Abstract
Ribosomal DNA sequence analysis, originally conceived as a way to provide a universal phylogeny for life forms, has proven useful in many areas of biological research. Some of the most promising applications of this approach are presently limited by the rate at which sequences can be analyzed. As a step toward overcoming this limitation, we have investigated the use of photolithography chip technology to perform sequence analyses on amplified small-subunit rRNA genes. The GeneChip (Affymetrix Corporation) contained 31,179 20-mer oligonucleotides that were complementary to a subalignment of sequences in the Ribosomal Database Project (RDP) (B. L. Maidak et al., Nucleic Acids Res. 29:173-174, 2001). The chip and standard Affymetrix software were able to correctly match small-subunit ribosomal DNA amplicons with the corresponding sequences in the RDP database for 15 of 17 bacterial species grown in pure culture. When bacteria collected from an air sample were tested, the method compared favorably with cloning and sequencing amplicons in determining the presence of phylogenetic groups. However, the method could not resolve the individual sequences comprising a complex mixed sample. Given these results and the potential for future enhancement of this technology, it may become widely useful.
Originally developed to better understand the evolution of life on our planet (11, 29), small-subunit (SSU) rRNA and ribosomal DNA (rDNA) sequence analysis has had many applications. The approach continues to be used to explore the biological diversity of our planet (1, 2, 20) and the biological composition of various ecosystems (23, 27). It has had a profound effect on microbial taxonomy (6, 28), has enabled the identification of uncultured pathogens (22, 25), and has been used in the setting of clinical microbiology (9). The Ribosomal Database Project (RDP) (16) has catalogued around 23,000 of these sequences, classifying them at various hierarchical phylogenetic levels (Fig. 1). The detailed classification can be downloaded from the website http://rdp.cme.msu.edu/download/SSU_rRNA/.
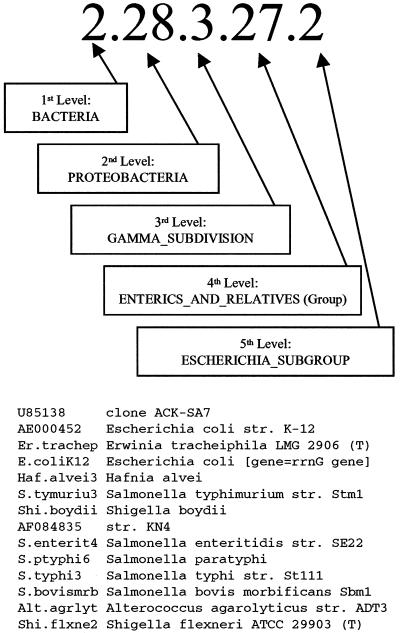
FIG. 1.
Example of phylogenetic placement of 14 SSU rDNA sequences included in RDP's Escherichia Subgroup. These sequences are contained in phylogenetic clusters at five hierarchical levels, each designated by a name and number as shown. Not all of these sequences are unique in the region represented on the chip.
There are several platforms currently used to analyze rRNA and rDNA sequences. By growing a pure culture of an organism or cloning mixed rDNAs, it is possible to obtain a pure template for dideoxy sequencing; sequences obtained in this way can be compared with sequences in large databases. Alternatively, oligonucleotide probes directed at phylogenetically specific sequences can be used to identify the SSU rDNA of phylogenetic clusters of organisms. Both of these approaches are for the most part serial, i.e., they can be used to identify only one organism at a time by detecting one sequence at a time, though it is possible to differentially label oligonucleotide probes for in situ hybridization (4). In addition, the approach of cloning amplicons and then sequencing them is laborious and time-consuming.
It has recently become possible to make arrays of oligonucleotide probes in various formats (10, 19). The photolithography chip is a particularly powerful approach, allowing simultaneous testing of over 250,000 oligonucleotide probes. By confirming the presence of relatively long DNA sequences, these chips are used for various applications, including rapid determination of mRNA expression profiles (15, 18) and detection of the rpoB gene sequence for simultaneous identification of the species and rifampin sensitivity of mycobacteria (12).
We report here the use of a photolithography chip that displays an array consisting of oligonucleotides complementary to rDNA sequences found in a subalignment of the RDP. SSU rDNA sequences are highly conserved, the overall degree of similarity between two sequences correlating with their distance from one another in the RDP hierarchical phylogenetic scheme. Because many organisms with sequences not included in RDP share sequence with organisms that were included, it was possible to glean information about unknown environmental organisms whose SSU rDNA sequences were neither included in RDP version 5.0 nor deliberately placed on the chip.
MATERIALS AND METHODS
Photolithography chip design.
RDP version 5.0 contained 3,286 aligned SSU rRNA sequences (17). The subalignment used for this work was defined by Escherichia coli positions 1409 to 1491. There were adequate data in this segment to include 1,945 prokaryotic sequences and 431 eukaryotic sequences on the chip. The region used is bounded on both ends by universally conserved segments that can be used as PCR priming sites to amplify any SSU rDNA (14, 26). Thus, primers amplified both prokaryotic and eukaryotic rDNA. Approximately half of the possible overlapping 20-mers were used, allowing extensive redundancy. Where the database contained an ambiguous base, the oligonucleotides with sites complementary to the ambiguously sequenced nucleotide were synthesized with the base omitted. Each array cell containing an rDNA probe (probe cell) was paired with a control cell in which the oligonucleotide was identical except that the 11th nucleotide consisted of a base that would mismatch the targeted rDNA sequence. Thus, the oligonucleotide in the control cell served as a mismatched control for nonspecific hybridization. The entire array consisted of 62,358 oligonucleotides synthesized on the chip, each with a copy number of around 107. The number of probes for individual sequences contained in RDP version 5.0 ranged from 0 to 70, depending on the completeness of sequence information between E. coli positions 1409 and 1491. All the synthesized probes that were designed to match one sequence were designated the probe set for that sequence.
Bacterial strains.
Most bacterial strains tested were type strains obtained from the American Type culture Collection (ATCC). These included Bacteroides vulgatus (ATCC 8482), Brucella abortus (ATCC 23448), Clostridium difficile (ATCC 9684), Acidobacterium capsulatum (ATCC 51196), Pseudomonas aeruginosa (ATCC 10145), Hafnia alvei (ATCC 13337), Legionella pneumophila (ATCC 33152), Campylobacter jejuni (ATCC 33560), Fusobacterium nucleatum (ATCC 25586), Staphylococcus aureus (ATCC 12600), Bacillus thuringiensis (ATCC 10792), Streptococcus salivarius (ATCC 7073), Mycoplasma pneumoniae (ATCC 15531), and Clostridium perfringens (ATCC 13124). The Sterne strain of Bacillus anthracis, cured of plasmid pX01, was obtained from the United States Army Medical Research Institute for Infectious Diseases, Frederick, Md. DNA from Rickettsia prowazekii and Coxiella burnetii was obtained from the Rocky Mountain Laboratory, Hamilton, Mont.
DNA preparation and PCR.
Bacteria were lysed and DNA was extracted by using a Mini-BeadBeater (Biospec Products, Inc., Bartlesville, Okla.) as described previously (26). PCR was carried out using primers able to amplify all SSU rDNAs, CcompLong (TTGTACACACCGCCCGTCA, E. coli positions 1390 to 1408) and PC5B (TACCTTGTTACGACTT, E. coli positions 1507 to 1492). The reaction mixtures contained 10 mM (total) deoxynucleoside triphosphates (dNTPs) and the Advantage 2 PCR enzyme system (Clontech, Palo Alto, Calif.), which included AdvanTaq DNA polymerase and TaqStart antibody (1.1 μg/μl) and the 10× Advantage 2 PCR buffer, consisting of 400 mM Tricine-KOH (pH 8.7), 150 mM potassium acetate, 35 mM magnesium acetate, 37.5 μg of bovine serum albumin (BSA) per ml, 0.05% Tween 20, and 0.05% Nonidet P-40 per reaction. PCR conditions were 1 cycle of 1 min at 95 C, followed by 35 cycles of 1 min at 94°C, 30 s at 45°C, and 45 s at 68°C. A 10-min incubation at 68°C was the final step.
Preparation of sample for GeneChip assay.
A fragmentation working mix was prepared of 0.25 U of fragmentation reagent (Affymetrix, Santa Clara, Calif.) with equal volume of 20 mM EDTA, 2.5 U of calf intestine alkaline phosphatase (Gibco-BRL Life Technologies, Rockville, Md.), and 0.5 mM Tris-acetate (pH 8.2). Concentrations of PCR products were determined on agarose gels with a DNA mass ladder (Life Technologies, Rockville, Md.). A 45-μl solution of 2 μg of unpurified amplicon DNA in distilled H2O was prepared, 5 μl of working mix was added, and fragmentation was performed on a PE 9600 thermocycler for 12 min at 25°C and 10 min at 95°C. Then 21.5 μl of fragmented products was biotin labeled for 2 h at 37°C in a reaction mix of 50 U of terminal transferase (Boehringer Mannheim), 1.8 mM CoCl2, and 57 μM Renaissance biotin-N6-ddATP (NEN Life Science Products) in 1× terminal transferase buffer, for a final volume of 35 μl.
GeneChip processing.
GeneChip probe arrays were prehybridized with 200 μl of RapidHyb buffer (Amersham, Arlington Heights, Ill.) containing 0.8 M tetramethylammonium chloride (TMAC) in a custom hybridization rotisserie oven (Stovall Life Science, Greensboro, N.C.) for 10 min at 45°C and 60 rpm. Target cocktail [Rapid Hyb/TMAC solution, 0.06 nM control oligonucleotide B1 (Affymetrix, Inc.) and 80 to 770 ng of biotin-labeled target sample] was heated for 5 min at 99°C and centrifuged for 5 min at 14,000 rpm. The cocktail supernatant fraction was equilibrated to 45°C and added to a GeneChip array that was drained of prehybridization solution.
Hybridization was performed in the rotisserie oven for 3 h at 45°C and 60 rpm. The microarray was washed and stained in a GeneChip fluidics station 400 (Affymetrix, Inc.). Four washes with 0.9 M NaCl-60 mM NaH2PO4-6 mM EDTA-6× SSPE)-0.005% Triton X-100, pH 7.4, were followed by two washes with 1× SSPE-0.005% Triton X-100, pH 7.4. The array was then stained with 2 μg of streptavidin phycoerythrin-R conjugate (SAPE) (Molecular Probes, Inc., Eugene, Oreg.) and 0.95 mg of acetylated BSA per ml in 6× SSPE-0.005% Triton X-100 for 15 min at 40°C, followed by three washes with 1× SSPE-0.005% Triton X-100 at 25°C.
Scanning and data analysis.
The array was subsequently scanned by a 488-nm argon-ion laser (GeneChip Scanner 50; Molecular Dynamics) with emission detection above 520 nm. The phycoerythrin emission was 576 nm. The scan was recorded as a pixel image. Data were analyzed using standard Affymetrix software (GeneChip Analysis Suite, version 3.3). Background hybridization and signal noise were treated as two separate phenomena and were calculated as suggested by the array manufacturer. Background cells were identified as those producing intensities in the lowest 2% of all intensities. The average intensity of the background cells was subtracted from the fluorescence intensity of all cells. The noise value (N) was the variation in pixel intensity signals observed by the scanner as it read the array surface. The standard deviation of the pixel intensities within each of the identified background cells was divided by the square root of the number of pixels comprising that cell. The average of the resulting quotients was used for N in the calculations described below.
In Affymetrix probe pair scoring, two thresholds must be surpassed for a probe to be scored as positive or negative. The empirically derived criteria for a probe to be scored as positive were as follows: (i) the intensity of fluorescence from the perfectly matched probe cell (PM) was greater than or equal to 1.25 times the intensity from the mismatched control cell (MM), and (ii) the difference in intensity, PM minus MM, was at least eightfold greater than the noise value (≥8 N). A negative PM required the MM intensity to exceed the PM according to the same two criteria. When intensities between probes in a pair did not differ sufficiently to exceed the thresholds in either direction, the pair was scored as indeterminate. Thus, a signal could be read as positive, negative, or indeterminate.
Our chip contained 62,358 unique oligonucleotides based on sequences in the RDP 5.0 database. We updated our analysis abilities by reassigning the PM probes using the sequences in the RDP 8.1 database. Positive signals were matched by computer to oligonucleotides found in rRNA sequences occurring in RDP version 8.1. As noted above, ambiguities in the database were handled by synthesizing oligonucleotides in which the ambiguous bases were deleted. In practice, unless these deleted bases occurred near the end of the oligonucleotide, many of the probes had a lower signal intensity than other probes in the probe set. For this reason, all potential probe sites containing these deletions were omitted from further analyses.
For amplicons derived from pure cultures, a positive result for an RDP sequence required that all probes for that sequence give a positive signal. Because some sequences were incomplete, another requirement was that at least 27 probes for a sequence be present on the chip. The sequences meeting these criteria were rank-ordered with respect to the mean difference in fluorescence intensity between all probe cells and matched control cells for each sequence. For analyses of hierarchical phylogenetic levels corresponding to chip data, the RDP sequence ranked first was used.
It was anticipated that in contrast to rDNA sequences from pure cultures, mixed rDNA sequences sampled directly from the environment would often fail to perfectly match an rDNA sequence in RDP. For that reason, software parameters were empirically loosened. A positive result for an RDP sequence required 24 probes to be present on the chip, of which 22 were scored as positive. Another new criterion was that no probe for that sequence could be negative, though probes could fall in the range between positive and negative.
Extraction of DNA from air sample.
Air was sampled outdoors at Lawrence Livermore National Laboratory, Livermore, Calif., on 15 August 2000 using a high-volume air filter system that routinely filters 60 m3 of air per h. The unit was run for 24 h and processed 1,411,000 liters of air, which was filtered through a 1-μm-pore-size 8-in. by 10-in. (ca. 20 by 25 cm) track-etched Poretics polyester filters (Osmonics, Westborough, Mass.). All particles >1 μm in diameter were deposited on the surface of the flat filter. The filter was cut into 20 to 30 strips using sterile scissors and forceps and placed in a 50-ml conical Falcon tube. To extract the bacteria and debris, the filter was washed twice with a phosphate buffer-Tween (PBT) solution of 17 mM KH2PO4, 72 mM K2HPO4 and 0.003% Tween 20. The eluted material contained in the Falcon tubes was centrifuged for 30 min at 4°C and 3,500 × g (4,000 rpm). The supernatant fraction was discarded, and the pellets were transferred to 1.5-ml microcentrifuge tubes and centrifuged again for 8 min at 16,000 × g. The supernatant fraction was discarded, and the two wash pellets were combined for a final volume of 200 μl. Genomic DNA was extracted directly from the filter concentrate using a derivation of the MoBio UltraClean Soil DNA kit (MoBio Laboratories, Solana Beach, Calif.) modified to use silica beads instead of garnet beads to lyse cells. DNA obtained in this way was amplified as described above.
Analysis of air sample.
The resulting air DNA amplicons were split into two samples, and microbial composition was analyzed by two methods. One sample was hybridized to three chips from different production lots on different days. The other sample was cloned using the pGEM-T Easy cloning vector system (Invitrogen, San Diego, Calif.) and transformed into Escherichia coli DH10B. Individual cloned rDNA fragments were sequenced using ABI 3700 instruments (Applied Biosystems, Foster City, Calif.) and assembled using Phred and Phrap (7, 8). The 368 sequenced clones passed quality tests of Phred 20 (base call error probability < 10−2.0) and were compared with sequences in the RDP 8.1 database (16) for phylogenetic identification by measuring sequence similarity with known rDNA sequences as described above. RDP level 3 codes were assigned.
RESULTS
Organisms in pure culture.
Table 1 shows the rank order of matching RDP sequences selected by the scanner's software for each of the bacterial species tested. Fifteen of 17 species were correctly ranked number one. In several instances, sequences from other species had an equal score with the one being tested. For all such species, the sequences in the subalignment were identical to the sequences of the test species. Bacillus anthracis was not included in the RDP version used to design the chip. However, B. anthracis has the same rDNA sequence (GenBank accession number AF290552) as the Bacillus cereus strain selected by the software. Thus, B. anthracis was not miscalled. The sequences of Clostridium perfringens and Eubacterium tarantellae in the region represented on the chip were identical except that an ambiguity occurred at the 3′ end of the C. perfringens sequence in RDP. For this reason, E. tarantellae had three more probes on the chip. Because these three probes gave signals higher than the average of the rest of the probes, the average signal was greater for E. tarantellae.
TABLE 1.
Ranking of positive probe sets for each test organism
| Test organism | Ranka | Matching RDP sequences |
|---|---|---|
| Clostridium difficile | 1 | C. difficile |
| 1 | L. pneumophila | |
| Legionella pneumophila | 2 | L. sainthelensi |
| 3 | L. anisa/L. cincinnatiensis/L. steigerwaltii/L. israelensisb | |
| 1 | S. aureus/S. sciuri | |
| Staphylococcus aureus | 2 | S. epidermidis/S. haemolyticus/S. hominis/S. muscae |
| 3 | Lactobacillus aviarius | |
| 1 | B. cereus/B. thuringiensis | |
| Bacillus anthracis | 2 | B. alcalophilus/B. firmus/B. cohnii and others |
| 3 | Sporolactobacillus dextrus | |
| Acidobacterium capsulatum | 1 | A. capsulatum |
| Coxiella burnetii | 1 | C. burnetii |
| 1 | Eubacterium tarantellae | |
| Clostridium perfringens | 2 | C. perfringens |
| 3 | C. perfringens CPN50 | |
| 1 | P. aeruginosa DSM 50071 | |
| Pseudomonas aeruginosa | 2 | P. aeruginosa ATCC 25330 |
| 3 | E. coli/Citrobacter freundii/Plesiomonas shigelloides and others | |
| Brucella abortus | 1 | B. melitensis/B. abortus/Mesorhizobium loti and others |
| 2 | Phyllobacterium myrsinacearum/Bartonella quintana/Bartonella vinsonii and others | |
| Rickettsia prowazekii | 1 | R. prowazekii/R. montanensis/R. amblyommii and others |
| Mycoplasma pneumoniae | 1 | M. pneumoniae |
| 1 | S. salivarius ATCC 13419/Enterococcus sulfureus | |
| Streptococcus salivarius | 2 | Bacillus sp. strain DSM 8725/Bacillus sp. strain DSM 8715 |
| 3 | Bacillus sp. strain DSM 8717/Bacillus sp. strain DSM 8714 | |
| Bacteroides vulgatus | 1 | B. vulgatus |
| Fusobacterium nucleatum | 1 | F. simiae |
| 1 | H. alvei/symbiont of Glossina/Pectobacterium carotovorum | |
| Hafnia alvei | 2 | Yersinia kristensenii/Y. mollaretii/Y. pseudotuberculosis |
| 3 | Serratia marcescens | |
| Campylobacter jejuni | 1 | C. jejuni/C. coli/C. lari and others |
| 1 | B. cereus/B. thuringiensis | |
| Bacillus thuringiensis | 2 | B. alcalophilus/B. firmus/B. cohnii and others |
| 3 | Sporolactobacillus dextrus | |
| 1 | M. pneumoniae | |
| Mycoplasma pneumoniae and Rickettsia prowazekii | 2 | R. typhi/Rickettsia sp. strain ELB agent |
| 3 | R. prowazekii/R. montanensis/R. amblyommii and others |
Sequences for which all probes on the chip were positive are listed in rank order of the mean signal intensity. Mean signal intensity refers to the mean difference of the signal from each perfectly matched probe minus the signal from the control mismatched probe. Only sequences corresponding to the three most intensely fluorescent probe sets are listed. If less than three lines are listed below a tested organism, less than three complete probe sets were positive for that test organism.
All sequences listed on the same line shared the same complete set of probes complementary to their rDNA sequence.
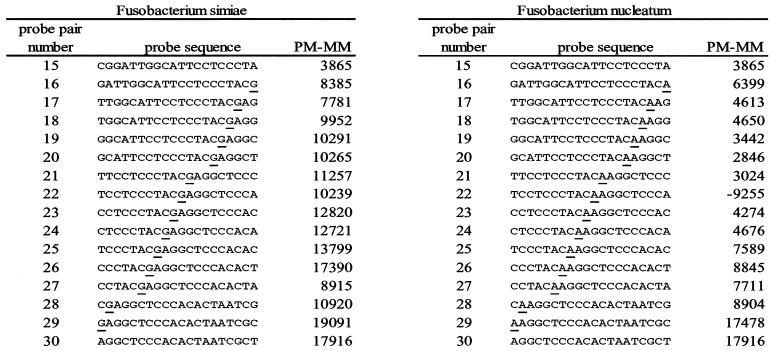
The software's failure to list Fusobacterium nucleatum came as a surprise. Analysis of the data from individual probes on the chip suggested that the amplicon's sequence differed from the RDP sequence for F. nucleatum at (E. coli) position 1443 (explained in Fig. 2). Therefore the PCR product was cloned, and six clones were sequenced. All gave the same result—as predicted from the chip data, there was a one-base discrepancy between the sequence in RDP and that of our amplicon, which had the same sequence as RDP's entry for Fusobacterium simiae. Figure 2 details hybridization data for probes involving this one-base mismatch.
FIG. 2.
Data from hybridization of amplified rDNA from F. nucleatum to probe sets for F. simiae and F. nucleatum. Probe sequences are shown 3′ to 5′, the order of synthesis on the chip. Probe pair numbers 1 through 15 and 30 are shared (identical) between the two probe sets. Underlined bases highlight the site of a sequence discrepancy between the RDP database used in the software analysis and the amplicon from F. nucleatum and the corresponding site in the F. simiae probe set. Signal intensity is measured as fluorescent intensity of the perfect match (PM) probe minus fluorescent intensity of the mismatch (MM) probe (PM − MM). Considering probe pairs 16 through 29, the amplicon from F. nucleatum hybridized better to the F. simiae probe set, which was actually complementary to its sequence. At position 22, the MM probe for F. nucleatum, incorporating a guanosine at the site of the discrepancy, hybridized much better than the PM probe. This finding led to the prediction that the amplicon actually contained a cytosine in this position. Sequencing confirmed this assessment.
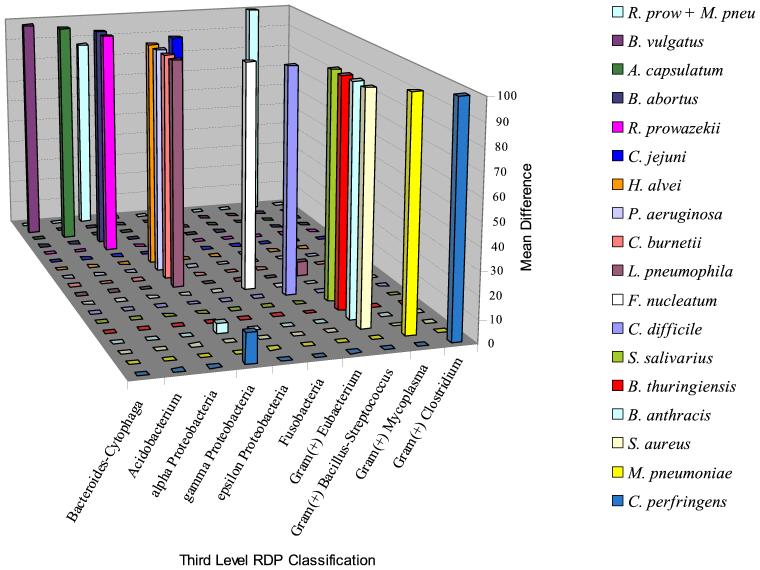
Table 2 shows the phylogenetic placement of the sequences ranked first for each of the DNA samples tested. The first-ranked sequence corresponded to the test organism's correct phylogenetic group within RDP in every case. Very few sequences in the database outside of the third-level phylogenetic designation of the test organism met criteria for a positive result, and in all cases, the mean difference in fluorescence was far lower than the mean difference for the correct group (Fig. 3).
TABLE 2.
Phylogenetic placement of sequences detected by GeneChip
| Organism(s)a | Phylocode (expected result) | Accuracy of RDP phylogenetic placement at each resolution levelb
|
|||||
|---|---|---|---|---|---|---|---|
| 1st | 2nd | 3rd | 4th | 5th | 6th | ||
| Single organism | |||||||
| B. vulgatus | 2.15.1.2.8 | + | + | + | + | + | na |
| A. capsulatum | 2.25.3.6 | + | + | + | + | na | na |
| B. abortus | 2.28.1.6.18 | + | + | + | + | + | na |
| R. prowazekii | 2.28.1.8.5.5 | + | + | + | + | + | + |
| L. pneumophila | 2.28.3.9.1.9 | + | + | + | + | + | + |
| C. burnetii | 2.28.3.9.2 | + | + | + | + | + | na |
| P. aeruginosa | 2.28.3.13.5 | + | + | + | + | + | na |
| H. alvei | 2.28.3.27.14.1 | + | + | + | +* | +* | +* |
| C. jejuni | 2.28.5.3.4 | + | + | + | + | + | na |
| F. nucleatum | 2.29.5 | + | + | + | na | na | na |
| S. aureus | 2.30.7.12.1 | + | + | + | + | + | na |
| B. anthracis | 2.30.7.12.4 | + | + | + | + | + | na |
| B. thuringiensis | 2.30.7.12.4 | + | + | + | + | + | na |
| S. salivarius | 2.30.7.21.6 | + | + | + | +* | +* | na |
| M. pneumoniae | 2.30.8.4.3 | + | + | + | + | + | na |
| C. difficile | 2.30.4.5.8 | + | + | + | + | + | na |
| C. perfringens | 2.30.9.2.11.3 | + | + | + | + | + | + |
| Two organisms | |||||||
| R. prowazekii | 2.28.1.8.5.5 | + | + | + | + | + | + |
| M. pneumoniae | 2.30.8.4.3 | + | + | + | + | + | na |
Organism(s) from which DNA was extracted for amplification of SSU rDNA.
Various levels of phylogenetic information concerning the corresponding sequence(s) rank-ordered by GeneChip software. +, correct call; +*, correct sequence corresponds to more than one phylocode; na, not applicable, taxon does not exist. Where applicable, all were identified correctly to the fourth-level grouping, although in the case of H. alvei and S. salivarius sequences outside of the expected fourth-level grouping gave as strongly positive a result as did sequences within the correct fourth-level grouping.
FIG. 3.
Average difference in intensity of fluorescence between probe cells and control cells for the phylogenetic groups identified by GeneChip software in experiments using amplicons derived from pure cultures. For each sample, the largest average difference was set at 100, and other peaks were adjusted relative to it.
In order to evaluate the usefulness of the chip for analysis of more complex samples, a mixture of DNA from Rickettsia prowazekii and Mycoplasma pneumoniae was tested. As shown in Table 1, each component was detected as accurately as when tested alone.
Testing of microbes filtered from air.
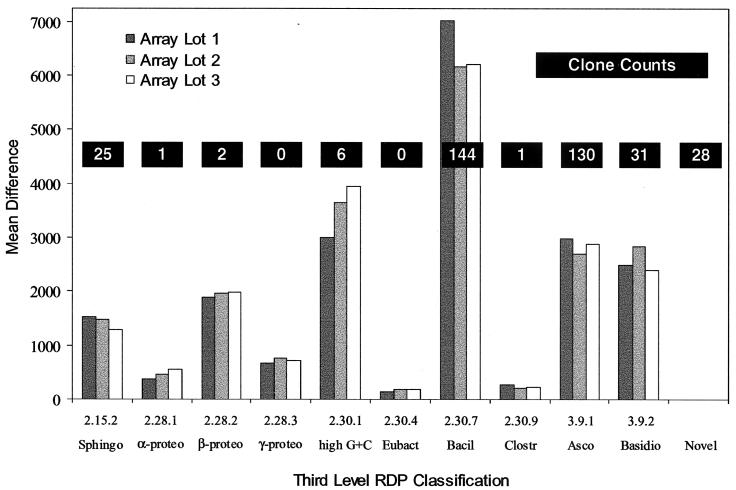
Figure 4 shows the reported relative abundance of sequences belonging to the various phylogenetic clusters detected by clone sequencing compared with groups found by chip analysis. As expected, both eukaryotic and bacterial sequences were detected by both methods. Overall, there was close agreement between the chip analysis and the sequencing results in detecting the presence of phylogenetic groups as defined by RDP. Eight of 10 phylogenetic clusters detected by the chip were confirmed as present when clones were sequenced, a result that was reproduced by all three production lots of the chip. Gamma proteobacteria and the group eubacteria were each detected in small amounts on the chip in all three array lots but were not sampled by cloning. Twenty-eight clones (7.6% of total) contained rDNA from organisms that could not be placed into phylogenetic groups based on similarity to any aligned sequence in RDP release 8.1 and were also not detected on the chip. Appropriate phylogenetic clusters for none of these sequences were represented in RDP release 5.0, from which the chip was designed. Most of these sequences had low similarity to all sequences in RDP release 8.1 as well and tended to be most similar to sequences that have yet to be placed into phylogenetic groups. Thus, they appear to be derived from novel organisms.
FIG. 4.
Relative abundance of microbes detected in an air sample as determined by rDNA amplification followed both by cloning with sequencing and by GeneChip analysis. rDNA sequences as determined by each method were sorted into taxa. Vertical bars represent mean difference of replicate hybridizations on three chips from different manufacturers' production lots. Bar height is proportional to the greatest mean difference in fluorescence found among the sequences in that group. Abbreviations for taxa and RDP phylogenetic numerical codes: Sphingobacterium group, Sphingo (2.15.2); α-proteobacteria subdivision, α-proteo (2.28.1); β-proteobacteria subdivision, β-proteo (2.28.2); γ-proteobacteria subdivision, γ-proteo (2.28.3); gram-positive high G+C bacteria, high G+C (2.30.1); gram-positive Eubacterium and relatives, Eubact (2.30.4); gram-positive Bacillus-Lactobacillus-Streptococcus subdivision, Bacil (2.30.7); gram-positive Clostridium and relatives, Clostr (2.30.9); Ascomycota fungi, Asco (3.9.1); and Basidiomycota fungi, Basidio (3.9.2). The 28 novel sequences did not correspond well to any phylogenetic group in the RDP database.
DISCUSSION
rRNA sequence analysis was initially conducted by cataloging short rRNA breakdown products (11). This approach led to startling revelations concerning the phylogeny of life forms on this planet (20, 29). The approach became more powerful with application of the Sanger method to sequence rRNA and rDNA by using conserved oligonucleotide primers (14). Sampling and sequencing were simplified considerably by the application of PCR to rDNA analysis (3, 26). PCR also made it possible to easily obtain complex samples of mixed rDNAs from natural environments (23, 24). Despite these advances, the use of rDNA analysis for microbial ecology remains problematic.
The complexity of microbial ecosystems and the number of unique ecosystems hinder efforts to explore biodiversity. For instance, it is estimated that one cubic millimeter of earth may contain as many as 10,000 different microbial types, and the microbial content of soil varies considerably from site to site (5). Furthermore, complex microbial ecosystems can change over time as environmental conditions vary. If these ecosystems are to be understood, they must be studied in the dimension of time as well as the dimensions of space. The need for increased throughput is obvious. By making it possible to conduct sequence analyses on many uncloned rDNAs at once, the photolithography chip has the potential to meet this need. The present study investigated the feasibility of this approach.
For samples known to consist of single organisms represented in RDP version 5.0, the approach rapidly determined the identity of most organisms tested, though sometimes more than one organism in the database had the same sequence. Occasionally the sequence of a close relative of the organism being tested was ranked above that of the test organism. In all instances the discrepancy could be explained on theoretical grounds. The chip was highly accurate at identifying the presence of sequences categorized at higher phylogenetic levels. Occasionally a low-amplitude signal was detected for an unrelated phylogenetic cluster. In these instances, it is unclear whether cross-hybridization occurred or there was contamination of the DNA sample or PCR product. When the chip results were compared with cloning and sequencing of rDNAs derived from an air sample, overall the two representations of phylogenetic groups occurring in the natural sample were highly similar. Cloned rDNAs not detected by the chip were all from novel organisms with no close relatives represented in RDP. Phylogenetic clusters detected on the chip but not sampled as rDNA clones appeared as low-amplitude fluorescent signals. In general, there was not a good correlation between the numbers of clones from a phylogenetic group and the intensity of the fluorescent signal from that group. Thus, the two methods may be complementary.
The photolithography chip reported here has several potential applications including the potential for public health applications, clinical microbiological testing, monitoring bioremediation, and monitoring for biological weapons. There is currently interest in microbiological observatories to monitor microbial populations around the world. While the present version of the SSU chip does not appear to be capable of differentiating mixed populations of highly similar organisms, it could still play a useful role in this endeavor. At the very least, it could probably determine whether or not the microbial communities found at a given site had changed over time and, if so, the probable makeup of the changes found.
The efficiency and usefulness of the SSU rDNA chip can be enhanced in several ways. Often, bacterial species have unique rDNA sequences at various sites (13, 21). A strong hybridization signal at such a site would have more than average significance. With further development of the software, it should be possible to weight such sites more highly than phylogenetically less informative sites. The subalignment used for the present chip is bounded by universally conserved sequences and is small enough that the segment can be amplified from even highly degraded DNA. However, it may not be realistic to expect always to identify species based on an 80- to 90-bp region of a conserved gene for which the reference database contains only one or two examples per species. Though the region studied is moderately variable, other areas are more variable and are worthy of consideration. As an alternative approach, it should be possible to amplify large segments of rDNA and probe only the most variable regions. Finally, the rDNA database is now much larger; the sequence information is more complete, and sequences contain fewer ambiguities than when this chip was designed. Given these areas for potential improvement and the fact that photolithography chips displaying over 250,000 oligonucleotides can now be produced, a vast enhancement of performance is theoretically attainable.
Acknowledgments
This work was supported by the Central Intelligence Agency, the Department of Veterans Affairs, the Hazardous Materials Response Unit of the Federal Bureau of Investigation, and the U.S. Department of Energy, NN-20, Chemical and Biological Non-Proliferation Program. This work was performed under the auspices of the U.S. Department of Energy by the University of California, Lawrence Livermore National Laboratory, under contract no. W-7405-Eng-48.
The chip studied in this paper was engineered by Rob Lipshutz and Tom Gingeras of Affymetrix, Inc., and by Don Morris. We thank Jenny Simchock (DUMC) for preparation of bacterial DNA samples.
REFERENCES
- 1.Barns, S. M., R. E. Fundyga, M. W. Jeffries, and N. R. Pace. 1994. Remarkable archaeal diversity detected in a Yellowstone National Park hot spring environment. Proc. Natl. Acad. Sci. USA 91:1609-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bowman, J. P., S. M. Rea, S. A. McCammon, and T. A. McMeekin. 2000. Diversity and community structure within anoxic sediment from marine salinity meromictic lakes and a coastal meromictic marine basin, Vestfold Hilds, Eastern Antarctica. Environ. Microbiol. 2:227-237. [DOI] [PubMed] [Google Scholar]
- 3.Chen, K., H. Neimark, P. Rumore, and C. R. Steinman. 1989. Broad range DNA probes for detecting and amplifying eubacterial nucleic acids. FEMS Microbiol. Lett. 57:19-24. [DOI] [PubMed] [Google Scholar]
- 4.DeLong, E. F., G. S. Wickham, and N. R. Pace. 1989. Phylogenetic stains: ribosomal RNA-based probes for the identification of single cells. Science 243:1360-1363. [DOI] [PubMed] [Google Scholar]
- 5.Dunbar, J., S. Takala, S. M. Barns, J. A. Davis, and C. R. Kuske. 1999. Levels of bacterial community diversity in four arid soils compared by cultivation and 16S rRNA gene cloning. Appl. Environ. Microbiol. 65:1662-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Everett, K. D., R. M. Bush, and A. A. Andersen. 1999. Emended description of the order Chlamydiales, proposal of Parachlamydiaceae fam. nov. and Simkaniaceae fam. nov., each containing one monotypic genus, revised taxonomy of the family Chlamydiaceae, including a new genus and five new species, and standards for the identification of organisms. Int. J. Syst. Bacteriol. 49:415-440. [DOI] [PubMed] [Google Scholar]
- 7.Ewing, B., and P. Green. 1998. Base-calling of automated sequencer traces using Phred. II. Error probabilities. Genome Res. 8:186-194. [PubMed] [Google Scholar]
- 8.Ewing, B., L. Hillier, M. C. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 9.Ferrero, D. V., H. N. Meyers, D. E. Schultz, and S. A. Willis. 1998. Performance of the Gen-Probe AMPLIFIED Chlamydia Trachomatis Assay in detecting Chlamydia trachomatis in endocervical and urine specimens from women and urethral and urine specimens from men attending sexually transmitted disease and family planning clinics. J. Clin. Microbiol. 36:3230-3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fodor, S. P., J. L. Read, M. C. Pirrung, L. Stryer, A. T. Lu, and D. Solas. 1991. Light-directed, spatially addressable parallel chemical synthesis. Science 251:767-773. [DOI] [PubMed] [Google Scholar]
- 11.Fox, G. E., K. Peckman, and C. R. Woese. 1977. Comparative cataloging of 16S ribosomal ribonucleic acid: molecular approach to prokaryotic systematics. Int. J. Syst. Bacteriol. 27:44-57. [Google Scholar]
- 12.Gingeras, T. R., G. Ghandour, E. Wang, A. Berno, P. M. Small, F. Drobniewski, D. Alland, E. Desmond, M. Holodniy, and J. Drenkow. 1998. Simultaneous genotyping and species identification using hybridization pattern recognition analysis of generic Mycobacterium DNA arrays. Genome Res. 8:435-448. [DOI] [PubMed] [Google Scholar]
- 13.Gobel, U., R. Maas, G. Havn, C. Vinge-Martins, and E. J. Stanbridge. 1987. Synthetic oligonucleotide probes complementary to rRNA for group and species-specific detection of mycoplasmas. Isr. J. Med. Sci. 23:742-746. [PubMed] [Google Scholar]
- 14.Lane, D. J., B. Pace, G. J. Olsen, D. A. Stahl, M. L. Sogin, and N. R. Pace. 1985. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analysis. Proc. Natl. Acad. Sci. USA 82:6955-6959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lockhart, D. J., H. Dong, M. C. Byrne, M. T. Follettie, M. V. Gallo, M. S. Chee, M. Mittmann, C. Wang, M. Kobayashi, H. Horton, and E. L. Brown. 1996. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat. Biotechnol. 14:1675-1680. [DOI] [PubMed] [Google Scholar]
- 16.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. J. Parker, P. R. Saxman, R. J. Farris, G. M. Garrity, G. J. Olsen, T. M. Schmidt, and J. M. Tiedje. 2001. The RDP-II (Ribosomal Database Project). Nucleic Acids Res. 29:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maidak, B. L., G. J. Olsen, N. Larsen, R. Overbeek, M. J. McCaughey, and C. R. Woese. 1996. The Ribosomal Database Project (RDP). Nucleic Acids Res. 24:82-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Notterman, D. A., U. Alon, A. J. Sierk, and A. J. Levine. 2001. Transcriptional gene expression profiles of colorectal adenoma, adenocarcinoma, and normal tissue examined by oligonucleotide arrays. Cancer Res. 61:3124-3130. [PubMed] [Google Scholar]
- 19.Okamoto, T., T. Suzuki, and N. Yamamoto. 2000. Microarray fabrication with covalent attachment of DNA using bubble jet technology. Nat. Biotechnol. 18:438-441. [DOI] [PubMed] [Google Scholar]
- 20.Pace, N. R. 1997. A molecular view of microbial diversity and the biosphere. Science 276:734-740. [DOI] [PubMed] [Google Scholar]
- 21.Paster, B. J., F. E. Dewhirst, W. G. Weisburg, L. A. Tordoff, G. J. Fraser, R. B. Hespell, T. B. Stanton, L. Zablen, L. Mandelco, and C. R. Woese. 1991. Phylogenetic analysis of the spirochetes. J. Bacteriol. 173:6101-6109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Relman, D. A., J. S. Loutit, T. M. Schmidt, S. Falkow, and L. S. Tompkins. 1990. The agent of bacillary angiomatosis. An approach to the identification of uncultured pathogens. N. Engl. J. Med. 323:1573-1580. [DOI] [PubMed] [Google Scholar]
- 23.Schmidt, T. M., E. F. DeLong, and N. R. Pace. 1991. Analysis of a marine picoplankton community by 16S rRNA gene cloning and sequencing. J. Bacteriol. 173:4371-4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weetall, H. H., and M. J. Lee. 1989. Antibodies immobilized on inorganic supports. Appl. Biochem. Biotechnol. 22:311-330. [DOI] [PubMed] [Google Scholar]
- 25.Wilson, K. H., R. Blitchington, R. Frothingham, and J. A. P. Wilson. 1991. Phylogeny of the Whipple's-disease-associated bacterium. Lancet 338:474-475. [DOI] [PubMed] [Google Scholar]
- 26.Wilson, K. H., R. Blitchington, and R. C. Greene. 1990. Amplification of bacterial 16S rRNA sequences with polymerase chain reaction. J. Clin. Microbiol. 28:1942-1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilson, K. H., and R. B. Blitchington. 1996. Human colonic biota studied by ribosomal DNA sequence analysis. Appl. Environ. Microbiol. 62:2273-2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wisotzkey, J. D., P. J. Jurtshuk, G. E. Fox, G. Deinhard, and K. Poralla. 1992. Comparative sequence analyses on the 16S rRNA (rDNA) of Bacillus acidocaldarius, Bacillus acidoterrestris, and Bacillus cycloheptanicus and proposal for creation of a new genus, Alicyclobacillus gen. nov. Int. J. Syst. Bacteriol. 42:263-269. [DOI] [PubMed] [Google Scholar]
- 29.Woese, C. R. 1987. Bacterial evolution. Microbiol. Rev. 51:221-271. [DOI] [PMC free article] [PubMed] [Google Scholar]