P. Kuipers, and W. M. de Vos. 1996. Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl. Environ. Microbiol. 62:3662-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fordyce, A. M., C. H. Moore, and G. C. Pritchard. 1982. Phosphofructokinase from Streptococcus lactis. Methods Enzymol. 90:77-82. [DOI] [PubMed] [Google Scholar]
- 8.Gasson, M. J. 1983. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 154:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hambraeus, G., M. Persson, and B. Rutberg. 2000. The aprE leader is a determinant of extreme mRNA stability in Bacillus subtilis. Microbiology 146:3051-3059. [DOI] [PubMed] [Google Scholar]
- 10.Hayes, F., C. Daly, and G. Fitzgerald. 1990. Identification of the minimal replicon of Lactococcus lactis subsp. lactis UC317 plasmid pCI305. Appl. Environ. Microbiol. 56:202-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heinrich, R., and T. A Rapoport. 1974. A linear steady-state treatment of enzymatic chains: general properties, control and effector-strength. Eur. J. Biochem. 42:89-95. [DOI] [PubMed] [Google Scholar]
- 12.Holo, H., and I. F. Nes. 1989. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl. Environ. Microbiol. 55:3119-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Israelsen, H., S. M. Madsen, A. Vrang, E. B. Hansen, and E. Johansen. 1995. Cloning and partial characterization of regulated promoters from Lactococcus lactis Tn917-lacZ integrants with the new promoter probe vector pAK80. Appl. Environ. Microbiol. 61:2540-2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jensen, P. R., and K. Hammer. 1993. Minimal requirements for exponential growth of Lactococcus lactis. Appl. Environ. Microbiol. 59:4363-4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jensen, P. R., H. V. Westerhoff, and O. Michelsen. 1993. The use of lac-type promotors in control analysis. Eur. J. Biochem. 211:181-191. [DOI] [PubMed] [Google Scholar]
- 16.Jensen, P. R., H. V. Westerhoff, and O. Michelsen. 1993. Excess capacity of H+-ATPase and inverse respiratory control in Escherichia coli. EMBO J. 12:1277-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jensen, P. R., and K. Hammer. 1998. The sequence of spacers between the consensus sequences modulates the strength of prokaryotic promoters. Appl. Environ. Microbiol. 64:82-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jensen, P. R., and K. Hammer. 1998. Artificial promotors for metabolic optimization. Biotechnol. Bioeng. 58:191-195. [PubMed] [Google Scholar]
- 19.Kacser, H., and J. A. Burns. 1973. The control of flux. Symp. Exp. Biol. 27:65-104. [PubMed] [Google Scholar]
- 20.Le Bourgeois, P., M. Lautier, M. Mata, and P. Ritzenthaler. 1992. New tools for the physical and genetic mapping of Lactococcus strains. Gene 111:109-114. [DOI] [PubMed] [Google Scholar]
- 21.Maguin, E., H. Prévost, S. D. Ehrlich, and A. Gruss. 1996. Efficient insertional mutagenesis in lactococci and other gram-positive bacteria. J. Bacteriol. 178:931-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Melin, L., H. Friden, E. Dehlin, L. Rutberg, and A. von Gabain. 1990. The importance of the 5′-region in regulating the stability of sdl mRNA in Bacillus subtilis. Mol. Microbiol. 4:1881-1889. [DOI] [PubMed] [Google Scholar]
- 23.Miller, J. H. 1972. Experiments in molecular genetics, p. 352-359. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 24.Persson, M., E. Glatz, and B. Ruthberg. 2000. Different processing of an mRNA species in Bacillus subtilis and Escherichia coli. J. Bacteriol. 182:689-695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Platteeuw, C., G. Simons, and W. M. de Vos. 1994. Use of the Escherichia coli beta-glucuronidase (gusA) gene as a reporter for analyzing promoters in lactic acid bacteria. Appl. Environ. Microbiol. 60:587-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ruijter, G. J. G., P. W. Postma, and K. van Dam. 1991. Control of glucose metabolism by enzyme IIGlc of the phosphoenolpyruvate-dependant phosphotransferase system in Escherichia coli. J. Bacteriol. 173:6184-6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Shaff, I., J. Heinish, and F. K. Zimmermann. 1989. Overproduction of glycolytic enzymes in yeast. Yeast 5:285-290. [DOI] [PubMed] [Google Scholar]
- 29.Snoep, J. L., L. P. Yomano, H. V. Westerhoff, and L. O. Ingram. 1995. Protein burden in Zymomonas mobilis: negative flux and growth control due to overproduction of glycolytic enzymes. Microbiology 141:2329-2337. [Google Scholar]
- 30.Terzaghi, B. E., and W. E. Sandine. 1975. Improved medium for lactic streptococci and their bacteriophages. Appl. Microbiol. 29:807-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walsh, K., and D. E. Koshland, Jr. 1985. Characterisation of rate controlling steps in vivo by use of adjustable expression vector. Proc. Natl. Acad. Sci. USA 82:3577-3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilson, K. J., K. E. Gilles, and R. A. Jefferson. 1991. Beta-glucuronidase (GUS) operon fusion as a tool for studying plant-microbe interactions, p. 226-229. In H. Hennecke and D. P. S. Verma (ed.), Advances in molecular genetics of plant-microbe interactions. Kluwer Academic Publishers, Dordrecht, The Netherlands.