Abstract
Impaired secretion of the hydrophobic CY028 cutinase invokes an unfolded protein response (UPR) in Saccharomyces cerevisiae cells. Here we show that the UPR in CY028-expressing S. cerevisiae cells is manifested as an aberrant morphology of the endoplasmic reticulum (ER) and as extensive membrane proliferation compared to the ER morphology and membrane proliferation of wild-type CY000-producing S. cerevisiae cells. In addition, we observed oxidative stress, which resulted in a 21-fold increase in carbonylated proteins in the CY028-producing S. cerevisiae cells. Moreover, CY028-producing S. cerevisiae cells use proteasomal degradation to reduce the amount of accumulated CY028 cutinase, thereby attenuating the stress invoked by CY028 cutinase expression. This proteasomal degradation occurs within minutes and is characteristic of ER-associated degradation (ERAD). Our results clearly show that impaired secretion of the heterologous, hydrophobic CY028 cutinase in S. cerevisiae cells leads to protein aggregation in the ER, aberrant ER morphology and proliferation, and oxidative stress, as well as a UPR and ERAD.
The yeast Saccharomyces cerevisiae is often used as a cell factory. When strong promoters and high gene copy numbers are used, high levels of heterologous proteins are obtained (14). To make such a process feasible for industrial applications, it is desirable that the cells secrete heterologous proteins into the culture medium, allowing easy purification. The secretion process, however, is complicated, and many factors are involved.
Secretion of heterologous proteins by S. cerevisiae has been studied by using cutinase as a model protein (21, 22). Cutinase is a lipase which originates from the fungus Fusarium solani subsp. pisi (15). Unlike other lipases, the catalytic serine of cutinase is not buried under surface loops but is accessible to solvents. Therefore, cutinase does not need interfacial activation (18), which makes it an important protein for industrial applications.
Sagt et al. found that CY000 cutinase was produced and secreted at a concentration of 30 mg/g of cells and had a specific activity of 325 specific lipase units (SLU)/mg of protein (21). To increase the specific activity of the cutinase, hydrophobic patches were introduced around the active site (G82A, A85F, V184I, A185L, L189F), which resulted in CY028 cutinase. The specific activity of CY028 cutinase was 1,093 SLU/mg of protein; however, this hydrophobic cutinase was produced at a concentration of only 10 mg/g of cells and was secreted at a concentration of 3 mg/g of cells. The difference in production between CY000 and CY028 in S. cerevisiae was not due to a difference in copy number or mRNA abundance. It has been shown that hydrophobic cutinase CY028 is retained in the endoplasmic reticulum (ER) in association with the upregulated molecular chaperone BiP (21), which results in impaired secretion. However, secretion of the hydrophobic cutinase could be increased by introduction of an N-glycosylation site (22).
Recently, Travers et al. (29) discovered that there is coordination between the unfolded protein response (UPR) and ER-associated degradation (ERAD) by performing functional and genomic analyses. In S. cerevisiae the UPR upregulates the genes involved in the secretory pathway, including the genes for protein folding, protein degradation, lipid and inositol metabolism, and other functions (29). During ERAD misfolded proteins are transported out of the ER by retrograde transport. For this transportation process ERAD requires an intact UPR pathway in S. cerevisiae (3). Once in the cytosol, these proteins are ubiquitinated and rapidly degraded by the proteasome (13).
To establish whether induction of CY028 cutinase production in S. cerevisiae also leads to a UPR and ERAD, we studied several parameters present in the pathways. We found by using immunogold labeling and transmission electron microscopy that CY028 cutinase is confined in the ER, resulting in ER membrane proliferation compared with the effect of wild-type CY000 in S. cerevisiae. Furthermore, the proliferation of ER membranes apparently results in oxidative stress, which results in carbonylation of proteins. Finally, CY028 cutinase is rapidly degraded by ERAD. S. cerevisiae cells degrade about 70% of the hydrophobic cutinase in a proteasome-dependent manner to downregulate the stress caused by CY028 cutinase expression. Based on our results we concluded that heterologous, hydrophobic protein production in S. cerevisiae cells results in protein aggregation, a change in ER membrane morphology, oxidative stress, a UPR, and ERAD.
MATERIALS AND METHODS
Media and growth conditions.
Defined Egli medium was used in a glucose-limited chemostat. The yeast was grown at 30°C in a Bioflow III fermentor (New Brunswick Scientific) essentially as described previously (25). After the batch phase, a continuous feed with 20 g of glucose per liter and 4 g of galactose per liter at a dilution rate of 0.07 h−1 was connected. After a steady state was reached, samples were taken and frozen quickly in liquid nitrogen.
Strains and genetic constructs.
Two different cutinase variants were expressed in S. cerevisiae VW cen.pk111-32D (leu2). The cutinase genes were placed under control of the GAL7 promoter and directed into the secretion route by the invertase signal sequence. To ensure genetic stability, the constructs were integrated at the chromosomal ribosomal DNA locus; each construct contained a leu2 gene, which enabled selection on agar plates lacking leucine (32). The Δubc7 and Δise1 mutants were obtained from Peter Kötter (Institut für Mikrobiologie, Johann Wolfgang Goethe-Universität, Frankfurt am Main, Germany) and were constructed in the isogenic VW strain.
Electron microscopy.
S. cerevisiae cells were taken up by capillary action in single dry cellulose capillary tubes about 10 mm long (11). The capillary tubes were submerged in 1-hexadecene, cut into pieces that were about 2 mm long, and subsequently sandwiched in aluminum specimen holders. After high-pressure freezing (Leica) each sandwich was put into liquid nitrogen (LN2). The two specimen holders were separated under LN2. The capillary tubes attached to one of the aluminum specimen holders were freed from adhering 1-hexadecene under LN2 with a fine needle. After this, the capillary tubes still attached to the specimen holder were transferred to a screw-cap vial, which contained frozen substitution medium consisting of 0.3% uranyl acetate and 0.01% glutaraldehyde in anhydrous methanol (CY028-producing cells were also put in a complex substitution medium consisting of 0.3% uranyl acetate, 0.01% glutaraldehyde, 0.3% osmium tetroxide, and 0.1% 3-amino-1,2,4-triazole in methanol) and a miniature transfer basket (11). The vial was placed in a CS-auto substitution apparatus (26) at −90°C for 36 h. Subsequently, the temperature was raised to −35°C at a rate of 5°C/h. The freeze-substitution medium was replaced with methanol. The specimen holders were removed from the vial. The capillary tubes were infiltrated with increasing concentrations of Lowicryl HM20, as follows: 25% for 2 h, 50% for 2 h, 75% for 16 h, and 100% for at least 24 h. After this, the capillary tubes were embedded in 100% Lowicryl HM20. Polymerization was done at −35°C for 24 h with a UV source attachment (26), followed by 1 day of curing under UV light at room temperature and sectioning with an ultramicrotome. Sections of both wild-type and mutant cells of S. cerevisiae were immunogold labeled to locate the cutinase or BiP or both, as described by Sagt et al. (21).
Determination of amounts of carbonylated proteins.
Amounts of carbonylated proteins were determined by a colorimetric procedure that measures binding of dinitrophenylhydrazine (DNPH), as described previously (17). The amount of protein-bound DNPH was also measured by using an anti-DNPH antibody (A-6435, lot 6241-1; diluted 1:4,000 in 0.4% Protifar in phosphate-buffered saline for 1 h; Molecular Probes Europe B.V.) with Western blotting as described previously (24).
Proteasomal inhibition.
Cells expressing CY028 were incubated at 30°C for 30 min on YNB medium containing 2% galactose with or without 25 μM clasto-lactacystin-β-lactone (CW8405-Z02185; Affinity Research Products Ltd.). Pulse-chase experiments were performed essentially as described by Sagt et al. (21). However, the pulse period was reduced to 1 min, and in order to obtain sufficient incorporation of radioactively labeled amino acids, the amount of 35S-labeled Met/Cys was increased to 150 μCi. The chase times were 0, 10, 30, 60, and 300 s. Excess label was washed away, and the cells were chased by taking samples at different times. The samples were subjected to immunoprecipitation with anti-cutinase and loaded onto sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels, and autoradiography was performed.
RESULTS
Membrane proliferation in CY028 cutinase-expressing S. cerevisiae cells.
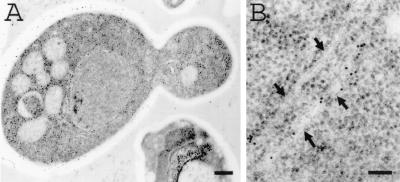
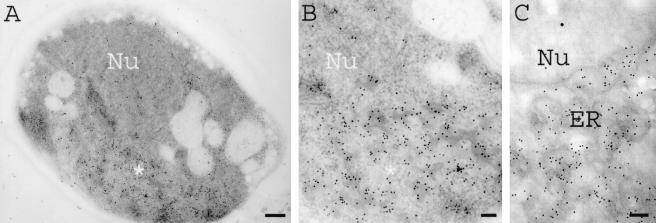
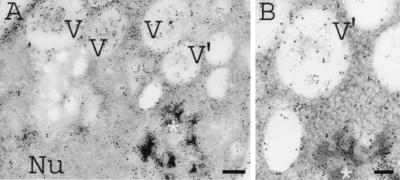
Recently, it was demonstrated that induction of synthesis of the hydrophobic CY028 cutinase resulted in aggregation of cutinase in the ER and increased expression of BiP (21), suggesting that there was a UPR. According to Travers et al. (29), a UPR induces upregulation of genes in the secretory pathway, including genes involved in lipid metabolism. This may result in increased lipid biosynthesis, manifested as proliferation of endomembranes. As shown in Fig. 1A, the ER of CY000-producing cells had a normal plate-like morphology, while no aggregation of CY000 cutinase was observed (Fig. 1B). However, in the CY028 cutinase-producing cells CY028 cutinase aggregated, as shown in Fig. 2A and B. To identify morphologically the confinement of aggregates in the ER, S. cerevisiae cells were grown in a complex substitution medium containing osmium tetroxide and an imidazole compound to improve endomembrane contrast. Figure 2C shows that the contrast of the ER membrane confining the CY028 cutinase was improved compared to the contrast of the aggregates shown in Fig. 2B. Proliferation of the ER membrane in CY028 cutinase-producing cells (Fig. 2B and C) coincided with an aberrant morphology compared to the ER morphology of CY000-producing cells (Fig. 1). In addition to ER membrane proliferation we observed increased vacuolization in some CY028 cutinase-producing cells, as shown in Fig. 3. We also observed in these cells CY028 cutinase, as well as BiP in the vacuoles, after immunogold labeling and electron microscopy. These results are consistent with the results of Travers et al. (29), who found that the UPR induces upregulation of genes in the vacuolar protein sorting system, as well as lipid metabolism.
FIG. 1.
CY000-producing S. cerevisiae cells under inducing conditions. Cells were cryofixed, freeze-substituted with 0.3% uranyl acetate-0.01% glutaraldehyde in methanol, and low-temperature embedded in Lowicryl HM20. After immunogold labeling with anti-cutinase (1:250) and goat anti-rabbit with conjugated 10-nm gold (1:10), CY000 cutinase was confined in the plate-like ER. (A) Low-power magnification of a CY000-producing cell. Bar = 500 nm. (B) High-power magnification of plate-like ER (arrows) with gold particles indicating the location of CY000 cutinase. Bar = 100 nm.
FIG. 2.
CY028-producing S. cerevisiae cells under inducing conditions. Cells were cryofixed, freeze-substituted with 0.3% uranyl acetate-0.01% glutaraldehyde in methanol (A and B) and with 0.3% uranyl acetate-0.01% glutaraldehyde-0.3% osmium tetroxide-0.1% aminotriazole in methanol (C), and subsequently low-temperature embedded in Lowicryl HM20. After immunogold labeling with anti-cutinase (1:250) and goat anti-rabbit with conjugated 10-nm gold (1:10), CY028 cutinase was confined in aggregates of the ER. (A) Low-power magnification of a CY028-producing cell, showing the labeled area of an aggregate (asterisk) that had a total area that was about half the area of the nucleus (Nu). Bar = 250 nm. (B) High-power magnification of the labeled aggregate (asterisk) with 10-nm gold particles indicating the location of CY028 cutinase. Bar = 100 nm. (C) High-power magnification of triazole-stained membranes of the ER and 10-nm gold particles indicating the location of CY028 cutinase in the aggregates. Bar = 100 nm.
FIG. 3.
CY028-producing S. cerevisiae cells under inducing conditions. Cells were cryofixed, freeze-substituted with 0.3% uranyl acetate-0.01% glutaraldehyde in methanol, and subsequently low-temperature embedded in Lowicryl HM20. After double immunogold labeling with anti-cutinase (1:250) and goat anti-rabbit with conjugated 6-nm gold (1:10) and with anti-BiP (1:200) and goat anti-rabbit with conjugated 10-nm gold (1:10), CY028 cutinase and BiP were located in aggregates, which were confined to the ER and the vacuoles. (A) Low-power magnification of a CY028-producing cell, showing the labeled area of aggregates (asterisk) and vacuoles (V and V′) containing cutinase and BiP. Bar = 250 nm. Nu, nucleus. (B) High-power magnification of a labeled aggregate (asterisk) and vacuole (V′) with 6-nm gold particles indicating the location of CY028 cutinase and with 10-nm gold particles indicating the location of BiP. Bar = 100 nm.
Oxidative stress in CY028 cutinase-expressing cells.
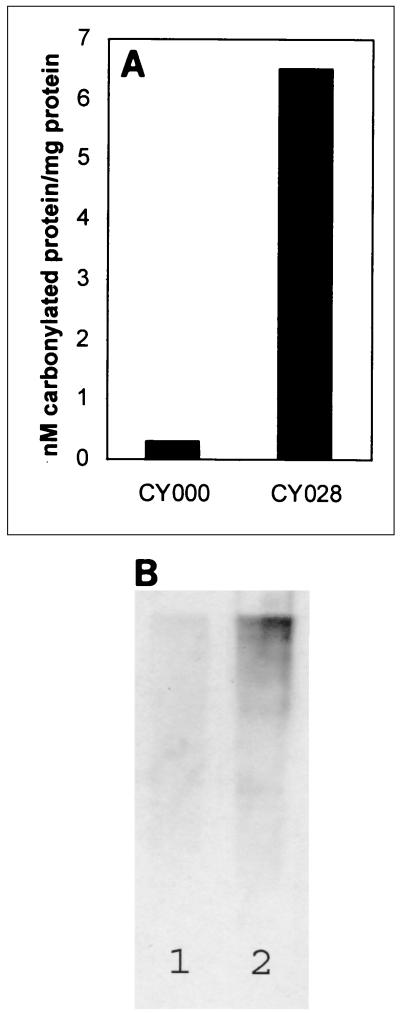
Travers et al. (29) observed upregulation of genes in phospholipid metabolism during a UPR. Our electron microscopy results revealed ER membrane proliferation, as well as aberrant ER morphology compared to the ER morphology of CY000-producing cells. Increased endomembrane synthesis may lead to reactive oxygen species (ROS), because biogenesis of membrane components involves several oxidative reactions. These free radicals take part in the damaging reactions (12). Once ROS production overrides the cellular antioxidant defense system, an oxidative stress situation arises. This may lead to damage of biomolecules (for example, proteins), which may result in the carbonylation of proteins. The amount of carbonylated proteins was determined colorimetrically and by Western blot analysis. Figure 4A shows that the amount of carbonylated proteins in yeast cells expressing CY028 cutinase (6.5 nmol of carbonylated proteins/mg of protein) was about 21-fold greater than the amount in the wild-type cells expressing CY000 cutinase (0.3 nmol of carbonylated protein/mg of protein). Figure 4B is the corresponding Western blot and also shows the increase in the amount of carbonylated proteins in the CY028 cutinase-producing cells compared to the amount in the CY000 cutinase-producing cells. These results indicate that oxidative stress occurs during impaired secretion of CY028 cutinase.
FIG. 4.
Cells producing CY028 cutinase have increased levels of carbonylated proteins. (A) Amount of carbonylated proteins, as determined by binding of DNPH and measured colorimetrically by using the method of Levine et al. (17). S. cerevisiae CY028-producing cells contained about 21-fold more carbonylated protein than wild-type S. cerevisiae CY000-producing cells. (B) Corresponding immunoblot, in which bound DNPH was identified by using anti-DNPB antibodies as described by Shacter et al. (24). S. cerevisiae CY028-producing cells (lane 2) contained more carbonylated protein than wild-type S. cerevisiae CY000-producing cells (lane 1).
Proteasomal degradation of CY028 cutinase.
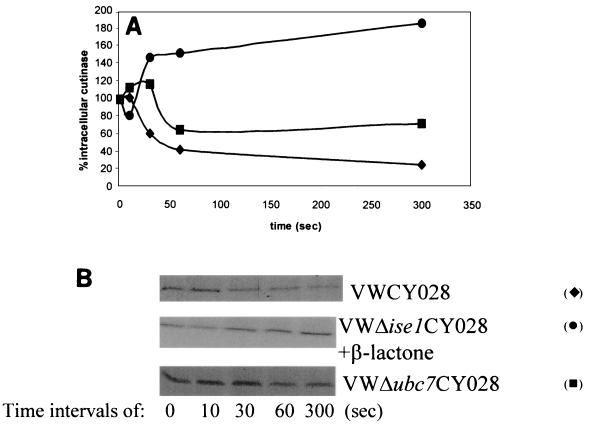
It was shown previously that the total amount of CY028 cutinase is about threefold less than the total amount of CY000 cutinase (21). Since the mRNA levels were comparable, the difference could be caused by rapid degradation of CY028 cutinase in a proteasome-dependent manner. As indicated above, essential coordination between the UPR and ERAD has been suggested (29). Therefore, we performed pulse-chase experiments with extremely short pulse periods. In these pulse-chase experiments a pulse of 1 min and even shorter chase times were used to detect possible rapid degradation of CY028 cutinase. As shown in Fig. 5, 50% of CY028 cutinase was degraded within 60 s and 70% was degraded in 5 min. To examine the involvement of proteasomes in this rapid degradation of CY028 cutinase, the effect of the proteasome inhibitor β-lactacystin was determined. Figure 5 shows that after inhibition of the proteasomes degradation of CY028 cutinase was completely blocked. This result supports the conclusion that the degradation of CY028 is proteasome dependent, which is typical for ERAD. Proteins that are preferred substrates for proteasomal degradation are conjugated with ubiquitin. To determine the involvement of ubiquitin in degradation of CY028 cutinase, the UBC7 gene, which encodes a ubiquitin-conjugating enzyme, was deleted, yielding strain VW ΔUBC7. Pulse-chase experiments in this ΔUBC7 background demonstrated that degradation of CY028 was delayed only slightly (Fig. 5) and that 30% of the intracellular CY028 cutinase was degraded within 5 min. This result indicates that the UBC7 gene plays a minor role in the rapid degradation of CY028 cutinase.
FIG. 5.
Degradation of CY028 cutinase. S. cerevisiae cells expressing CY028 were incubated at 30°C for 30 min on YNB medium containing 2% galactose with or without the proteasomal inhibitor clasto-lactacystin-β-lactone at a concentration of 25 μM. The cells were pulse-labeled for 1 min with 150 μCi of [35S]Met/Cys. After washing, the cells were chased by taking samples at different times. The samples were then subjected to immunoprecipitation with anti-cutinase and loaded on SDS-PAGE gels, and autoradiography was performed. (A) Scanning values obtained with a personal densitometer from Molecular Dynamics. At 5 min no degradation was found after treatment with the proteasomal inhibitor (•), in contrast to the untreated S. cerevisiae CY028-expressing cells (⧫). After deletion of the UBC7 gene, degradation was delayed (▪. All values were normalized by using the cutinase level at zero time. (B) Corresponding autoradiographs after immunoprecipitation on the SDS-PAGE gels for different times (0, 10, 30, 60, and 300 s).
DISCUSSION
Travers et al. revealed the essential coordination of the UPR and ERAD by performing functional and genomic analyses (29). Recently, Sagt et al. demonstrated that induction of synthesis of the hydrophobic CY028 cutinase in S. cerevisiae cells resulted in aggregation of cutinase in the ER and increased expression of BiP (21), suggesting that there is a UPR. To establish whether induction of CY028 cutinase production in S. cerevisiae cells indeed leads to a UPR and ERAD, we studied several parameters present in these pathways, including endomembrane proliferation, oxidative stress, and proteasomal degradation.
Here we clearly show that in CY028 cutinase-producing S. cerevisiae cells there is significant irregular ER membrane proliferation, as revealed by electron microscopic studies. This is consistent with the results of a microarray study performed by Travers et al. (29), with the work of Cox et al. (5), who found that a UPR coordinates the production of ER proteins and ER membrane, and with the studies of Umebayashi et al. (30) with yeast, in which accumulation of misfolded protein aggregates affected the ER morphology drastically. In addition, Koudinova et al. (16) showed in a study of Alzheimer's disease, which is accompanied by aggregation of certain proteins, that Alzheimer's Abeta1-40 peptide increased the synthesis of different classes of lipids in brain tissue. Furthermore, Sagt et al. showed that BiP is upregulated in CY028-expressing cells, indicating that there is a UPR (21); therefore, our electron microscopic results confirmed the link between membrane proliferation and the occurrence of a UPR in S. cerevisiae.
Membrane proliferation, however, may result in an increase in ROS taking part in damaging reactions (12). The presence of ROS results in oxidative stress. In our study we indeed observed an increase in the amount of the carbonylated proteins, indicating that oxidative stress occurs when CY028 cutinase accumulates in the ER. This indicates that S. cerevisiae is not able to neutralize ROS completely. Since oxidative stress-related genes do not play a direct role in the UPR (29), this stress is probably a secondary effect. However, the presence of carbonylated proteins in CY028 cutinase-producing cells could induce proteasomal activity (8).
The link between the UPR and ERAD in S. cerevisiae has been described by Travers et al. (29). These workers used a genomics approach with S. cerevisiae mutants deficient in UPR signaling. Here we confirmed this link in an industrially important biological system in which the UPR and ERAD have been identified as the main pathways by which the yeast cell attempts to decrease the accumulation of CY028 cutinase in the ER. Since the amount of CY028 cutinase produced by S. cerevisiae was threefold smaller than the amount of CY000 cutinase produced by S. cerevisiae, we performed pulse-chase experiments. Previously, Sagt et al. performed similar experiments with pulse periods of 10 min (21); however, in order to be able to study processes on a short time scale, we used 1-min pulse periods. These experiments revealed that there was very rapid degradation of CY028 cutinase. Rapid proteasomal degradation of misfolded proteins has also been found in Arabidopsis seedlings (8-min half time) (20) and in human astrocytoma cells transfected with a human cytomegalovirus gene (US11), which resulted in histocompatibility complex class 1 molecules with a half time of less than 1 min (33). Such rapid degradation of secretory proteins is characteristic of ERAD.
In proteasomal degradation (23) the aberrant proteins confined in the lumen of the ER are translocated across the ER membrane (2) and ubiquitinated mainly by the ubiquitin-conjugating enzyme Ubc7p (6). Although the degradation of CY028 cutinase was proteasome dependent, the role of ubiquitination could not be clearly established in our study. Although the UBC7 gene, which encodes the ubiquitin-conjugating enzyme Ubc7p, was deleted, yielding strain VW ΔUBC7, pulse-chase experiments in the ΔUBC7 background demonstrated that the degradation was delayed and that only 30% of the intracellular CY028 cutinase was degraded in 5 min, while in the CY028-producing strain 70% was degraded. This indicates that during the delay in degradation of CY028 cutinase in strain VW ΔUBC7, other conjugating enzymes, such as UBC1p, UBC4p, UBC5p, and UBC6p, are involved in the ubiquitinaton of CY028 cutinase (4, 7, 9, 10, 27, 28). Most likely, the stress-related ubiquitin conjugation enzymes UBC4p and UBC5p take part in this ubiquitination process (1). Alternatively, an ill-defined proteolytic system (31), which is involved in degradation of misfolded proteins, might play a role in the degradation of CY028 cutinase in ΔUBC7 cells. Another possibility is that the vacuolar degradation system functions in concert with, or is an alternative for, ubiquitin-dependent degradation. However, as shown in our study, vacuolar degradation of CY028 cutinase is a slow process and not as specific as ERAD for several reasons, such as the fact that vacuoles nonspecifically engulf the cytoplasmic contents (10). In addition, Ng et al. (19) have shown that the UPR also has connections to other regulatory pathways, such as the transcriptional regulator for genes encoding subunits of the proteasome (SON1/RPN4). In view of the results of Ng et al. (19) and the results of Travers et al. (29) concerning the role of UPR upregulated genes in ubiquitin-proteasome protein degradation, the results of our proteasomal inhibition studies warrant the conclusion that the rapid degradation of hydrophobic cutinase CY028 in S. cerevisiae cells is not vacuolar degradation but rather is ubiquitin-proteasome protein degradation.
Impaired secretion of CY028 cutinase invokes a complex stress response in S. cerevisiae cells. After translocation and folding of CY028 cutinase, BiP is still able to bind to the exposed hydrophobic patches (21; this study). The binding of BiP to hydrophobic cutinase CY028 results in keeping the CY028 cutinase in the ER, which results in a high concentration of CY028 cutinase. Therefore, CY028 cutinase forms aggregates with BiP (21; this study) in the ER lumen. Moreover, the yeast cell uses retrograde translocation through Sec61p and proteasomes to degrade CY028 cutinase, which is kept in a retrograde transport-competent state by BiP (2). These events in the ER, which determine the secretion efficiency of CY028 cutinase, lead to a variety of subcellular responses to circumvent accumulation of pathological amounts of protein in the ER lumen. Based on the results of this study, we concluded that induction of the hydrophobic CY028 cutinase in S. cerevisiae cells leads to CY028 cutinase aggregation in the lumen of the ER, membrane proliferation and aberrant morphology in the ER, oxidative stress as a secondary effect, a UPR, and ERAD.
Acknowledgments
We thank Chris Visser (Unilever Research Laboratory Vlaardingen, Vlaardingen, The Netherlands) for constructing the mutant cutinase CY028; Han Peeters (Unilever Research Laboratory Vlaardingen) for molecular modeling of cutinase; M. C. D. van der Burg-Koorevaar (Unilever Research Laboratory Vlaardingen) for purification of cutinase; Peter Kötter (Institut für Mikrobiologie, Johann Wolfgang Goethe-Universität, Frankfurt am Main, Germany) for providing the ΔUBC7 and ΔISE1 mutants; and Yvonne Thomassen, René Verwaal, and Jan Andries Post (Department of Molecular Cell Biology and Institute of Biomembranes, Utrecht University, Utrecht, The Netherlands) for discussions.
REFERENCES
- 1.Arnason, T., and M. J. Ellison. 1994. Stress resistance in Saccharomyces cerevisiae is strongly correlated with assembly of a novel type of multiubiquitin chain. Mol. Cell. Biol. 14:7876-7883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brodsky, J. L., E. D. Werner, M. E. Dubas, J. L. Goeckeler, K. B. Kruse, and A. A. McCracken. 1999. The requirement for molecular chaperones during endoplasmic reticulum-associated protein degradation demonstrates that protein export and import are mechanistically distinct. J. Biol. Chem. 274:3453-3460. [DOI] [PubMed] [Google Scholar]
- 3.Casagrande, R., P. Stern, M. Diehn, C. Shamu, M. Osaria, M. Zuniga, P. O. Brown, and H. Ploegh. 2000. Degradation of proteins from the ER of S. cerevisiae requires an intact unfolded protein response pathway. Mol. Cell 5:729-735. [DOI] [PubMed] [Google Scholar]
- 4.Chen, P., P. Johnson, T. Sommer, S. Jentsch, and M. Hochstrasser. 1993. Multiple ubiquitin-conjugating enzymes participate in the in vivo degradation of the yeast MAT alpha 2 repressor. Cell 74:357-369. [DOI] [PubMed] [Google Scholar]
- 5.Cox, J. S., R. E. Chapman, and P. Walter. 1997. The unfolded protein response coordinates the production of endoplasmic reticulum protein and endoplasmic reticulum membrane. Mol. Biol. Cell 8:1805-1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deak, P. M., and D. H. Wolf. 2001. Membrane topology and function for Der3/Hrd1p as a ubiquitin-protein ligase (E3) involved in endoplasmic reticulum degradation. J. Biol. Chem. 276:10663-10669. [DOI] [PubMed] [Google Scholar]
- 7.Frielander, R., E. Jarosch, J. Urban, C. Volkwein, and T. Sommer. 2000. A regulatory link between ER-associated protein degradation and the unfolded-protein response. Nat. Cell Biol. 2:379-384. [DOI] [PubMed] [Google Scholar]
- 8.Grune, T., T. Reinheckel, M. Joshi, and K. J. A. Davies. 1995. Proteolysis in cultured liver epithelial cells during oxidative stress. J. Biol. Chem. 270:2344-2351. [DOI] [PubMed] [Google Scholar]
- 9.Hiller, M. M., A. Finger, M. Schweiger, and D. H. Wolf. 1996. ER degradation of a misfolded luminal protein by the cytosolic ubiquitin-proteasome pathway. Science 273:1725-1728. [DOI] [PubMed] [Google Scholar]
- 10.Hochstrasser, M. 1996. Ubiquitin-dependent protein degradation. Annu. Rev. Genet. 30:405-439. [DOI] [PubMed] [Google Scholar]
- 11.Hohenberg, H., K. Mannweiter, and M. Müller. 1994. High-pressure freezing of cell suspensions in celllulose capillary tubes. J. Microsc. 175:34-43. [DOI] [PubMed] [Google Scholar]
- 12.Hooper, D. C., S. Spitsin, R. B. Kean, J. M. Champion, G. M. Dickson, I. Chaudhry, and H. Koprowski. 1998. Uric acid, a natural scavenger of peroxynitrite, in experimental allergic encephalomyelitis and multiple sclerosis. Proc. Natl. Acad. Sci. USA 95:675-680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson, E. S., P. C. M. Ma, I. M. Ota, and A. Varshavsky. 1995. A proteolytic pathway that recognizes ubiquitin as a degradation signal. J. Biol. Chem. 270:17442-17456. [DOI] [PubMed] [Google Scholar]
- 14.Kingsman, S. M., A. J. Kingsman, M. J. Dobson, J. Mellor, and N. A. Roberts. 1985. Heterologous gene expression in Saccharomyces cerevisiae. Biotechnol. Genet. Eng. Rev. 3:337-416. [DOI] [PubMed] [Google Scholar]
- 15.Kolattukudy, P. E. 1984. Cutinases from fungi and pollen, p. 471-504. In B. Borgstrom and H. Brockman (ed.), Lipases. Elsevier Science, Amsterdam, The Netherlands.
- 16.Koudinova, N. V., A. R. Koudinov, and E. Yavin. 2000. Alzheimer's Abeta1-40 peptide modulates lipid synthesis in neuronal cultures and intact rat fetal brain under normoxic and oxidative stress conditions. Neurochem. Res. 25:653-660. [DOI] [PubMed] [Google Scholar]
- 17.Levine, R. L., D. Garland, C. N. Oliver, A. Amici, I. Climents, A. Lenz, B. Ahn, S. Shalteil, and E. R. Stadtman. 1990. Determination of carbonyl content in oxidatively modified proteins. Methods Enzymol. 186:464-478. [DOI] [PubMed] [Google Scholar]
- 18.Martinez, C., P. de Geus, M. Lauwereys, G. Matthysens, and C. Cambillau. 1992. Fusarium solani cutinase is a lipolytic enzyme with a catalytic serine accessible to solvent. Nature 356:615-618. [DOI] [PubMed] [Google Scholar]
- 19.Ng, D. T. W., E. D. Spear, and P. Walter. 2000. The unfolded protein response regulates multiple aspects of secretory and membrane protein biogenesis and endoplasmic reticulum quality control.J. Cell Biol. 150:77-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramos, J. A., N. Zenser, O. Leyser, and J. Callis. 2001. Rapid degradation of auxin/indoleacetic acid proteins requires conserved amino acids of domain II and is proteasome dependent. Plant Cell 13:2349-2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sagt, C. M. J., W. H. Müller, J. Boonstra, A. J. Verkleij, and C. T. Verrips. 1998. Impaired secretion of a hydrophobic cutinase by Saccharomyces cerevisiae correlates with an increased association with immunoglobulin heavy-chain binding protein (BiP). Appl. Environ. Microbiol. 64:316-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sagt, C. M. J., B. Kleizen, R. Verwaal, M. D. M. De Jong, W. H. Müller, A. Smits, C. Visser, J. Boonstra, A. J. Verkleij, and C. T. Verrips. 2000. Introduction of an N-glycosylation site increases secretion of heterologous proteins in yeasts. Appl. Environ. Microbiol. 66:4940-4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seufert, W., and S. Jentsch. 1990. Ubiquitin-conjugating enzymes UBC4 and UBC5 mediate selective degradation of short-lived and abnormal proteins. EMBO J. 9:543-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shacter, E., J. A. Williams, M. Lim, and R. L. Levine. 1994. Oxidative modification of fibrinogen inhibits thrombin-catalyzed clot formation. Free Radic. Biol. Med. 18:815-821. [DOI] [PubMed] [Google Scholar]
- 25.Sillje, H. H., E. G. ter Schure, A. J. Verkleij, J. Boonstra, and C. T. Verrips. 1996. The Cdc25 protein of Saccharomyces cerevisiae is required for normal glucose transport. Microbiology 142:1765-1773. [DOI] [PubMed] [Google Scholar]
- 26.Sitte, H., K. Neuman, and L. Edelman. 1985. Cryo-fixation and cryo-substitution for routine work in transmission electron microscopy, p. 103-118. In M. Müller, R. P. Becker. A. Boyde, and J. J. Wolosewick (ed.), Science of biological specimen preparation. SEM Inc., AMF O'Hare, Chicago, Ill.
- 27.Sommer, T., and W. Seufert. 1992. Genetic analysis of ubiquitin-dependent protein degradation. Experientia 48:172-178. [DOI] [PubMed] [Google Scholar]
- 28.Sommer, T., and S. Jentsch. 1993. A protein translocation defect linked to ubiquitin conjugation at the endoplasmic reticulum. Nature 365:176-179. [DOI] [PubMed] [Google Scholar]
- 29.Travers, K. J., C. K. Patil, L. Wodicka, D. J. Lockhart, J. S. Weissman, and P. Walter. 2000. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 101:249-258. [DOI] [PubMed] [Google Scholar]
- 30.Umebayashi, K., A. Hirata, R. Fuduka, H. Horiuchi, A. Ohta, and M. Takagi. 1997. Accumulation of misfolded protein aggregates leads to the formation of Russell body-like dilated endoplasmic reticulum in yeast. Yeast 13:1009-1020. [DOI] [PubMed] [Google Scholar]
- 31.Umebayashi, K., R. Fuduka, A. Hirata, H. Horiuchi, A. Nakano, A. Ohta, and M. Takagi. 2001. Activation of the Ras-cAMP signal pathway inhibits the proteasome-independent degradation of misfolded protein aggregates in the endoplasmic reticulum lumen. J. Biol. Chem. 276:41444-41454. [DOI] [PubMed] [Google Scholar]
- 32.van Gemeren, I. A., W. Musters, C. A. M. J. J. van den Hondel, and C. T. Verrips. 1995. Construction and heterologous expression of a synthetic copy of the cutinase cDNA from Fusarium solani pisi. J. Biotechnol. 40:155-162. [DOI] [PubMed] [Google Scholar]
- 33.Wiertz, E. J. H. J., T. R. Jones, L. Sun, M. Bogyo, H. J. Geuze, and H. L. Ploegh. 1996. The human cytomegalovirus US11 gene product dislocates MHC class I heavy chains from the endoplasmic reticulum to the cytosol. Cell 84:769-779. [DOI] [PubMed] [Google Scholar]