Abstract
Mutants of Corynebacterium glutamicum were made and enzymatically characterized to clone ilvD and ilvE, which encode dihydroxy acid dehydratase and transaminase B, respectively. These genes of the branched-chain amino acid synthesis were overexpressed together with ilvBN (which encodes acetohydroxy acid synthase) and ilvC (which encodes isomeroreductase) in the wild type, which does not excrete l-valine, to result in an accumulation of this amino acid to a concentration of 42 mM. Since l-valine originates from two pyruvate molecules, this illustrates the comparatively easy accessibility of the central metabolite pyruvate. The same genes, ilvBNCD, overexpressed in an ilvA deletion mutant which is unable to synthesize l-isoleucine increased the concentration of this amino acid to 58 mM. A further dramatic increase was obtained when panBC was deleted, making the resulting mutant auxotrophic for d-pantothenate. When the resulting strain, C. glutamicum 13032ΔilvAΔpanBC with ilvBNCD overexpressed, was grown under limiting conditions it accumulated 91 mM l-valine. This is attributed to a reduced coenzyme A availability and therefore reduced flux of pyruvate via pyruvate dehydrogenase enabling its increased drain-off via the l-valine biosynthesis pathway.
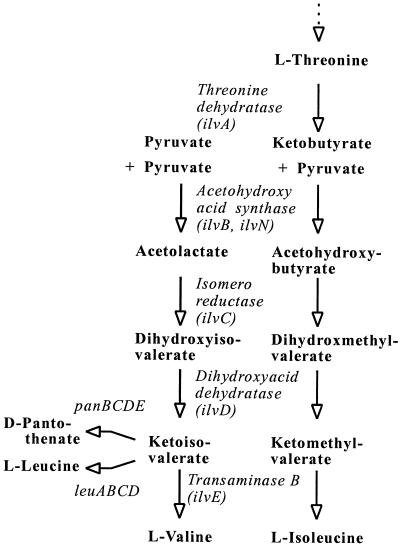
Metabolites of the aspartate family like l-lysine, l-threonine, the branched-chain amino acids, and d-pantothenate are of particular industrial interest since they are essential for vertebrates. We have already developed l-isoleucine-producing strains from Corynebacterium glutamicum by redirecting the carbon flux in an l-lysine producer to l-isoleucine (11, 21, 22). This was done by the use of the biosynthesis genes thrB, thrC (13, 30), ilvBN, and ilvC (5, 15) together with feedback-resistant variants of the key enzymes encoded by hom (29) and ilvA (19). In a further development we used the ilvBNC genes together with panBC to make d-pantothenate (32). This is possible because the acetohydroxy acid synthase encoded by ilvBN catalyzes not only the conversion of ketobutyrate plus pyruvate but also that of pyruvate plus pyruvate to make ketomethyl valerate and ketoiso valerate, respectively (Fig. 1). Whereas the former keto acid is the precursor for l-isoleucine only, the latter is the precursor for d-pantothenate, l-leucine, and l-valine as well. This therefore represents an opportunity to use the existing information and genes to study l-valine formation.
FIG. 1.
Linked synthesis of l-valine with those of d-pantothenate, l-leucine, and l-isoleucine.
Wild-type isolates of various bacteria accumulating l-valine have already been described (4, 28, 39). By undirected mutagenesis, strains of C. glutamicum excreting up to 10 g of l-valine liter−1 have been obtained (23). A mutant of C. glutamicum subsp. lactofermentum accumulates as much as 31 g liter−1 (38). Through an optimum supply of oxygen, accumulation with this strain is increased to about 40 g liter−1 (1). Mutants excreting l-valine are also known from Serratia marcescens (16). In general, it is poorly understood why the excretion of l-valine is possible since all strains described so far have been made by undirected mutagenesis. However, at least in some cases l-valine excretion seems to be correlated to acetohydroxy acid synthase activity (37) or the lack of allosteric control of this enzyme (16).
The aim of this work was to study l-valine formation with C. glutamicum. To this end the as yet not available biosynthesis genes of the branched-chain amino acid synthesis ilvD (dihydroxy acid dehydratase) and ilvE (transaminase) needed to be made available. Since the carbon skeleton of l-valine originates exclusively from pyruvate, we were furthermore very interested in discovering whether an increased availability of this precursor, important for many metabolites, can be achieved intracellularly and in testing the effect this would have on l-valine formation.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Unless otherwise specified, C. glutamicum was cultured at 30°C in complex medium CGIII or salt medium CGIX (15). Escherichia coli was grown at 30°C in Luria-Bertani medium. When appropriate chloramphenicol (5 mg liter−1) or kanamycin (25 mg liter−1) was used.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or description | Reference or source |
|---|---|---|
| R127 | Restriction deficient | 18 |
| R127-7 | Dehydratase negative | This work |
| R127-12 | Transaminase negative | This work |
| 13032 | Wild type | ATCC |
| 13032ΔilvA | Wild type with ilvA deleted | 32 |
| 13032ΔpanBC | Wild type with panBC deleted | This work |
| 13032ΔilvAΔpanBC | Wild type with ilvA and panBC deleted | This work |
| 13032pk18mob′panC′ | Wild type with panC disrupted | This work |
| pRV | pJC1::4.2-kb genomic Sau3A fragment of wild type | 41 |
| pJCilvE1 | pJC1::6.2-kb genomic Sau3A fragment of wild type | 41 |
| pUC18ilvE | pUC18::3.8-kb Nael fragment with ilvE | This work |
| pJC1ilvD | pJC1::2.9-kb XbaI fragment | This work |
| pJC1ilvBNCD | pKK5::2.6-kb XbaI fragment with ilvD | 32 |
| pECM3ilvD | pECM3::2.9-kb XbaI fragment | This work |
| pECM3ilvBNCD | pECM3::5.7-kb XbaI fragment with ilvNBC and 3.1-kb XbaI fragment with ilvD | 32 |
| pJC4 | Shuttle vector, derived from pJC1; Kanr | 6 |
| pJC4ilvBNC | pJC4::5.9-kb EcoRI-HindIII fragment with ilvBNC from pCC2-42 | This work |
| pJC4ilvBNCE | pJC4ilvBNC::1.6-kb BglI-NaeI fragment with ilvE | This work |
| pk18mob′panC′ | Mobilizable vector with 168-bp internal panC fragment | 32 |
For accumulation of amino acids, cells were pregrown overnight in CGIII. After washing, they were inoculated into CGXII (22) to give an initial optical density at 600 nm of 1.0. In the case of strains with ilvA deleted, CGXII contained 2 mM l-isoleucine.
Cloning of genes.
The restriction-deficient strain R-127 was mutagenized as described by Vrljić et al. (41), and auxotrophic mutants were isolated by replica plating on minimal medium CGIX with or without amino acids or keto acids (each 300 mg liter−1). The gene bank of the genomic DNA of wild-type C. glutamicum ATCC 13032 is also described by Vrljić et al. (41). It was used to transform strain R127-7 and R127-12 to kanamycin resistance (18). Prototrophic clones were identified after replica plating on CGIX without supplements.
Plasmid constructions.
In previous work focusing on d-pantothenate accumulation we had already constructed vectors to enable the overexpression of different combinations of ilv genes (32). To include studies on the influence of ilvE on l-valine accumulation, plasmid pJC4ilvBNCE was constructed. For this purpose, plasmid pJC4 was cleaved with XbaI and blunted. ilvBNC was isolated as a 5.9-kb EcoRI-HindIII fragment from pCC2-42 (5) and, after treatment with Klenow enzyme, used for ligation. The resulting vector pJC4ilvBNC was restricted with BamHI, blunted, and ligated with the ilvE-carrying fragment derived from pUC18ilvE. The ilvE fragment was 1.57 kb in size (BglI-NaeI) and was Klenow treated before ligation. The final plasmid pJC4ilvBNCE of 13.4 kb was characterized by restriction analysis and transaminase activity determination.
Enzyme assays.
The transaminase activity was determined with crude extract passed over a PD10 column, with 20 mM Tris-HCl, pH 8, used for equilibration. The assay system contained (in 1 ml) 200 mM Tris-HCl (pH 8), 0.25 mM pyridoxal-5′-phosphate, 4 mM ketoisocaproate, 50 mM sodium glutamate, and crude extract. Samples (50 μl) were taken at several time points over a period of 20 min, and the reaction was terminated by addition of 30 μl of stop reagent consisting of 40 ml of ethanol, 6.7 ml of perchloric acid (70%), and 43 ml of water. Samples were subsequently neutralized, and l-leucine formation was quantified by high-performance liquid chromatography.
The dihydroxy acid dehydratase activity was determined in a system containing (per milliliter) 50 mM Tris-HCl (pH 8), 10 mM MgCl2, 10 mM dihydroxymethyl valerate (pH 4), and 50 μl of extract. Samples (200 μl) were taken at several time points to quantify the ketomethylvalerate formed according to the method of Hara et al. (14). The samples were mixed with 200 μl of 4,5-dimethoxy-1,2-diaminobenzene solution (0.4 mM 4,5-dimethoxy-1,2-diaminobenzene and 0.21 mM β-mercaptoethanol, in 0.5 mM hydrochloric acid). The samples were incubated for 2 h at 104°C and afterwards were separated by reverse-phase chromatography and quantified by fluorescence detection. The mobile phase consisted of a gradient of 20 mM phosphate buffer (pH 3.2) and methanol.
Nucleotide sequence accession numbers.
The nucleotide sequence accession numbers of the sequences derived are AJ012293 for ilvD and AF424637 for ilvE.
RESULTS
Identification of ilvD.
We first isolated 10 mutants of C. glutamicum R127 requiring the three branched-chain amino acids for growth. Enzyme tests showed that two of these strains did not have any detectable dihydroxy acid dehydratase activity, whereas with extracts of the initial strain a specific activity of 0.001 μmol min−1 mg (dry weight)−1 was obtained. However, the acetohydroxy acid synthase and isomeroreductase activities were unchanged (not shown). Using the gene bank of genomic DNA of the wild type described by Vrljić et al. (41), we transformed one of the mutants (strain R127-7) to kanamycin resistance. In this way, a prototrophic clone was obtained which contained a recombinant plasmid with an insert of 4.2 kb. This was subcloned as a 2.9-kb ScaI-XhoI fragment. Sequencing revealed that the open reading frame (ORF) termed ilvD encodes a polypeptide of 611 aminoacyl residues (Mr = 64,899). It exhibits high identities over its entire length with similar peptides present in many bacteria and archaea. For instance, that of S. coelicolor shares 63% identical amino acyl residues, that of Neisseria meningitidis shares 62% identical amino acyl residues, and that of Methanococcus jannaschii share 42% identical amino acyl residues. To check for the function of the gene a 2.9-kb XbaI fragment was subcloned in pJC1 and pECM3. The resulting plasmids were introduced into the wild-type ATCC 13032. If ATCC 13032 had a specific activity of 0.008 μmol min−1 mg of protein−1, then through pJCilvD and pECM3ilvD, activities of 0.051 and 0.126 μmol min−1 mg of protein−1 resulted. This shows that the cloned fragment codes for active dehydratase.
Identification of ilvE.
Mutants obtained as above and requiring the three branched-chain amino acids but unable to grow upon supply of ketomethylvalerate plus ketoisovalerate plus ketoisocaproate were assayed for transaminase B activity. Mutant R127-12 showed the expected very low specific transaminase activity of less than 0.0005 μmol min−1 mg of protein−1, in comparison to 0.017 μmol min−1 mg of protein−1 in R127. The growth defect of the mutant was complemented by plasmid pJCilvE1. With this plasmid, the resulting specific activity in R127-12 was 0.21 μmol min−1 mg (dry weight)−1. Sequencing the corresponding insert of 6,185 bp yielded an ORF with the expected identities to several transaminase B genes. The gene ilvE encodes a polypeptide of 367 aminoacyl residues (Mr = 40,363). A 1.57-kb BglI-NaeI fragment of pUCilvE was subcloned in pJC4ilvBNC. With the resulting vector pJC4ilvBNCE, the specific activity in strain R127 was 0.27 μmol min−1 mg of protein−1, thus confirming the functional identity and the integrity of the cloned gene.
l-Valine accumulation with overexpressed ilv genes.
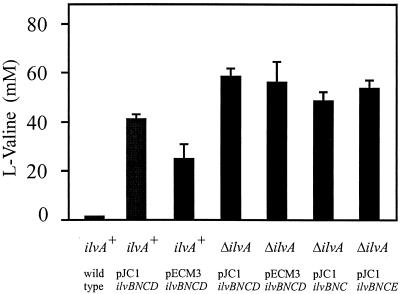
In previous work focusing on d-pantothenate accumulation we had already found l-valine accumulation upon overexpression of ilvBNC (32). Furthermore, we had found that the deletion of ilvA, which codes for threonine dehydratase, favorably influences l-valine formation, possibly due to an increased pyruvate availability (Fig. 1). In order to study the effect of overexpressing ilvD and ilvE in combination with other genes, plasmid pJC1ilvBNCE was used. This plasmid was transferred into the wild-type ATCC 13032 as well as into strain 13032ΔilvA. The resulting strains were subsequently cultivated in minimal medium CGXII together with controls (22). Three independent fermentations were carried out, with the results shown in Fig. 2. Obviously, with ilvA deleted, l-valine accumulates in higher concentrations than when ilvA is present. Moreover, the use of pJC1 and the ilvBNCD genes is superior to the same genes in pECM3, at least in the ilvA+ background. A further advantage of the genes in pJC1 is the reduced variation of l-valine which accumulated in the different experiments.
FIG. 2.
l-Valine accumulation with isogenic C. glutamicum strains. Below the columns the genotype of each strain, either ilvA+ or ΔilvA, is given. The strains additionally carry the plasmid pECM3 or pJC1 carrying ilvBNC, ilvBNCD, or ilvBNCE. Means are from three independent experiments. Error bars show standard deviations.
In the ΔilvA background a possible effect of ilvE was studied (Fig. 2). With plasmid pJC1ilvBNCE an increased l-valine accumulation of 53.7 mM was obtained compared to that obtained with pJC1ilvBNC (48.0 mM). However, the effect of pJC1ilvBNCE is not as high as that of pJC1ilvBNCD. We did not attempt to assay for a combined effect of ilvE and ilvD due to the large size of the vectors already made and the absence of appropriate cloning sites.
d-Pantothenate-dependent product formation.
Since l-valine originates from two pyruvate molecules we looked for ways to increase pyruvate availability. It is known that by limiting a C. glutamicum subsp. lactofermentum mutant for d-pantothenate its l-lysine synthesis (which requires one pyruvate molecule) can be increased (2). Since d-pantothenate is a constituent of coenzyme A (CoA) it might be possible to attribute this effect to its reduced availability and thus to the reduced flux from pyruvate to acetyl-CoA. In order to verify this directly we inactivated panC in the wild-type ATCC 13032 with an internal fragment of this pantothenate biosynthesis gene (32). Salt medium CGXII with or without 0.001 mM d-pantothenate was inoculated with the resulting strain, 13032pk18mob′panC′, and after 30 h the cell density was determined. The optical density without d-pantothenate was 18, compared to 26 in the control. As shown in independent experiments, growth observed without vitamin addition is apparently due to sufficient d-pantothenate still being present in the cells used for inoculation, since growth depends on inoculation density and the number of washing steps (not shown). Without any d-pantothenate addition, 6.3 mM pyruvate, 11.3 mM l-alanine, and 5.6 mM l-valine accumulated, whereas in the culture with the vitamin added, no pyruvate and only a maximum of 0.1 mM amino acid in each case were formed.
Construction of a panBC deletion mutant.
Based on this promising information, we constructed a strain with no panBC sequences present to have the kanamycin resistance gene available for plasmid selection. For this purpose, the 1,293-bp EcoRV-NruI fragment, encompassing panBC sequences, was deleted from plasmid pUR1.2 (32). After religation PCR was used to amplify the deletion fragment. This fragment, with EcoRI and SalI sites attached during the PCR, was isolated and ligated with pK19mobsacB. Taking advantage of intergeneric gene transfer (33), homologous recombination, and two rounds of positive selection, the wild-type derivative C. glutamicum 13032ΔpanBC was obtained, and the chromosomal deletion of this derivative was verified by PCR. C. glutamicum 13032ΔilvAΔpanBC was made in a similar manner, using 13032ΔilvA as a recipient during gene transfer. These two strains were transformed with the pJC-based vectors containing ilv genes and then used to assay for product accumulation.
l-Valine accumulation with ΔpanBC strains.
The fermentations were carried out in salt medium CGXII (22), with the ΔpanBC strains receiving 0.001 mM d-pantothenate, and the ΔilvA strains receiving 150 mg of l-isoleucine liter−1. As expected from the previous experiment, the d-pantothenate limitation led to a considerable accumulation of l-valine and l-alanine synthesized directly from pyruvate (Table 2). More l-alanine than l-valine is obtained, which may be due to the one-step synthesis pathway for forming l-alanine. Interestingly, with simultaneous ilvA deletion the product spectrum shifts toward l-valine. This is probably due to increased acetohydroxy acid synthase and isomeroreductase activity as a consequence of ilvA deletion, since the ilvBNC operon encoding these enzymes (20) is controlled by a translationally coupled attenuation mechanism involving l-isoleucine (7). If ilvBNCD was overexpressed in the strain with the double deletion, then after 48 h a very high l-valine concentration of more than 90 mM was obtained. l-Alanine could only be detected in low concentrations. In comparison to the strain overexpressing ilvBNCD, the strain overexpressing ilvBNCE displayed a reduced accumulation of l-valine. This is in line with an elevated concentration of l-alanine. Together with the analysis of product formation in strains without panBC deletion (Fig. 2), this leads one to assume that the major factor for the high l-valine accumulation in the constructed wild-type derivatives of C. glutamicum is the d-pantothenate limitation.
TABLE 2.
Amino acid accumulation with recombinant wild-type derivatives of C. glutamicum
| Strain | Amino acid concn (mM) at time
|
|||
|---|---|---|---|---|
| 24 h
|
48 h
|
|||
| l-Ala | l-Val | l-Ala | l-Val | |
| 13032 | 9.7 | 1.2 | 0 | 0 |
| 13032ΔpanBC | 27.2 | 6.7 | 50.1 | 7.6 |
| 13032ΔpanBCΔilvA | 19.9 | 4.0 | 29.1 | 20.5 |
| 13032ΔpanBCΔilvA pJC1ilvBNCD | 3.8 | 43.1 | 1.3 | 91.9 |
| 13032ΔpanBCΔilvA pJC1ilvBNCE | 9.7 | 49.4 | 7.1 | 81.2 |
DISCUSSION
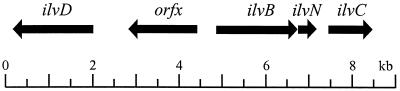
The gene ilvE is the only transaminase in C. glutamicum of relevance for the transamination step within the biosynthesis of the three branched-chain amino acids. This has to be concluded from the ilvE deletion mutant's requirement for the branched-chain amino acids and confirms previous results obtained with strains derived by undirected mutagenesis (7). This is therefore different from E. coli, an ilvE mutant of which requires l-isoleucine only (3). This is attributed to an overlapping specificity of the avtA-encoded alanine valine transaminase, which might supply l-valine and l-leucine and, in the case of its overexpression, might even make the ilvE mutant independent of l-isoleucine. Inspection of the genome sequence of C. glutamicum suggests that ilvE, like ilvA encoding the threonine dehydratase (5), is not part of an operon or clustered with other genes of the pathway. However, ilvD (dihydroxy acid dehydratase) is separated by just one ORF from the ilvBNC operon of C. glutamicum (Fig. 3). As in the case of ilvBNC (15), there are no other known genes or corresponding enzyme activities for ilvE (transaminase B) either. Therefore, in contrast to the Enterobacteriaceae, for instance, C. glutamicum does not have isoenzymes. Thus, together with the genes of the l-leucine synthesis (25, 26), all biosynthesis genes of the branched-chain amino acids are known.
FIG. 3.
The ilvBNCD gene locus in C. glutamicum ATCC 13032. Within the locus of about 9 kb is located the ilvBNC operon (5, 15), as well as ilvD. These are separated by orfx, an ORF of unknown function.
Upon overexpression of ilvBNCD the wild type already accumulates 42 mM l-valine. Therefore, the known inhibition of the ilvBN-encoded acetohydroxy acid synthase is not a major problem, probably due to the only partial inhibition of the enzyme (7). A further increase in the l-valine concentration up to 58 mM is present in the case of the strain from which ilvA, the threonine dehydratase-encoding gene, is deleted and which, therefore, requires l-isoleucine for growth. This could be due to several effects. First, since acetohydroxy acid synthase and isomeroreductase are involved in l-isoleucine and l-valine synthesis too (Fig. 1), the entire catalytic activity is now available for the initial steps of l-valine synthesis. Second, since ilvBNC expression is increased upon l-isoleucine shortage by an attenuation mechanism (20), an increased enzyme level might also result in increased pathway flux. A third possibility would be a general growth limitation introduced by the l-isoleucine auxotrophy. Several amino acid producer strains have auxotrophies (2, 23, 37, 39), often for an amino acid whose pathway is linked to that of the amino acid synthesized (17). It is therefore difficult to separate regulatory effects from others, such as, for instance, an increased precursor availability. However, in the case of l-lysine formation with C. glutamicum, graded growth limitation introduced by graded gene expression has demonstrated that improved lysine formation is based on increased precursor availability (12).
Whereas the effects discussed so far focus on the biosynthesis genes of the branched-chain amino acids, panBC deletion does not. Although the general principles of engineering the biosynthesis paths for increased pathway flux are comparatively well established (8), considerably less information is available to link further cellular activities like central metabolism (31), energy metabolism (24), or even cell wall structures to cellular flux properties (9, 10, 35). One example in which the precursor availability was engineered is the increased availability of erythrose-4-phosphate and phosphoenolpyruvate for aromatic amino acid synthesis (27). As shown by the panC inactivation mutant in this study, reduced d-pantothenate availability leads to pyruvate excretion, meaning that increased quantities of precursor are directly available for l-valine synthesis. Reduced CoA formation and thus a reduced flux through the pyruvate dehydrogenase is probably due to the d-pantothenate limitation, because there is no acceptor for the dihydrolipoyl transacetylase activity of the pyruvate dehydrogenase complex. If the pyruvate dehydrogenase activity in E. coli is controlled by lipoic acid deficiency, pyruvate excretion may result (42). Pyruvate is also a precursor of l-lysine. In successful attempts to influence l-lysine formation by reduced pyruvate dehydrogenase activity, an additional formation of the amino acid l-alanine, formed directly from pyruvate, was observed (36). Furthermore, l-lysine formation can be influenced with a C. glutamicum mutant requiring d-pantothenate through the availability of d-pantothenate (2).
Engineering pyruvate dehydrogenase activity itself is an attractive proposition. Due to its large number of coenzymes and domains (34, 40), this enzyme offers a wide range of approaches to its engineering. On the whole, the decisive factor is naturally the proper integration of activities in the central metabolism and the biosynthesis pathway. Since the strain currently available still excretes some l-alanine at the start of the fermentation, this strain has the potential for improving the l-valine accumulation by further increasing the flux via subsequent “bottlenecks,” such as the synthesis pathway or the cell wall.
Acknowledgments
We thank the Hermann Schlosser Stiftung for financial support of A. Vaitsikova.
We thank J. Carter-Sigglow for help with the language of the manuscript.
REFERENCES
- 1.Akashi, K., H. Shibai, and Y. Hirose. 1977. Effect of oxygen supply on L-valine fermentation. J. Ferment. Technol. 55:364-368. [Google Scholar]
- 2.An, G.-H., K. B. Song, and A. J. Sinskey. 1999. Redirection of carbon flux to lysine in a recombinant of Corynebacterium lactofermentum ATCC 21799 by limited supply of pantothenate. J. Biosci. Bioeng. 88:168-172. [DOI] [PubMed] [Google Scholar]
- 3.Berg, C. M., M. Wang, N. B. Vartak, and L. Liu. 1988. Acquisition of new metabolic capabilities: multicopy suppression by cloned transaminase genes in Escherichia coli K-12. Gene 65:195-202. [DOI] [PubMed] [Google Scholar]
- 4.Chattopadhyay, S. P., and A. K. Banerjee. 1978. Production of L-valine by a Bacillus sp. Zeitschrift Allgemeine Mikrobiologie 18:243-254. [DOI] [PubMed] [Google Scholar]
- 5.Cordes, C., B. Möckel, L. Eggeling, and H. Sahm. 1992. Cloning, organization and functional analysis of ilvA, ilvB and ilvC genes from Corynebacterium glutamicum. Gene 112:113-116. [DOI] [PubMed] [Google Scholar]
- 6.Cremer, J., L. Eggeling, and H. Sahm. 1990. Cloning the dapA dapB cluster of the lysine-secreting bacterium Corynebacterium glutamicum. Mol. Gen. Genet. 220:478-480. [Google Scholar]
- 7.Eggeling, I., C. Cordes, L. Eggeling, and H. Sahm. 1987. Regulation of acetohydroxy acid synthase in Corynebacterium glutamicum during fermentation of α-ketobutyrate to L-isoleucine. Appl. Microbiol. Biotechnol. 25:346-351. [Google Scholar]
- 8.Eggeling, L., and H. Sahm. 1999. Amino acid production: principles of metabolic engineering, p. 153-176. In S. Y. Lee and E. T. Papoutsakis (ed.), Metabolic engineering. Marcel Dekker, Inc., New York, N.Y.10935928
- 9.Eggeling, L., and H. Sahm. 2001. The cell wall barrier of Corynebacterium glutamicum and amino acid efflux. J. Biosci. Bioeng. 92:201-213. [Google Scholar]
- 10.Eggeling, L., K. Krumbach, and H. Sahm. 1999. L-Glutamate efflux with Corynebacterium glutamicum: why is penicillin treatment or Tween addition doing the same? J. Mol. Microbiol. Biotechnol. 3:67-68. [PubMed] [Google Scholar]
- 11.Eggeling, L., S. Morbach, and H. Sahm. 1997. The fruits of molecular physiology: engineering the L-isoleucine biosynthesis pathway in Corynebacterium glutamicum. J. Biotechnol. 56:167-182. [Google Scholar]
- 12.Eggeling, L., S. Oberle, and H. Sahm. 1998. Improved L-lysine yield with Corynebacterium glutamicum: use of dapA resulting in increased flux combined with growth limitation. Appl. Microbiol. Biotechnol. 49:24-30. [DOI] [PubMed] [Google Scholar]
- 13.Eikmanns. B. J., M. Metzger, D. J. Reinscheid, M. Kircher, and H. Sahm. 1991. Amplification of three threonine biosynthesis genes in Corynebacterium glutamicum and its influence on carbon flux in different strains. Appl. Microbiol. Biotechnol. 34:617-622. [DOI] [PubMed] [Google Scholar]
- 14.Hara, S., Y. Takemori, T. Iwata, M. Yamaguchi, and M. Nakamura. 1985. Fluorometric determination of α-keto acids with 4,5-dimethoxy-1,2-diaminobenzene and its application to high-performance liquid chromatography. Anal. Chim. Acta 172:167-173. [Google Scholar]
- 15.Keilhauer, C., L. Eggeling, and H. Sahm. 1993. Isoleucine synthesis in Corynebacterium glutamicum: molecular analysis of the ilvB-ilvN-ilvC operon. J. Bacteriol. 175:5595-5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kisumi, M., S. Komatsubara, and I. Chibata. 1971. Valine accumulation by α-aminobutyric acid-resistant mutants of Serratia marcescens. J. Bacteriol. 106:493-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Konstantinov, K. B., N. Nishio, T. Seki, and T. Yoshida. 1991. Physiologically motivated strategies for control of the fed-batch cultivation of recombinant Escherichia coli for phenylalanine production. J. Ferment. Bioeng. 71:350-355. [Google Scholar]
- 18.Liebl, W., A. Bayerl, U. Stillner, and K. H. Schleifer. 1989. High efficiency electroporation of intact Corynebacterium glutamicum cells. FEMS Microbiol. Lett. 65:299-304. [DOI] [PubMed] [Google Scholar]
- 19.Möckel, B., L. Eggeling, and H. Sahm. 1994. Threonine dehydratases of Corynebacterium glutamicum with altered allosteric control: their generation and biochemical and structural analysis. Mol. Microbiol. 13:833-842. [DOI] [PubMed] [Google Scholar]
- 20.Morbach, S., C. Junger, H. Sahm, and L. Eggeling. 2000. Attenuation control of ilvBNC in Corynebacterium glutamicum: evidence of leader peptide formation without the presence of a ribosome binding site. J. Biosci. Bioeng. 90:501-507. [DOI] [PubMed] [Google Scholar]
- 21.Morbach, S., L. Eggeling, and H. Sahm. 1995. Use of feedback-resistant threonine dehydratases of Corynebacterium glutamicum to increase carbon flux towards l-isoleucine. Appl. Environ. Microbiol. 61:4315-4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morbach, S., R. Kelle, S. Winkels, L. Eggeling, and H. Sahm. 1996. Engineering the homoserine dehydrogenase and threonine dehydratase control points to analyse flux towards L-isoleucine in Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 45:612-620. [Google Scholar]
- 23.Nakayama, K., S. Kitada, and S. Kinoshita. 1961. L-Valine production using microbial auxotroph. J. Gen. Appl. Microbiol. Tokyo 7:52-69. [Google Scholar]
- 24.Niebisch, A., and M. Bott. 2001. Molecular analysis of the cytochrome bc1-aa3 branch of the Corynebacterium glutamicum respiratory chain containing an unusual diheme cytochrome c1. Arch. Microbiol. 175:282-294. [DOI] [PubMed] [Google Scholar]
- 25.Pátek, M., J. Hochmannová, M. Jelínková, J. Nesvera, and L. Eggeling. 1998. Analysis of the leuB gene from Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 50:42-47. [DOI] [PubMed] [Google Scholar]
- 26.Pátek, M., K. Krumbach, L. Eggeling, and H. Sahm. 1994. Leucine synthesis in Corynebacterium glutamicum: structure of leuA and effect of leuA inactivation on lysine synthesis. Appl. Environ. Microbiol. 60:133-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patnaik, R., R. G. Spitzer, and J. C. Liao. 1995. Pathway engineering for production of aromatics in Escherichia coli: confirmation of stoichiometric analysis by independent modulation of AroG, TktA, and Pps activities. Biotech. Bioeng. 46:361-370. [DOI] [PubMed] [Google Scholar]
- 28.Plachy, J. 1975. The effect of medium composition on the production of valine by Corynebacterium 9366-EMS/184. Folia Microbiol. 20:346-350. [DOI] [PubMed] [Google Scholar]
- 29.Reinscheid, D. J., B. J. Eikmanns, and H. Sahm. 1991. Analysis of a Corynebacterium glutamicum hom gene coding for a feedback-resistant homoserine dehydrogenase. J. Bacteriol. 173:3228-3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reinscheid, D. J., W. Kronemeyer, L. Eggeling, B. J. Eikmanns, and H. Sahm. 1994. Stable expression of hom-1-thrB in Corynebacterium glutamicum and its effects on the carbon flux to threonine and related amino acids. Appl. Environ. Microbiol. 60:126-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Riedel, C., D. Rittmann, P. Dangel, B. Möckel, S. Petersen, H. Sahm, and B. J. Eikmanns. 2001. Characterization of the phosphoenolpyruvate carboxykinase gene from Corynebacterium glutamicum and significance of the enzyme for growth and amino acid production. J. Mol. Microbiol. Biotechnol. 3:573-583. [PubMed] [Google Scholar]
- 32.Sahm, H., and L. Eggeling. 1999. d-Pantothenate synthesis in Corynebacterium glutamicum and use of panBC and genes of l-valine synthesis for its overproduction. Appl. Environ. Microbiol. 65:1973-1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schäfer, A., A. Tauch, W. Jäger, J. Kalinowski, G. Thierbach, and A. Pühler. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 34.Schwinde, J. W., P. F. Hertz, H. Sahm, B. J. Eikmanns, and A. Guyonvarch. 2001. Lipoamide dehydrogenase from Corynebacterium glutamicum: molecular and physiological analysis of the lpd gene and characterization of the enzyme. Microbiology 147:2223-2231. [DOI] [PubMed] [Google Scholar]
- 35.Simic, P. L. Eggeling, and H. Sahm. 2001. l-Threonine export: use of peptides to identify a new translocator from Corynebacterium glutamicum. J. Bacteriol. 183:5317-5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tosaka, O., Y. Yoshihara, S. Ikeda, and K. Takinami. 1985. Production of L-lysine by fluoropyruvate-sensitive mutants of Brevibacterium lactofermentum. Agric. Biol. Chem. Tokyo 49:1305-1312. [Google Scholar]
- 37.Tsuchida, T., and H. Momose. 1975. Genetic changes of regulatory mechanisms occurred in leucine and valine producing mutants derived from Brevibacterium lactofermentum 2257. Agric. Biol. Chem. Tokyo 39:2193-2198. [Google Scholar]
- 38.Tsuchida, T., F. Yoshinaga, and K. Kubota. 1975. Production of L-valine by 2-thiazolealanine resistant mutants derived from glutamic acid bacteria. Agric. Biol. Chem. Tokyo 39:1319-1322. [Google Scholar]
- 39.Udaka, S., and S. Kinoshita. 1960. The fermentative production of L-valine by bacteria. J. Gen. Appl. Microbiol. Tokyo 5:160-174. [Google Scholar]
- 40.Usuda, Y., N. Tujimoto, C. Abe, Y. Asakura, E. Kimura, Y. Kawahara, O. Kurahashi, and H. Matsui. 1996. Molecular cloning of the Corynebacterium glutamicum (Brevibacterium lactofermentum AJ12036) odhA gene encoding a novel type of 2-oxoglutarate dehydrogenase. Microbiology 142:3347-3354. [DOI] [PubMed] [Google Scholar]
- 41.Vrljić, M., L. Eggeling, and H. Sahm. 1996. A new type of transporter with a new type of cellular function: l-lysine export from Corynebacterium glutamicum. Mol. Microbiol. 22:815-826. [DOI] [PubMed] [Google Scholar]
- 42.Yokota, A., Shimizu, H., Terasawa, Y., Takaoka, N., and F. Tomita. 1994. Pyruvic acid production by a lipoic acid auxotrophic of Escherichia coli W1485. Appl. Microbiol. Biotechnol. 41:638-643. [DOI] [PubMed] [Google Scholar]