Abstract
The culturability of bacteria in the bulk soil of an Australian pasture was investigated by using nutrient broth at 1/100 of its normal concentration (dilute nutrient broth [DNB]) as the growth medium. Three-tube most-probable-number serial dilution culture resulted in a mean viable count that was only 1.4% of the mean microscopically determined total cell count. Plate counts with DNB solidified with agar and with gellan gum resulted in viable counts that were 5.2 and 7.5% of the mean microscopically determined total cell count, respectively. Prior homogenization of the soil sample with an ultrasonic probe increased the viable count obtained by using DNB solidified with gellan gum to 14.1% of the mean microscopically determined cell count. A microscopic examination of the cell aggregates that remained after sonication revealed that the potential CFU count was only 70.4% of the total cell count, due to cells occurring as pairs or in clumps of three or more cells. Staining with SYTO 9 plus propidium iodide indicated that 91.3% of the cells in sonicated soil samples were potentially viable. Together, these findings suggest that the maximum achievable CFU count may be as low as 64.3% of the total cell count. Thirty isolates obtained from plate counting experiments performed with DNB as the growth medium were identified by comparative analysis of partial 16S rRNA gene sequences. A large proportion of these isolates represent the first known isolates of globally distributed groups of soil bacteria belonging to novel lineages within the divisions Actinobacteria, Acidobacteria, Proteobacteria, and Verrucomicrobia.
It has been established that the genetic diversity of soil bacteria is high (7, 34, 48) and that soils contain many bacterial species of lineages for which no known cultivated isolates are available (14, 25). Many soil bacteria are referred to as uncultured or even nonculturable. A range of methods have been developed to study these organisms directly in their habitats (2, 10, 35). These methods are extremely useful for studying the ecology of microorganisms as parts of communities, but initial physiological and genetic studies of pure cultures should greatly facilitate such synecological studies. We believe that many of these bacteria are in fact culturable using relatively simple technologies. To test our hypothesis, we used a simple growth medium to investigate the culturability of soil bacteria. We reasoned that if we could increase the culturability of soil bacteria above the level of 5% that often appears to be the upper limit for cultivation studies, we should begin to cultivate bacteria that belong to some of the uncultured groups.
MATERIALS AND METHODS
Soil and sampling.
The soil used in this study was collected from a rotationally grazed pasture dominated by Lolium perenne and Trifolium repens at the Dairy Research Institute, Ellinbank, Victoria, Australia. The soil is a krasnozem clay loam (Gn4.11 [32]; basaltic clay loam; Ferrosol [15]). The management regime, consisting of two cows per ha and 35 kg of P per ha per year, and characteristics of the soil have been described elsewhere (39, 42).
A 25-mm-diameter metal corer was used to obtain 100-mm-long soil cores, which were transported intact at the ambient temperature in sealed polyethylene bags and processed within 3 h of collection. The upper 30 mm of each core was discarded, and large roots and stones were removed from the remainder, which was then sieved through a brass sieve with a 2-mm aperture size (Endecotts Ltd., London, United Kingdom) and used immediately for dry weight determination, microscopic investigations, or cultivation experiments.
Aliquots of freshly sieved soil were accurately weighed and then dried at 105°C for 3 days. The samples were then reweighed after they were first allowed to cool to room temperature in a desiccator. The factor for conversion of fresh weight to dry weight of soil was calculated, and all results were expressed per gram (dry weight) of soil.
Total cell counts.
Freshly sieved soil samples (0.1 g) were fixed for 16 h in phosphate-buffered saline (0.13 M NaCl, 7 mM Na2HPO4, 3 mM NaH2PO4; pH 7.2) containing 4% (wt/vol) paraformaldehyde. The samples were then concentrated by centrifugation at 16,000 × g for 5 min at room temperature and washed once in phosphate-buffered saline before being resuspended in 1 ml (final total volume) of phosphate-buffered saline. The samples were dispersed by treatment (sonication) with a titanium ultrasonic needle probe (4 mm in diameter, 127 mm long) by using a Labsonic 2000 ultrasonic homogenizer (B. Braun AG, Melsungen, Germany) at 260 W/cm2 for 15 s with the probe tip positioned at two-thirds of the liquid depth in a 1.5-ml polypropylene microcentrifuge tube, and then they were stained at 4°C for 1 h with 4′,6-diamidino-2-phenylindole (DAPI) at a final concentration of 100 μg/ml. The samples were diluted 10-fold in phosphate-buffered saline, and 10-μl samples of the dilutions were applied to gelatin-coated microscope slides (45) and spread evenly under coverslips (22 by 22 mm). The preparations were examined at a magnification of ×1,000 with a Diaplan microscope (Ernst Leitz GmbH, Wetzlar, Germany) under UV illumination (excitation filter BP340-380, dichromatic mirror RKP400, suppression filter LP430). At least 30 fields, evenly distributed over the area of the coverslip, were examined for each sample, and the blue-fluorescing cells were counted.
Live-dead staining.
Freshly sieved soil samples (0.1 g) were suspended by sonication in 1.0 ml of filtered (pore size, 0.2 μm) distilled water as described above, and the soil was collected by centrifugation at 16,000 × g for 5 min at room temperature. The supernatant was removed, and 1 ml of filtered distilled water was added. One microliter each of the SYTO 9 and propidium iodide dye preparations of a LIVE/DEAD BacLight bacterial viability kit (Molecular Probes, Inc., Eugene, Oreg.) was added, and the samples were stained for 5 min. Aliquots (10 μl) were placed on gelatin-coated microscope slides and examined at a magnification of ×1,000 under UV illumination (excitation filter BP450-490, dichromatic mirror RKP510, suppression filter LP520).
Media.
Dilute nutrient broth (DNB) consisted of Difco nutrient broth (BD Diagnostic Systems, Sparks, Md.) at a concentration of 0.08 g per liter of distilled water. For solid media, 15 g of washed (49) Difco technical agar (BD Diagnostic Systems) or 8 g of gellan gum (Sigma-Aldrich Pty. Ltd., Castle Hill, New South Wales, Australia) plus 0.6 mmol of CaCl2 was added per liter of medium, which was then dispensed into sterile polystyrene petri dishes that were 90 mm in diameter. The final pH was approximately 6.0.
Cultivation experiments.
Accurately weighed samples consisting of about 1 g of freshly sieved soil were added to 100-ml aliquots of sterile distilled water in 150-ml conical flasks and dispersed by stirring with Teflon-coated magnetic bars (8 mm in diameter, 50 mm long) for 15 min at approximately 200 rpm. When required, 1-ml portions of the dispersed soil suspension were transferred to 1.5-ml plastic tubes, and the samples were dispersed by sonication as described above. Unless noted otherwise, samples used for cultivation experiments were not treated by sonication.
One-milliliter aliquots of soil suspension, sonicated or not, were added to 9-ml portions of DNB in borosilicate glass tubes (internal diameter, 14 mm; length, 150 mm) capped with Kim-Kap polypropylene closures (Kimble-Kontes, Vineland, N.J.). These preparations were mixed with a vortex mixer at approximately 150 rpm for 10 s, and then 1-ml aliquots were rapidly transferred to other 9-ml aliquots of DNB. Dilution series were prepared by further transfers and either used as inocula for plate count experiments (see below) or incubated for three-tube most-probable-number (MPN) counting experiments (i.e., three tubes for each dilution step). For plate count experiments, 200-μl aliquots from different dilutions were transferred to petri dishes (plates) containing the desired solid medium and spread over the surface with a sterile glass spreading rod. Each dilution series was used to inoculate a series of plates, with three plates at each dilution level. In all cultivation experiments the preparations were incubated at 25°C in the dark.
Growth in liquid cultures was scored as positive if visible turbidity developed. The MPN was calculated from the dry weight of soil, the dilution factor, and tables of three parallel dilution series based on a statistical treatment of such counting methods (3). The numbers of colonies appearing on solid media were determined by examining the plates at a magnification of ×10 with a stereomicroscope. Each count represents the mean for a series of dilution steps with three plates at each dilution and was calculated based on the dry weight of the soil and dilution factors. Each type of cultivation experiment was carried out 12 or 21 times by using soil from four separate soil cores.
Student's t test was performed with Excel 2001 software (Microsoft Corp., Redmond, Wash.), employing a two-tailed test and assuming that the two samples had unequal variance.
Identification of isolates.
Partial sequencing of the 16S rRNA genes of new isolates was carried out as essentially described by Janssen and O'Farrell (16), after the 16S rRNA gene was amplified by PCR with oligonucleotide primers 27f (5′-GAGTTTGATCMTGGCTCAG-3′) and either 1492r or 1525r (21), using sequencing primer 519r (21). The sequences were compared with those in the GenBank databases (www.ncbi.nlm.nih.gov/blast) by using the BLAST program (1). Sequence similarities were determined by using the software package ClustalX (47). The reference sequences (GenBank accession numbers given in parentheses) used for comparison were sequences from Acidobacterium capsulatum (D26171), Rubrobacter radiotolerans (AJ243870), Rubrobacter xylanophilus (AJ243871), Verrucomicrobium spinosum (X90515), Prosthecobacter spp. (U60012, U60013, U60014, and U60015), and six other verrucomicrobial isolates (X99390, X99391, X99392, AJ229325, AJ229239, and AJ229246).
Nucleotide sequence accession numbers.
The partial 16S rRNA gene sequences obtained in this study have been deposited in the GenBank databases under accession numbers AF432233 to AF432262.
RESULTS
Microscopic examination of soil microorganisms.
The number of microbial cells, as determined by DAPI staining and examination by epifluorescence microscopy, was 1.33 × 109 cells per g (dry weight) of soil (standard deviation [SD] = 2.44 × 108 cells per g). A total of three to five samples from each of nine soil cores were examined. Very few fungal hyphae were observed. The majority of the cells were very small coccobacilli or short rods.
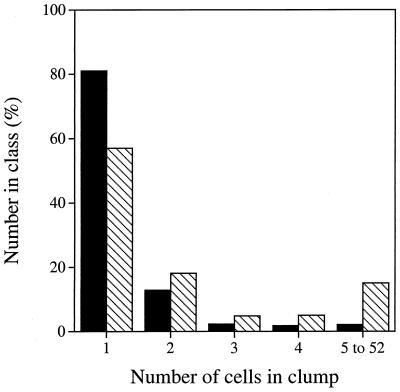
After soil samples were disrupted by sonication, the numbers and compositions of the clumps of cells remaining were determined microscopically after staining with DAPI. A total of 4,642 cells in seven soil samples were examined (Fig. 1). The majority of the cells (57%) were present as single cells, and a further 18% of the cells were in pairs. The largest clumps of cells observed contained 22, 43, 47, and 52 cells. Within each pair or clump, the cells had the same morphology. The total number of single cells, pairs, and clumps of three or more cells (i.e., the total potential CFU count) was 3,270.
FIG. 1.
Numbers of cells observed as single cells, in pairs, or in clumps of three, four, five, or more cells after treatment of soil samples by sonication. The cross-hatched bars represent the percentage of cells occurring in each category, and the solid bars represent the percentage of potential CFU in each category.
The BacLight bacterial viability staining system was used to estimate the proportion of cells with an intact cytoplasmic membrane. A total of 2,299 cells in three soil samples were stained and counted, and 91.3% (SD = 3.8%) of these cells fluoresced green, indicating that they were alive. The other 8.7% of the stained cells fluoresced red, indicating that they had compromised cytoplasmic membranes. Cells in pairs or clumps tended to be either all alive or all dead.
Viable counts of soil bacteria.
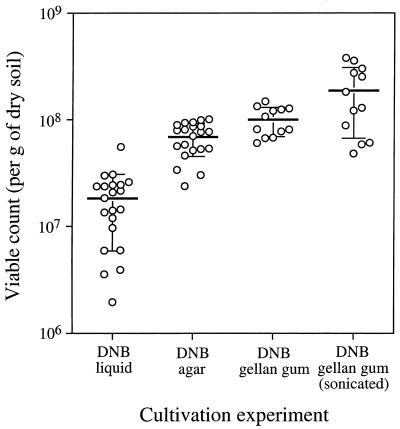
Viable counts were determined by using DNB as the growth medium. Three-tube MPN counts with liquid medium gave viable counts of 1.96 × 106 to 5.56 × 107 per g (dry weight) of soil (mean = 1.83 × 107 per g; SD = 1.24 × 107 per g; n = 21 three-tube counting series) (Fig. 2). The same medium, solidified with agar and used in plate count experiments, yielded higher viable counts, 2.39 × 107 to 1.01 × 108 CFU per g (dry weight) of soil (mean = 6.88 × 107 CFU per g; SD = 2.35 × 107 CFU per g; n = 21 three-plate counting series) (Fig. 2). When agar was replaced with gellan gum in the presence of 0.6 mM CaCl2, the plate counts were even higher, 6.03 × 107 to 1.49 × 108 CFU per g (dry weight) of soil (mean = 1.00 × 108 CFU per g; SD = 3.07 × 107 CFU per g; n = 12 three-plate counting series) (Fig. 2). Student's t test showed that the differences between these three means were statistically significant (MPN count versus agar count, P = 9 × 10−10; agar count versus gellan gum count, P = 0.006). All of these experiments were carried out with samples that were not treated by sonication.
FIG. 2.
Viable counts obtained with DNB in different counting experiments. See text for details. Each point represents the result of one three-tube or three-plate counting experiment. The horizontal lines are the means, and the vertical bars indicate one SD from the mean.
When the soil samples were treated by sonication prior to dispersal, the viable count increased almost twofold, but the variability also increased. We found that sonication times of 15 or 20 s resulted in the highest viable counts, with longer or shorter times yielding lower counts (data not shown). The viable counts from sonicated samples ranged from 4.82 × 107 to 3.79 × 108 CFU per g (dry weight) of soil (mean = 1.88 × 108 CFU per g; SD = 1.21 × 108 CFU per g; n = 12 three-plate counting series) (Fig. 2). Student's t test showed that the difference in the viable count achieved by sonication was statistically significant (P = 0.030).
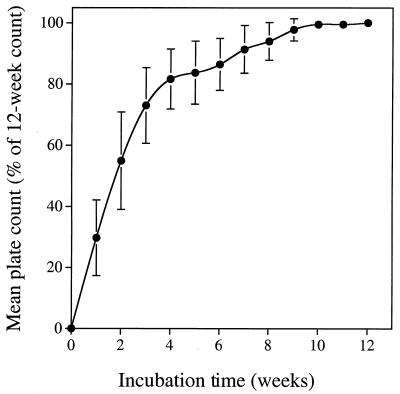
The plate count experiment preparations were incubated for 12 weeks. The viable counts, determined on a weekly basis, continued to increase until about week 10 (Fig. 3).
FIG. 3.
Effect of incubation time on the viable count on DNB solidified with agar, expressed as a percentage of the count obtained after 12 weeks of incubation. The points are the means of 12 three-plate counting experiments, and the vertical bars indicate one SD from the mean.
Isolation and identification of soil bacteria.
The colonies growing on the plates with the lowest CFU counts (i.e., the colonies on the plates that received the most dilute inoculum and yielded colonies) were generally small (approximately 1 mm in diameter) and well separated. There was no evidence of halos of similar colonies appearing around a mother colony that might have been suggestive of spore dispersal and germination during the incubation period that yielded new colonies derived from one original colony. A total of 74 colonies from a number of the terminal positive plates of dilution series on DNB solidified with gellan gum were subcultured onto the same medium. Eleven of these colonies did not grow after transfer, even after 3 months of incubation, while a further 16 subcultures were lost to fungal contamination. The remaining 47 transfers yielded bacterial cultures, 3 of which were mixed and were subsequently separated into two isolates each, which gave a total of 50 isolates.
Thirty of the 50 isolates were identified by comparative analysis of partial 16S rRNA gene sequences (Table 1). All but 8 (Ellin408, Ellin413, Ellin420, Ellin433, Ellin438, Ellin442, Ellin452, and Ellin457) of these 30 were selected from experiments in which the soil sample was treated by sonication prior to plating. The sequence of approximately 400 to 480 bp of the 5′ region of the 16S rRNA gene was determined and compared to previously reported sequences in the GenBank database by using the BLAST program (1). The isolates were assigned to the divisions Proteobacteria, Actinobacteria, Acidobacteria, Verrucomicrobia, and low-G+C-content gram-positive bacteria (Table 1). On the basis of 16S rRNA gene sequence similarities, some of these isolates are members of lineages of bacterial descent for which pure-culture isolates have not previously been recognized.
TABLE 1.
Analysis of isolates on the basis of partial 16S rRNA gene sequences, including the closest relatives as identified by using the BLAST program in the GenBank databases
| Phylogenetic group (division, subdivision) | Isolate | Sequence length (bp) | Top match [GenBank accession no.]a | Nucleotide identity (%) |
|---|---|---|---|---|
| Acidobacteria, subdivision 1 | Ellin408 | 438 | env. WR805 [AJ292771] | 98.4 |
| Ellin457 | 429 | env. WR805 [AJ292771] | 96.7 | |
| Actinobacteria, Actinobacteridae | Ellin402 | 412 | (Dactylosporangium sp.), str. SF2809 [AB017374]b | 97.8 |
| Ellin406 | 400 | Micromonospora peucetica, str. DSM 43363 [X92603] | 93.1 | |
| Ellin410 | 400 | Micromonospora fulvoviolaceus, str. 71-m115 [X92619] | 98.4 | |
| Ellin412 | 402 | Micromonospora fulvoviolaceus, str. 71-m115 [X92619] | 99.0 | |
| Ellin422 | 403 | env. WR169 [AJ233561] | 99.5 | |
| Ellin423 | 436 | Actinoplanes sp., str. IM-3145 [AF131348] | 94.3 | |
| Ellin419 | 421 | Leifsonia poae, str. VKM Ac-1401 [AF116342] | 99.7 | |
| Ellin432 | 447 | Leifsonia poae, str. VKM Ac-1401 [AF116342] | 98.9 | |
| Ellin416 | 435 | env. WR160 [AJ233560] | 93.1 | |
| Ellin420 | 407 | env. WR160 [AJ233560] | 96.3 | |
| Ellin424 | 424 | env. WR160 [AJ233560] | 94.8 | |
| Ellin425 | 446 | env. WR160 [AJ233560] | 93.3 | |
| Ellin438 | 455 | env. WR160 [AJ233560] | 93.8 | |
| Actinobacteria, Rubrobacteridae | Ellin404 | 478 | env. S12_712 [AF078396] | 93.8 |
| Ellin442 | 433 | env. Ac18 [AF388343] | 92.2 | |
| Proteobacteria, α subclass | Ellin413 | 410 | (Methylobacterium sp.), str. 6c [AJ223453] | 99.8 |
| Ellin414 | 401 | (Methylobacterium sp.), str. 6c [AJ223453] | 98.5 | |
| Ellin426 | 400 | (Sphingomonas sp.), env. D64 [AF337832] | 96.8 | |
| Ellin431 | 426 | Afipia sp., str. G9018 [U87769] | 99.8 | |
| Ellin435 | 430 | Nitrobacter sp., str. LL [L11662] | 99.5 | |
| Ellin441 | 406 | Phenylobacterium immobile, str. E [Y18216] | 94.2 | |
| Ellin429 | 407 | env. wr0111 [AJ295550] | 98.3 | |
| Ellin452 | 414 | env. wr0111 [AJ295550] | 95.4 | |
| Proteobacteria, γ subclass | Ellin405 | 480 | env. Ac53 [AF388360] | 95.9 |
| Ellin433 | 468 | env. BIfci40 [AJ318117] | 98.7 | |
| Low-G+C-content gram-positive bacteria | Ellin411 | 432 | (Bacillus), str. G11014 [AB011787] | 98.8 |
| Ellin427 | 465 | env. C006 [AY037665] | 93.2 | |
| Verrucomicrobia, subdivision 2 | Ellin428 | 481 | env. CO19 [AF013522] | 94.1 |
Top match as determined by the BLAST method. The BLAST program was described by Altschul et al. (1); str. denotes 16S rRNA gene sequences from cultivated bacteria, while env. denotes sequences detected as PCR-amplified products in complex microbial communities.
Parentheses indicate that a genus affiliation is tentative based on 16S rRNA gene sequence similarities.
DISCUSSION
Soils contain a great diversity of bacteria, with many of the organisms belonging to groups for which no cultivated representatives are known. It has often been claimed that plate count methodologies are not suitable for cultivation of soil bacteria and that the members of groups without cultivated representatives are somehow nonculturable. We reasoned that by increasing the culturable fraction of soil bacteria above that generally reported, we should be able to isolate some members of groups that to date have not been grown in pure culture.
The use of serial dilution liquid culture techniques has proved to be effective for isolating numerically significant members of the community in anoxic rice paddy soil (6, 11, 38). However, when these techniques were used to cultivate bacteria from the Ellinbank soil, the mean viable count with DNB was only 1.4% of the mean microscopically determined total cell count. Plate counting with DNB solidified with agar produced better counts (5.2% of the mean microscopically determined total cell count). DNB solidified with gellan gum was an even better medium for cultivating soil bacteria, yielding viable counts without and with sonication that were 7.5 and 14.1%, respectively, of the mean microscopically determined total cell count. We found that incubation for at least 10 weeks was required to allow maximum colony development. Dispersion of the soil by sonication increased the mean viable count but also resulted in increased variability between replicates.
A closer examination of the occurrence of cell clumps that remained after sonication revealed that the potential CFU count was only 70.4% of the total cell count, due to the cells occurring in pairs or as clumps of three or more cells. A clump of cells consisting of two or more species may give rise to a colony dominated by only the fastest growing species, so that the presence of the other species goes unrecognized once a pure culture is obtained. However, within the pairs and clumps, the cells seemed to have the same morphology, meaning that although the CFU count may be reduced compared with the total cell count, the diversity of isolates may not be as greatly affected.
The majority of cells appeared to be viable, as judged by the BacLight bacterial viability staining system. Live cells accounted for 91.3% of the cells stained in the soil. If this finding is combined with the mean total microscopic count and the number of potential CFU after sonication, the maximum achievable CFU count may be as low as 8.55 × 108 CFU per g (dry weight) of soil, which is 64.3% of the total cell count. In this light, the mean viable count obtained with DNB solidified with gellan gum combined with prior sonication of the soil suspension may be as high as 22% of the maximum CFU count that could be expected with the samples and treatments used.
This level of culturability allowed the isolation of previously uncultivated bacteria, representing in some cases the first known isolates of globally distributed groups of soil bacteria. We selected a modest number of isolates and identified these by comparative analysis of partial 16S rRNA gene sequences. These isolates were identified as members of five of the major divisions of the domain Bacteria. We choose to use the term division, in agreement with Hugenholtz et al. (14), as the major lineages of the bacteria are variously termed phyla or classes and hierarchical equivalence has not been finalized. In addition, the taxonomic rank of the subdivisions within the divisions Acidobacteria and Verrucomicrobia (14) has not been clarified.
Of the 30 isolates identified and classified, 15 belonged to the division Actinobacteria. Eight of these could be assigned to well-studied lineages within the subclass Actinobacteridae. None of these had identical 16S rRNA gene sequences. Five other isolates had high 16S rRNA gene sequence similarity to a group of the suborder Frankineae detected to date only as cloned, PCR-amplified 16S rRNA genes in the rhizosphere of Alnus viridis (31) and in a polluted moorland soil in Germany (29, 30). Their 16S rRNA gene sequences had similarities of 91.6 to 98.2% to each other.
Two of our isolates (Ellin404 and Ellin442) are members of the subclass Rubrobacteridae, which is one of the five major lineages (subdivisions) of the division Actinobacteria (44). Previously, only two species of this subclass have been isolated in pure culture. Rubrobacter radiotolerans and Rubrobacter xylanophilus are moderate thermophiles with temperature optima of 48 and 60°C, respectively (5, 50), and are unlikely to be good models for members of this group in soils. The two isolates obtained in our study are most closely related to members of groups within the subclass Rubrobacteridae that have been detected only as PCR-amplified 16S rRNA genes, within the radiations termed Group I actinobacteria by Rheims and Stackebrandt (36) and Group 2 rubrobacteria by Holmes et al. (13). Our new strains are most closely related, on the basis of partial 16S rRNA gene sequences, to uncultivated bacteria detected in a polluted moorland soil in Germany (29, 30), a peat bog in Germany (37), a beech forest soil in Germany (36), a pasture soil in the United Kingdom (28), the rhizosphere of L. perenne in Switzerland (27), a cropped soil in Sweden (41), a desert soil in Australia (13), and a forest soil in Australia (43). The two new isolates are members of different lineages within the subclass Rubrobacteridae, as their 16S rRNA gene sequences were only 88.5% similar over the regions sequenced. The sequence similarities to the 16S rRNA genes of the cultivated Rubrobacter spp. were between 76.5 and 80.3%.
Six isolates assigned to the α subclass (subdivision) of the division Proteobacteria belonged to lineages with known and well-studied pure-culture isolates. None of these isolates had 16S rRNA gene sequences that were identical. Two other α-proteobacterial isolates (Ellin429 and Ellin452) were most closely related to members of a radiation within the α subclass of the division Proteobacteria to date detected only as cloned, PCR-amplified 16S rRNA genes, termed cluster 6 by McCaig et al. (28). Members of this radiation have been detected in a pasture soil in the United Kingdom (28), in a peat bog in Germany (37), and on the rhizoplane of Brassica napus in Germany (18). The two strains belonging to this group had 97.3% sequence identity in the region of the 16S rRNA genes for which sequence information was obtained.
Two isolates each could be assigned to the γ subclass of the division Proteobacteria and to the Bacillus group of the division of low-G+C-content gram-positive bacteria.
Two of our isolates (Ellin408 and Ellin457) are members of subdivision 1 (sensu Hugenholtz et al. [14]) of the division Acidobacteria. Previously, only one species in subdivision 1, the aerobic heterotroph Acidobacterium capsulatum, has been isolated in pure culture (19). This species was isolated from acidic mineral environments. Two further species of the division Acidobacteria are members of subdivision 8 and are only distantly related to A. capsulatum (14). These are the strict anaerobes Holophaga foetida (24) and Geothrix fermentans (8). The two isolates obtained in our study are most closely related to members of groups within subdivision 1 of the division Acidobacteria that have been detected only as PCR-amplified 16S rRNA genes. In the regions for which 16S rRNA gene sequence information was obtained, isolates Ellin408 and Ellin457 shared 98.6% sequence identity, while the sequence identities to the 16S rRNA gene of A. capsulatum were 88.9 and 88.0%, respectively. Our new isolates are most closely related, on the basis of partial 16S rRNA gene sequences, to uncultivated bacteria detected in forest and pasture soil in Brazil (4), a polluted moorland soil in Germany (29, 30), a peat bog in Germany (37), a forest soil in Hawaii (33), and an oak forest soil in the United Kingdom (35).
One of our isolates (Ellin 428) is a member of subdivision 2 (sensu Hugenholtz et al. [14]) of the division Verrucomicrobia. Bacteria belonging to subdivisions 1 and 4 of the division Verrucomicrobia have been obtained in pure culture (6, 12, 17, 40), but members of subdivision 2 have been detected only as PCR-amplified 16S rRNA genes in a wide range of different soils and other habitats. The 16S rRNA gene of isolate Ellin428 had sequence identities of 81.8 to 86.4% to 16S rRNA genes of the cultivated members of subdivisions 1 and 4 of the division Verrucomicrobia in the region analyzed. Our new isolate is most closely related, on the basis of partial 16S rRNA gene sequences, to uncultivated bacteria detected in a pinyon-juniper forest soil in the United States (20), a pasture soil in the United Kingdom (28), a cropped soil in Sweden (41), the rhizosphere of B. napus in the United Kingdom (26), a grassland soil in The Netherlands (9), a pasture soil in the United States (22), and a forest soil in Australia (23).
At present, the reason for the success of our cultivation experiments to isolate representatives of groups of so-called nonculturable soil bacteria is not known. Determining the reason will require a systematic investigation. Although sonication of the soil sample resulted in increased viable counts, our sample of identified isolates is too small to uncover trends in the recovery of organisms from different phylogenetic groups as a function of this treatment. We speculate that the extended incubation period (12 weeks) may be significant, but we have not systematically tested if isolates from the previously uncultured groups appeared late in the incubation period. Other factors to be tested include the use of gellan gum instead of agar as the gelling agent in the solid medium and the significance of the added CaCl2. Gellan gum has been reported to be a useful gelling agent that overcomes some of the toxic effects that agar has on some groups of microorganisms (25). We added CaCl2 to the solid medium containing gellan gum because divalent cations are required to facilitate gel formation, but CaCl2 has also been reported to be a useful medium additive for the cultivation of soil bacteria (46). Our experiments did not separately address the use of gellan gum as the gelling agent and the addition of CaCl2.
Our experiments resulted in isolation of soil bacteria belonging to both known and new groups, suggesting that our methods extend the range of culturability among soil bacteria. We obtained pure-culture isolates for the first time of a number of groups of globally distributed soil bacteria, which should now allow physiological and genetic characterization of representatives of these soil bacterial groups and help elucidate their roles in the soil. We are currently carrying out more detailed phenotypic and phylogenetic characterizations of our novel isolates to begin to characterize them.
Acknowledgments
We thank Cameron Gourley and Sharon Aarons (Dairy Research Institute, Ellinbank) for help with access to the sampling site.
This work was supported by a grant from the Australian Research Council.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Amann, R., and W. Ludwig. 2000. Ribosomal RNA-targeted nucleic acid probes for studies in microbial ecology. FEMS Microbiol. Rev. 24:555-565. [DOI] [PubMed] [Google Scholar]
- 3.Beliaeff, B., and J. Y. Mary. 1993. The “most probable number” estimate and its confidence limits. Water Res. 27:799-805. [Google Scholar]
- 4.Borneman, J., and E. W. Triplett. 1997. Molecular microbial diversity in soils from eastern Amazonia: evidence for unusual microorganisms and microbial population shifts associated with deforestation. Appl. Environ. Microbiol. 63:2647-2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carreto, L., E. Moore, M. F. Nobre, R. Wait, P. W. Riley, R. J. Sharp, and M. S. da Costa. 1996. Rubrobacter xylanophilus sp. nov., a new thermophilic species isolated from a thermally polluted effluent. Int. J. Syst. Bacteriol. 46:460-465. [Google Scholar]
- 6.Chin, K.-J., D. Hahn, U. Hengstmann, W. Liesack, and P. H. Janssen. 1999. Characterization and identification of numerically abundant culturable bacteria from the anoxic bulk soil of rice paddy microcosms. Appl. Environ. Microbiol. 65:5042-5049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clegg, C. D., K. Ritz, and B. S. Griffith. 1998. Broad-scale analysis of soil microbial community DNA from upland grasslands. Antonie Leeuwenhoek 73:9-14. [DOI] [PubMed] [Google Scholar]
- 8.Coates, J. D., D. J. Ellis, C. V. Gaw, and D. R. Lovley. 1999. Geothrix fermentans gen. nov., sp. nov., a novel Fe(III)-reducing bacterium from a hydrocarbon-contaminated aquifer. Int. J. Syst. Bacteriol. 49:1615-1622. [DOI] [PubMed] [Google Scholar]
- 9.Felske, A., A. Wolterink, R. van Lis, and A. D. L. Akkermans. 1998. Phylogeny of the main bacterial 16S rRNA sequences in Drentse A grassland soils (The Netherlands). Appl. Environ. Microbiol. 64:871-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gray, N. D., and I. M. Head. 2001. Linking genetic identity and function in communities of uncultured bacteria. Environ. Microbiol. 3:481-492. [DOI] [PubMed] [Google Scholar]
- 11.Großkopf, R., P. H. Janssen, and W. Liesack. 1998. Diversity and structure of the methanogenic community in anoxic rice paddy soil microcosms as examined by cultivation and direct 16S rRNA gene sequence retrieval. Appl. Environ. Microbiol. 64:960-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hedlund, B. P., J. J. Gosink, and J. T. Staley. 1997. Verrucomicrobia div. nov., a new division of the Bacteria containing three new species of Prosthecobacter. Antonie Leeuwenhoek 72:29-38. [DOI] [PubMed] [Google Scholar]
- 13.Holmes, A. J., J. Bowyer, M. P. Holey, M. O'Donoghue, M. Montgomery, and M. R. Gillings. 2000. Diverse, yet-to-be-cultured members of the Rubrobacter subdivision of the Actinobacteria are widespread in Australian arid soils. FEMS Microbiol. Ecol. 33:111-120. [DOI] [PubMed] [Google Scholar]
- 14.Hugenholtz, P., B. M. Goebel, and N. R. Pace. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180:4765-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Isbell, R. F. 1996. Australian soil classification. CSIRO Publishing, Collingwood, Victoria, Australia.
- 16.Janssen, P. H., and K. A. O'Farrell. 1999. Succinispira mobilis gen. nov., sp. nov., a succinate-decarboxylating anaerobic bacterium. Int. J. Syst. Bacteriol. 49:1009-1013. [DOI] [PubMed] [Google Scholar]
- 17.Janssen, P. H., A. Schuhmann, E. Mörschel, and F. A. Rainey. 1997. Novel anaerobic ultramicrobacteria belonging to the Verrucomicrobiales lineage of bacterial descent isolated by dilution culture from anoxic rice paddy soil. Appl. Environ. Microbiol. 63:1382-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaiser, O., A. Pühler, and W. Selbitschka. 2001. Phylogenetic analysis of microbial diversity in the rhizoplane of oilseed rape (Brassica napus cv. Westar) employing cultivation-dependent and cultivation-independent approaches. Microb. Ecol. 42:136-149. [DOI] [PubMed] [Google Scholar]
- 19.Kishimoto, N., Y. Kosako, and T. Tano. 1991. Acidobacterium capsulatum gen. nov., sp. nov.: an acidophilic chemoorganotrophic bacterium containing menaquinone from an acidic mineral environment. Curr. Microbiol. 22:1-7. [DOI] [PubMed] [Google Scholar]
- 20.Kuske, C. R., S. M. Barns, and J. D. Busch. 1997. Diverse uncultivated bacterial groups from soils of the arid southwestern United States that are present in many geographic regions. Appl. Environ. Microbiol. 63:3614-3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Chichester, United Kingdom.
- 22.Lee, S.-Y., J. Bollinger, D. Bezdicek, and A. Ogram. 1996. Estimation of the abundance of an uncultured soil bacterial strain by a competitive quantitative PCR method. Appl. Environ. Microbiol. 62:3787-3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liesack, W., and E. Stackebrandt. 1992. Occurrence of novel groups of the domain Bacteria as revealed by analysis of genetic material isolated from an Australian terrestrial environment. J. Bacteriol. 174:5072-5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liesack, W., F. Bak, J.-U. Kreft, and E. Stackebrandt. 1994. Holophaga foetida gen. nov., sp. nov., a new, homoacetogenic bacterium degrading methoxylated aromatic compounds. Arch. Microbiol. 162:85-90. [DOI] [PubMed] [Google Scholar]
- 25.Liesack, W., P. H. Janssen, F. A. Rainey, N. L. Ward-Rainey, and E. Stackebrandt. 1997. Microbial diversity in soil: the need for a combined approach using molecular and cultivation techniques, p. 375-439. In J. D. van Elsas, J. T. Trevors, and E. M. H. Wellington (ed.), Modern soil microbiology. Marcel Dekker, New York, N.Y.
- 26.Macrae, A., D. L. Rimmer, and A. G. O'Donnell. 2000. Novel bacterial diversity recovered from the rhizosphere of oilseed rape (Brassica napus) determined by the analysis of the 16S ribosomal DNA. Antonie Leeuwenhoek 78:13-21. [DOI] [PubMed] [Google Scholar]
- 27.Marilley, L., and M. Aragno. 1999. Phylogenetic diversity of bacterial communities differing in degree of proximity of Lolium perenne and Trifolium repens roots. Appl. Soil Ecol. 13:127-136. [Google Scholar]
- 28.McCaig, A. E., L. A. Glover, and J. I. Prosser. 1999. Molecular analysis of bacterial community structure and diversity in unimproved and improved upland grass pastures. Appl. Environ. Microbiol. 65:1721-1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nogales, B., E. R. B. Moore, W. R. Abraham, and K. N. Timmis. 1999. Identification of the metabolically active members of a bacterial community in a polychlorinated biphenyl-polluted moorland soil. Environ. Microbiol. 1:199-212. [DOI] [PubMed] [Google Scholar]
- 30.Nogales, B., E. R. B. Moore, E. Llobet-Brossa, R. Rossello-Mora, R. Amann, and K. N. Timmis. 2001. Combined use of 16S ribosomal DNA and 16S rRNA to study the bacterial community of polychlorinated biphenyl-polluted soil. Appl. Environ. Microbiol. 67:1874-1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Normand, P., and C. Chapelon. 1997. Direct characterization of Frankia and of close phyletic neighbors from an Alnus viridis rhizosphere. Physiol. Plant. 99:722-731. [Google Scholar]
- 32.Northcote, K. H. 1971. A factual key for the recognition of Australian soils, 3rd ed. Rellim Technical Publications, Glenside, South Australia, Australia.
- 33.Nüsslein, K., and J. M. Tiedje. 1999. Soil bacterial community shift correlated with change from forest to pasture vegetation in a tropical soil. Appl. Environ. Microbiol. 65:3622-3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Øvreås, L., and V. Torsvik. 1998. Microbial diversity and community structure in two different agricultural soil communities. Microb. Ecol. 36:303-315. [DOI] [PubMed] [Google Scholar]
- 35.Radajewski, S., P. Ineson, N. R. Parekh, and J. C. Murrell. 2000. Stable-isotope probing as a tool in microbial ecology. Nature 403:646-649. [DOI] [PubMed] [Google Scholar]
- 36.Rheims, H., and E. Stackebrandt. 1999. Application of nested polymerase chain reaction for the detection of as yet uncultured organisms of the class Actinobacteria in environmental samples. Environ. Microbiol. 1:137-143. [DOI] [PubMed] [Google Scholar]
- 37.Rheims, H., F. A. Rainey, and E. Stackebrandt. 1996. A molecular approach to search for diversity among bacteria in the environment. J. Ind. Microbiol. 17:159-169. [Google Scholar]
- 38.Rosencrantz, D., F. A. Rainey, and P. H. Janssen. 1999. Culturable populations of Sporomusa spp. and Desulfovibrio spp. in the anoxic bulk soil of flooded rice microcosms. Appl. Environ. Microbiol. 65:3526-3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sargeant, I. J., and J. K. M. Skene. 1970. Some properties of the krasnozems of southern Victoria, Australia. Aust. J. Soil Res. 8:281-295. [Google Scholar]
- 40.Schlesner, H. 1992. The genus Verrucomicrobium, p. 3806-3808. In A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K. H. Schleifer (ed.), The prokaryotes, 2nd ed. Springer Verlag, Berlin, Germany.
- 41.Sessitsch, A., A. Weilharter, M. H. Gerzabek, H. Kirchmann, and E. Kandeler. 2001. Microbial population structures in soil particle size fractions of a long-term fertilizer field experiment. Appl. Environ. Microbiol. 67:4215-4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Singh, D. K., P. W. G. Sale, C. J. P. Gourley, and C. Hasthorpe. 1999. High phosphorus supply increases persistence and growth of white clover in grazed dairy pastures during dry summer conditions. Aust. J. Exp. Agric. 39:579-585. [Google Scholar]
- 43.Stackebrandt, E., W. Liesack, and B. M. Goebel. 1993. Bacterial diversity in a soil sample from a subtropical Australian environment as determined by 16S rDNA analysis. FASEB J. 7:232-236. [DOI] [PubMed] [Google Scholar]
- 44.Stackebrandt, E., F. A. Rainey, and N. L. Ward-Rainey. 1997. Proposal for a new hierarchic classification system, Actinobacteria classis nov. Int. J. Syst. Bacteriol. 47:479-491. [Google Scholar]
- 45.Stahl, D., and R. Amann. 1991. Development and application of nucleic acid probes, p. 205-248. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Chichester, United Kingdom.
- 46.Taylor, C. B. 1951. Nature of the factor in soil-extract responsible for bacterial growth-stimulation. Nature 168:115-116. [DOI] [PubMed] [Google Scholar]
- 47.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Torsvik, V., J. Goksøyr, and F. L. Daae. 1990. High diversity in DNA of soil bacteria. Appl. Environ. Microbiol. 56:782-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Widdel, F., and F. Bak. 1992. Gram-negative mesophilic sulfate-reducing bacteria, p. 3352-3378. In A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K. H. Schleifer (ed.), The prokaryotes, 2nd ed. Springer-Verlag, New York, N.Y.
- 50.Yoshinaka, T., K. Yano, and H. Yamaguchi. 1973. Isolation of a highly radioresistant bacterium, Arthrobacter radiotolerans nov. sp. Agric. Biol. Chem. 37:2269-2275. [Google Scholar]