Abstract
Isolates of the toxic, N2-fixing species Cylindrospermopsis raciborskii from various geographic locations were analyzed with respect to their genetic diversity based on the nifH and cpcBA-IGS genes. Gene sequences clustered according to their geographic origin, with the nifH sequences separating into European, Australian, and American groups and the cpcBA-IGS sequences separating into American and European or Australian groups. PCR primers for both genes were designed to exclusively amplify DNA from Cylindrospermopsis species, and an additional primer set for cpcBA-IGS was designed to specifically amplify the American C. raciborskii strains.
Cylindrospermopsis raciborskii is a cosmopolitan, nitrogen (N2)-fixing cyanobacterial species found in temperate to tropical freshwater habitats. The widespread proliferation of C. raciborskii in some drinking and recreational water supplies has caused international public health concerns (11). This concern is due to the potential for some strains to produce the alkaloid hepatotoxin cylindrospermopsin. Cyanobacterial toxins have been implicated in a range of animal and human health issues (1, 11, 13, 14, 21, 22, 32, 36). Regional characterization of C. raciborskii isolates is necessary for the detection of these strains at prebloom densities in areas susceptible to toxic cyanobacterial growth. Furthermore, analyzing strains from geographically diverse origins may help elucidate the mechanisms of their expansion and dispersal.
Molecular approaches are particularly useful in the detection and identification of specific strains, especially those that are morphologically identical at the species level. Genetic identification has been used to discriminate nuisance species in cyanobacterial genera, including Microcystis, Anabaena, Nodularia, and Cylindrospermopsis (5, 8, 10, 23-26, 28, 40). This information can also be used to characterize the degree of genetic similarity among populations. For example, C. raciborskii strains originating from different parts of Australia have been compared based on genetic analysis of the 16S rRNA gene (32, 33) and the rpoC1 (RNA polymerase) gene (39). This is the first study to compare C. raciborskii isolates originating from a wider geographic area.
Genetic differences between C. raciborskii cultures were identified with two environmentally relevant genes. One of the genes utilized was nifH, a highly conserved gene that encodes dinitrogenase reductase, a protein subunit in the nitrogenase complex involved in N2 fixation. Common to all N2 fixers, the 324-bp nifH fragment is useful in characterizing diazotrophic communities and for differentiating cyanobacterial genera (6-7, 40). The other genetic locus used in this survey was cpcBA-IGS, which includes the highly variable intergenic spacer (IGS) region between two phycobilisome (bilin) subunits (cpcB and cpcA) within the phycocyanin operon (24). Phycocyanin is an accessory pigment that gives cyanobacteria their characteristic blue-green color and, together with chlorophyll a, is contained in the photosynthetic apparatus (17). A 685-bp fragment within the phycocyanin operon was chosen because of its potential for greater variability that may be useful in differentiating cyanobacterial strains (8, 24). Both cpcBA-IGS and nifH appear to be more useful in discriminating between strains than the commonly employed 16S rRNA gene, which exhibits low intrageneric variability in many cyanobacteria (20, 30).
PCR amplification and sequencing.
C. raciborskii cultures from Australia (northern Queensland and Sydney), Germany, Portugal, Hungary (Lake Balaton), Brazil, and the United States (Florida) (12, 19, 31) were analyzed. The origin, morphology, and GenBank accession number for nifH and cpcBA-IGS sequences of each isolate used are given in Table 1. DNA extraction was performed by the XS method (37). A negative control in which no culture was added was run with each extraction set. From each of the C. raciborskii DNA isolates, the 324-bp nifH and the 685-bp cpcBA-IGS gene fragments were PCR amplified (in a 20-μl reaction volume containing 1× manufacturer's buffer [Fisher Biotech, Perth, Australia], 2.5 mM MgCl2, 200 μM concentrations of each deoxyribonucleoside triphosphate, 10 pmol of each primer, 1 U of Taq DNA polymerase [Fisher Biotech], and 1 μl [ca. 10 ng] of isolated DNA). The primers used were the cyanobacterial nifH primers (27) and cyanobacterial cpcBA-IGS primers (24). The amplification parameters for nifH were 94°C for 5 min, with 30 cycles of 94°C for 10 s, 55°C for 20 s, 72°C for 1 min, and then an extension at 72°C for 7 min. PCR parameters were the same for cpcBA-IGS, except that a 52°C annealing temperature was used. The presence of the PCR product was confirmed on a 1% agarose gel (Progen, Brisbane, Australia) run at 75 V with 3 μl of PCR product and 100 ng of a φX174/HaeIII molecular size marker. The PCR products were ethanol precipitated and sequenced in both the forward and reverse directions with Big Dye terminators according to the manufacturer's instructions (Applied Biosystems Inc., Foster City, Calif.) and with an automated sequencer (PRISM cycle sequencing system and the ABI 373 sequencer [Applied Biosystems Inc.]). Sequences were aligned with the SeqLab program and were checked manually. Phylogenetic trees were generated with the Dayhoff PAM matrix and neighbor-joining algorithm with PHYLIP software (University of Wisconsin Genetic Computer Group). Consensus sequences were identified from these alignments by using the CONSENSE protocol in PHYLIP (16).
TABLE 1.
Origins, morphologies, and GenBank accession numbers of the C. raciborskii cultures used in this study
| Strain | Origin
|
Date isolated | Morphology | nifH accession no. | cpcBA-IGS accession no. | |
|---|---|---|---|---|---|---|
| Country | Water body | |||||
| Aqc | Australia | Aquaculture pond, Townsville | 1997 | Coiled | AF426782 | AF426788 |
| Aqs | Australia | Aquaculture pond, Townsville | 1997 | Straight | AF426783 | AF426789 |
| Sdc | Australia | Solomon Dam | 1996 | Coiled | AF426780 | AF426803 |
| Sds | Australia | Solomon Dam | 1996 | Straight | AF426781 | AF426804 |
| Mk | Australia | Lake McKinley | 1997 | Straight | NTb | AF426802 |
| Goon | Australia | Goonyella Dam | 1998 | Straight | NT | AF426799 |
| LJ | Australia | Lake Julius | 1995 | Straight | AF426778 | AF426800 |
| A205 | Australia | Ornamental pond, Sydney | NA | Straight | AF426768 | AF426787 |
| Germany 1 | Germany | NAa | NA | Straight | AF426776 | AF426797 |
| Germany 2 | Germany | NA | NA | Straight | AF426777 | AF426798 |
| Bal 5 | Hungary | Lake Balaton | 1984 | Straight | AF426769 | AF426790 |
| Bal 6 | Hungary | Lake Balaton | 1994 | Straight | AF426770 | AF426791 |
| Caia | Portugal | Caia reservoir | NA | Straight | AF426773 | AF426794 |
| 4799 | Portugal | NA | NA | Straight | AF426767 | AF426786 |
| Marau 1 | Portugal | Marau reservoir | NA | Straight | AF426779 | AF426801 |
| Brazil 1 | Brazil | Paranoa Lake | NA | Straight | AF426771 | AF426792 |
| Brazil 2 | Brazil | Bilings reservoir | NA | Straight | AF426772 | AF426793 |
| Florida D | United States | Lake Dora, Fla. | 1999 | Coiled | AF426774 | AF426795 |
| Florida F | United States | Lake Dora, Fla. | 1999 | Straight | AF426784 | AY078437 |
| Florida G | United States | Lake Dora, Fla. | 1999 | Straight | AF426775 | AF426796 |
| Florida I | United States | Lake Dora, Fla. | 1999 | Coiled | AF426785 | AY078438 |
NA, information not available for this isolate.
NT, not enough DNA was available to sequence nifH for this isolate.
Primer design.
Primers specific to C. raciborskii were designed based on sequences amplified from the cultures used in this study. Oligonucleotides were synthesized by Genset Oligos Pty. Ltd (Lismore, Australia). For nifH, the following primers were used to amplify Cylindrospermopsis species to the exclusion of all other heterocystous cyanobacteria and resulted in a 225-bp PCR product: cylnif F (5′-TAARGCTCAAACTACCGTAT) and cylnif R (5′-ATTTAGACTTCGTTTCCTAC). For cpcBA-IGS, two forward primers were designed and used with the general cyanobacterial reverse primer from the original amplifications. One forward primer, cylcpc F (5′-GGCTTACGCGAAACCTATATA) (a 638-bp PCR product), was genus specific, and the other, FBcpc F (5′-AGCAGCAGCTGTTGCATAGTCCA) (a 464-bp PCR product), was specific to Florida and Brazil isolates. The specificity of these primers was tested against the isolates listed in Table 1 as well as against other heterocystous and nonheterocystous cyanobacteria.
Phylogenetic analysis of the nifH and cpcBA-IGS sequences.
A 324-bp fragment from the nifH gene and a 685-bp fragment of the phycocyanin operon (cpcBA-IGS) were amplified from C. raciborskii cultures isolated from Australia, Europe, and the Americas (Table 1). PCR amplification products were detected for both genes from all isolates analyzed. Sequencing these products revealed phyletically significant differences in the nucleotide sequences for C. raciborskii from different regions.
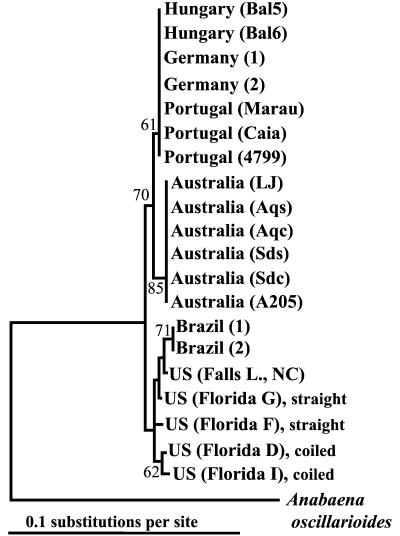
Variation within the nifH gene reflected a distinct geographic grouping of isolates. All the C. raciborskii nifH nucleotide sequences from Europe were identical and were 100% similar to the consensus sequence. The Australian sequences were also 100% similar to each other and deviated from the European sequences by <0.7%. The Brazilian sequences were identical to each other and contained four sites where they were distinct from the consensus nucleotide sequence (1.34% sequence dissimilarity), two of which were shared with the Florida isolates. The Florida isolates were 99% similar to each other and displayed a 2% overall divergence from the consensus sequence. Phylogenetic analysis of these nifH nucleotide sequences confirmed a distinct clustering of C. raciborskii based on geographic origin. The six sequences from Australian isolates formed one cluster, the sequences from European isolates (Germany, Hungary, and Portugal) formed a second cluster, and the sequences from American isolates (Brazil, Florida, and North Carolina) formed a third cluster (Fig. 1).
FIG. 1.
Phylogenetic tree based upon nifH sequences of C. raciborskii isolates originating from different geographic locations. Bootstrap values (>50) are given by the corresponding nodes and were generated with distance methods. Anabaena oscillarioides (GenBank accession no. M63686) was used as the outgroup.
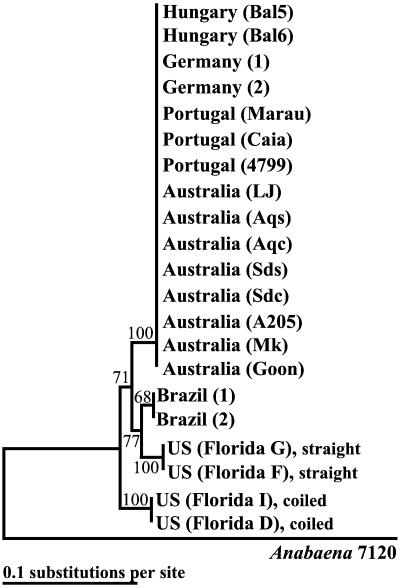
For cpcBA-IGS, there was more genetic variation between the Florida isolates of C. raciborskii but less variation in isolates from other locations. Sequences from the European and Australian isolates were >99.8% similar to each other at the nucleotide level. Brazilian sequences were 100% similar to each other, but only 97.8% similar to the sequences from European and Australian isolates. Florida sequences had much higher genetic variability (96.2% similarity to each other) and were only 94.5% similar to those from Australia and Europe at the nucleotide level. There were nine distinct sites shared by Florida and Brazilian cpcBA-IGS nucleotide sequences to the exclusion of those originating from other regions. The phylogenetic analysis based upon cpcBA-IGS for these isolates did not delineate European and Australian sequences due to the high percentage of similarity, but they did separate sequences from American isolates into a distinct cluster (Fig. 2).
FIG. 2.
Phylogenetic tree based upon cpcBA-IGS sequences of C. raciborskii isolates originating from different geographic locations. Bootstrap values (>50) are given by the corresponding nodes and were generated with distance methods. Anabaena 7120 (GenBank accession no. X05239) was used as the outgroup.
Thus, the variation among global C. raciborskii populations was reflected differently in the nifH and cpcBA-IGS nucleotide sequences. Although nifH was the smaller of the two gene fragments sequenced, regional distinctions could be made based on these sequences. C. raciborskii isolates originating from Europe, Australia, or the Americas were separated phylogenetically into distinct clusters based upon the nifH sequence data. While there were distinctions between regions revealed by the nifH sequence data (variation of up to 2.5%), the amount of variation within each of these three geographic regions was very low (<1%). Phylogenetic analyses based upon cpcBA-IGS nucleotide sequences only differentiated American C. raciborskii isolates. The higher degree of similarity between European and Australian isolates within this part of the phycocyanin operon did not allow distinctions to be made among these groups. In previous studies, variation in cpcBA-IGS has been sufficient to differentiate strains of Nodularia spumigena, Anabaena circinalis, and Microcystis aeruginosa originating from the same geographic region (i.e., Australia) as well as those from other regions (such as North America and the Baltic Sea) (8). However, the delineating power of cpcBA-IGS does not appear uniform for all cyanobacterial species or strains (8), as is evident from the C. raciborskii sequences of this study. Bolch et al. (10) found a sequence dissimilarity of less than 1% in cpcBA-IGS between Nodularia strains and suggested that this indicates a single morphospecies. Using this criteria, the <1% variation between the Australian and European nifH and cpcBA-IGS sequences indicates these two C. raciborskii populations may be defined as a single species cluster.
The American C. raciborskii isolates displayed greater genetic variability, and it appears that within some Florida lakes there are multiple distinct genotypes. The relatively low similarity between C. raciborskii isolates within a single Florida lake is noteworthy, especially when compared to the high similarities between isolates originating from different European countries or Australian states. Morphologically similar but genetically distinct cyanobacteria may coexist in one body of water and form separate blooms, or there may be a mixture of cyanobacteria within one bloom (e.g., Nodularia [10], Synechococcus [15], Microcystis [9, 35, 38], and Cylindrospermopsis [4, 34, 39]). A genetically mixed population, such as those examined from the Florida lakes, may be the result of accelerated molecular evolution in highly favorable environmental conditions (warm temperatures, abundant nutrients, and sufficient light year-round) or the more recent introduction of new strains. Possible mechanisms for the introduction and spread of this species include transport on the feet or in the guts of migratory birds that would be present in lakes and reservoirs and would travel long distances (2-3), human-related transport (i.e., recreational boats, commercial ship ballast water, or aquaria), or oceanic currents. C. raciborskii would most likely be transported as an akinete that would then germinate when suitable growth conditions were encountered (18, 29). The high degree of genetic similarity between C. raciborskii collected from Brazil and Florida could potentially be explained if one of these mechanisms allowed the two populations to be continually mixed or if they originated from the same source population relatively recently.
C. raciborskii isolates from Florida had two morphological variations, coiled and straight. Previous genetic comparisons of these two morphologies in Australian isolates have shown them to be nearly genetically identical (99.8%) based on 16S rRNA sequences (33), and this study demonstrated that they were genetically identical based on nifH and cpcBA-IGS sequences. However, for the Florida isolates there was a genetic distinction between these two morphologies. Sequences from the two coiled isolates (Florida D and I) clustered separately from isolates with straight morphologies (Florida G and F) in both nifH and cpcBA-IGS phylogenies (Fig. 1 and 2). While morphology may not be strictly controlled by genetics (33), it is interesting that it appears to be reflected in the nucleotide sequences for two separate genes. The presence of this distinction only in the more genetically diverse Florida isolates and not in those from Australia suggests that there may be other factors besides morphology being regulated. Analysis of additional C. raciborskii strains with coiled morphologies is necessary to determine the extent of this relationship between genetic and morphological variation.
Specificity of designed primers.
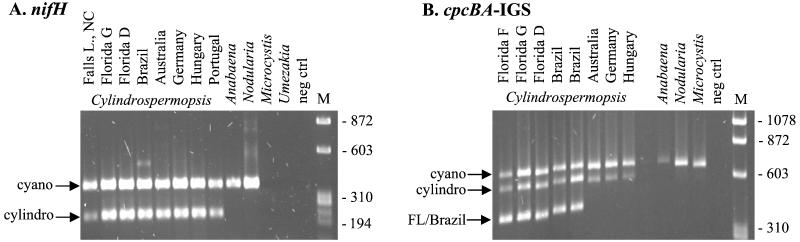
The specificity and accuracy of the primers designed to amplify nifH from Cylindrospermopsis spp. and cpcBA-IGS from Cylindrospermopsis spp. and American strains were tested by using other cyanobacterial isolates. The 324-bp PCR product for nifH was seen in all cyanobacterial N2 fixers by the general cyanobacterial primer set (positive control), while the 225-bp band amplified by the Cylindrospermopsis-specific primer set was seen only in C. raciborskii isolates (and excluded the closely related species Anabaena circinalis and Nodularia spumigena) (Fig. 3A). These primer sets did not amplify nifH from the non-N2 fixers (Microcystis aeriginosa and Umezakia natans). For cpcBA-IGS, the general cyanobacterial primers amplified a 685-bp band in all the cyanobacterial isolates. The Cylindrospermopsis-specific forward primer in combination with the general cyanobacterial reverse primer amplified a 638-bp PCR product from just the C. raciborskii cultures and not from the other cyanobacterial cultures. The American strain-specific primer (FBcyl) in combination with the general cyanobacterial reverse primer successfully amplified a 464-bp cpcBA-IGS band exclusively from C. raciborskii isolates from Florida and Brazil (Fig. 3B). Thus, the designed primer sets appeared to yield the desired specificity. The nifH Cylindrospermopsis-specific primer set was tested on DNA extracted from environmental water samples from the Falls Lake reservoir in North Carolina to evaluate its effectiveness in a mixed phytoplankton population. The sequence of the amplified PCR product was unique but was grouped within the C. raciborskii cluster. The Falls Lake sequence was most similar to the American (Florida and Brazil) C. raciborskii nifH sequences.
FIG. 3.
PCR products amplified with the nifH and cpcBA-IGS primer sets. Each set of primers was used in a separate PCR, and then the two (for nifH) or three (for cpcBA-IGS) PCR products from each isolate were run together in the same lane of the gel. A φX174/HaeIII molecular size marker (M) was run on each gel. (A) For nifH, the 324-bp product was amplified by the general cyanobacterial primer set (cyano) which had been used in the original sequencing of the C. raciborskii isolates. Each of these samples was also amplified with the Cylindrospermopsis-specific primer set (cylindro), which resulted in a 225-bp product exclusively from C. raciborskii samples. The C. raciborskii isolates used were the following: Falls Lake NC, Florida G, Florida D, Brazil 1, A205, Germany 1, Bal 6, and Marau 1. (B) For cpcBA-IGS, the 685-bp product was amplified by the general cyanobacterial primer set (cyano) which had been used in the original sequencing of the C. raciborskii isolates. Each of these samples was also amplified with the Cylindrospermopsis-specific primer set (cylindro), which resulted in a 638-bp product exclusively from C. raciborskii samples, and an American strain-specific primer set (FL/Brazil) which amplified a 464-bp band exclusively from the Florida and Brazil isolates. The C. raciborskii isolates used were Florida F, Florida G, Florida D, Brazil 1, Brazil 2, A205, Germany 1, and Bal 6.
Prior to this study, primer sequences had been reported for the PCR amplification of cyanobacterial nifH and cpcBA-IGS, but none was designed specifically for Cylindrospermopsis spp. The development of both genus-specific and region-specific primers is useful for two reasons. First, it confirms that the regional origin of isolates is evident at the genetic level and C. raciborskii from specific geographic regions can be selectively amplified. The high specificity of the designed primers verifies that they have close homology with C. raciborskii and confirms that there is sufficient variation within these two genes for the distinction of the Cylindrospermopsis genus. Secondly, since these primers have been demonstrated to amplify C. raciborskii to the exclusion of other closely related heterocystous cyanobacterial species, they can be used for rapid, unequivocal detection of C. raciborskii in water supplies, particularly when there is a mix of cyanobacterial species present. The use of highly sensitive and specific molecular methods in conjunction with traditional microscopic screening will improve the speed and accuracy of detecting C. raciborskii and other toxic species in waters prone to cyanobacterial blooms.
Nucleotide sequence accession numbers.
The GenBank database accession numbers for C. raciborskii nifH sequences from isolates in this study are AF426767 to AF426785 and for the cpcBA-IGS sequences are AF726786 to AF426804 and AY078437 and AY078438.
Acknowledgments
This study was financially supported by the St. Johns River Water Management District, National Science Foundation, U.S. Department of Agriculture, and the Australian Research Council. J.D. was supported by a National Science Foundation Graduate Research Fellowship.
We thank T. Steppe and M. Piehler for critically reading the manuscript, and we give special thanks to colleagues who contributed cultures for this study.
REFERENCES
- 1.Anderson, R. J., H. A. Luu, D. Z. Chen, C. F. B. Holmes, M. L. Kent, M. LeBlanc, F. J. R. Taylor, and D. E. Williams. 1993. Chemical and biological evidence links microcystins to salmon ‘netpen liver disease.’ Toxicon 31:1315-1323. [DOI] [PubMed] [Google Scholar]
- 2.Atkinson, K. M. 1972. Birds as transporters of algae. Br. Phycol. J. 7:319-321. [Google Scholar]
- 3.Atkinson, K. M. 1980. Experiments in dispersal of phytoplankton by ducks. Br. Phycol. J. 15:49-58. [Google Scholar]
- 4.Baker, P. D., and A. R. Humpage. 1994. Toxicity associated with commonly occurring cyanobacteria in surface waters of the Murray-Darling Basin, Australia. Aust. J. Mar. Freshwater Res. 45:773-786. [Google Scholar]
- 5.Beltran, E. C., and B. A. Neilan. 2000. Geographical segregation of the neurotoxin-producing cyanobacterium Anabaena circinalis. Appl. Environ. Microbiol. 66:4468-4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ben-Porath, J., E. J. Carpenter, and J. P. Zehr. 1993. Genotypic relationships in Trichodesmium (Cyanophyceae) based on nifH sequence comparisons. J. Phycol. 29:806-810. [Google Scholar]
- 7.Ben-Porath, J., and J. P. Zehr. 1994. Detection and characterization of cyanobacterial nifH genes. Appl. Environ. Microbiol. 60:880-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bolch, C. J., S. I. Blackburn, B. A. Neilan, and P. M. Grewe. 1996. Genetic characterization of strains of cyanobacteria using PCR-RFLP of the cpcBA intergenic spacer and flanking regions. J. Phycol. 32:445-451. [Google Scholar]
- 9.Bolch, C. J., S. I. Blackburn, G. J. Jones, P. T. Orr, and P. M. Grewe. 1997. Plasmid content and distribution in Australian isolates of Microcystis Kützing ex Lemmermann (Cyanobacteria: Chroococcales). Phycologia 36:6-11. [Google Scholar]
- 10.Bolch, C. J. S., P. T. Orr, G. J. Jones, and S. I. Blackburn. 1999. Genetic, morphological, and toxicological variation among globally distributed strains of Nodularia (Cyanobacteria). J. Phycol. 35:339-355. [Google Scholar]
- 11.Carmichael, W. W. 1997. The cyanotoxins. Adv. Bot. Res. 27:211-256. [Google Scholar]
- 12.Chapman, A. D., and C. L. Schelske. 1997. Recent appearance of Cylindrospermopsis (cyanobacteria) in five hypereutrophic Florida lakes. J. Phycol. 33:191-195. [Google Scholar]
- 13.Codd, G. A. 1984. Toxins of freshwater cyanobacteria. Microbiol. Sci. 1:48-52. [PubMed] [Google Scholar]
- 14.Edwards, C., K. A. Beattie, C. M. Scrimgeour, and G. A. Codd. 1992. Identification of anatoxin-a in benthic cyanobacteria (blue-green algae) and in associated dog poisonings at Lock Insh, Scotland. Toxicon 30:1165-1175. [DOI] [PubMed] [Google Scholar]
- 15.Ernst, A., P. Marshcall, and C. Postius. 1995. Genetic diversity among Synechococcus spp. (cyanobacteria) isolated from the pelagial Lake Constance. FEMS Microbiol. Ecol. 17:197-204. [Google Scholar]
- 16.Felsenstein, J. 1989. PHYLIP: phylogeny inference package. Cladistics 5:258-266. [Google Scholar]
- 17.Glazer, A. N. 1984. Phycobilisome: a macro molecular complex optimized for light energy transfer. Biochim. Biophys. Acta 768:29-52. [Google Scholar]
- 18.Isvánovics, V., H. M. Shafik, M. Présing, and S. Juhos. 2000. Growth and phosphate uptake kinetics of the cyanobacterium, Cylindrospermopsis raciborskii (Cyanophyceae) in throughflow cultures. Freshwater Biol. 43:257-275. [Google Scholar]
- 19.Moisander, P. H., E. McClinton, and H. W. Paerl. Salinity effects on growth, nitrogenase activity, and photosynthetic parameters of estuarine, planktonic cyanobacteria. Microb. Ecol., in press. [DOI] [PubMed]
- 20.Moore, L. R., G. Rocap, and S. W. Chisholm. 1998. Physiology and molecular phylogeny of coexisting Prochlorococcus ecotypes. Nature 393:464-467. [DOI] [PubMed] [Google Scholar]
- 21.Negri, A. P., and G. J. Jones. 1995. Bioaccumulation of paralytic shellfish poisoning (PSP) toxins from the cyanobacterium Anabaena circinalis by the freshwater mussel Alathryia condola. Toxicon 33:667-678. [DOI] [PubMed] [Google Scholar]
- 22.Negri, A. P., G. J. Jones, and M. Hindmarsh. 1995. Sheep mortality associated with paralytic shellfish poisons from the cyanobacterium Anabaena circinalis. Toxicon 33:1321-1329. [DOI] [PubMed] [Google Scholar]
- 23.Neilan, B. A. 1995. Identification and phylogenetic analysis of toxigenic cyanobacteria by multiplex randomly amplified polymorphic DNA PCR. Appl. Environ. Microbiol. 61:2286-2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neilan, B. A., D. Jacobs, and A. E. Goodman. 1995. Genetic diversity and phylogeny of toxic cyanobacteria determined by DNA polymorphisms within the phycocyanin locus. Appl. Environ. Microbiol. 61:3875-3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neilan, B. A., D. Jacobs, T. Del Dot, L. L. Blackall, P. R. Hawkins, P. T. Cox, and A. E. Goodman. 1997. rRNA sequences and evolutionary relationships among toxic and nontoxic cyanobacteria of the genus Microcystis. Int. J. Syst. Bacteriol. 47:693-697. [DOI] [PubMed] [Google Scholar]
- 26.Neilan, B. A., P. R. Hawkins, P. T. Cox, and A. E. Goodman. 1994. Towards a molecular taxonomy for the bloom-forming cyanobacteria. Aust. J. Mar. Freshwater Res. 45:869-873. [Google Scholar]
- 27.Olson, J. B., T. F. Steppe, R. W. Litaker, and H. W. Paerl. 1998. N2-fixing microbial consortia associated with the ice cover of Lake Bonney, Antarctica. Microbiol. Ecol. 36:231-238. [DOI] [PubMed] [Google Scholar]
- 28.Otsuka, S., S. Suda, R. Li, M. Watanabe, H. Oyaizu, S. Matsumoto, and M. M. Watanabe. 1999. Phylogenetic relationships between toxic and non-toxic strains of the genus Microcystis based on 16S and 23S internal transcribed spacer sequences. FEMS Microbiol. Lett. 172:15-21. [DOI] [PubMed] [Google Scholar]
- 29.Padisak, J. 1997. Cylindrospermopsis raciborskii (Woloszynska) Seenayya et Subba Raju, an expanding, highly adaptive cyanobacterium: worldwide distribution and review of its ecology. Arch. Hydrobiol. 107:563-593. [Google Scholar]
- 30.Rudi, K., O. M. Skulberg, and K. S. Jakobsen. 1998. Evolution of cyanobacteria by exchange of genetic material among phyletically related strains. J. Bacteriol. 180:3453-3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saker, M. L. 2000. Ph.D. dissertation. James Cook University, Queensland, Australia.
- 32.Saker, M. L., and B. A. Neilan. 2001. Varied diazotrophies, morphologies, and toxicities of genetically similar isolates of Cylindrospermopsis raciborskii (Nostocales, Cyanophyceae) from Northern Australia. Appl. Environ. Microbiol. 67:1839-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saker, M. L., B. A. Neilan, and D. J. Griffiths. 1999. Two morphological forms of Cylindrospermopsis raciborskii (Cyanobacteria) isolated from Solomon Dam, Palm Island, Queensland. J. Phycol. 35:599-606. [Google Scholar]
- 34.Schembri, M. A., B. A. Neilan, and C. P. Saint. 2001. Identification of genes implicated in toxin production in the cyanobacterium Cylindrospermopsis raciborskii. Environ. Toxicol. 16:413-421. [DOI] [PubMed] [Google Scholar]
- 35.Schwabe, W., A. Weihe, T. Börner, M. Henning, and J.-G. Kohl. 1988. Plasmids in toxic and non-toxic strains of the cyanobacterium Microcystis aeruginosa. Curr. Microbiol. 17:133-137. [Google Scholar]
- 36.Thomas, A., M. L. Saker, J. H. Norton, and R. D. Olsen. 1998. Cyanobacterium Cylindrospermopsis raciborskii as a probable cause of death in cattle in northern Queensland. Aust. Vet. J. 76:592-594. [DOI] [PubMed] [Google Scholar]
- 37.Tillett, D., and B. A. Neilan. 2000. Xanthogenate nucleic acid isolation from cultured and environmental cyanobacteria. J. Phycol. 35:1-8. [Google Scholar]
- 38.Tillett, D., D. L. Parker, and B. A. Neilan. 2001. Detection of toxigenicity by a probe for the microcystin synthetase A gene (mcyA) of the cyanobacterial genus Microcystis: comparison of toxicities with 16S rRNA and phycocyanin operon (phycocyanin intergenic spacer) phylogenies. Appl. Environ. Microbiol. 67:2810-2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilson, K. M., M. A. Schembri, P. D. Baker, and C. P. Saint. 2000. Molecular characterization of the toxic cyanobacterium Cylindrospermopsis raciborskii and design of a species-specific PCR. Appl. Environ. Microbiol. 66:332-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zehr, J. P., M. T. Mellon, and W. D. Hiorns. 1997. Phylogeny of cyanobacterial nifH genes: evolutionary implications and potential applications to natural assemblages. Microbiology 143:1443-1450. [DOI] [PubMed] [Google Scholar]