Abstract
We calculate room temperature thermal fluctuational base pair opening probability of a daunomycin-poly d(GCAT).poly d(ATGC) complex. This system is constructed at an atomic level of detail based on x-ray analysis of a crystal structure. The base pair opening probabilities are calculated from a modified self-consistent phonon approach of anharmonic lattice dynamics theory. We find that daunomycin binding substantially enhances the thermal stability of one of the base pairs adjacent the drug because of strong hydrogen bonding between the drug and the base. The possible effect of this enhanced stability on the drug inhibition of DNA transcription and replication is discussed. We also calculate the probability of drug dissociation from the helix based on the selfconsistent calculation of the probability of the disruption of drug-base H-bonds and the unstacking probability of the drug. The calculations can be used to determine the equilibrium drug binding constant which is found to be in good agreement with observations on similar daunomycin-DNA systems.
Full text
PDF
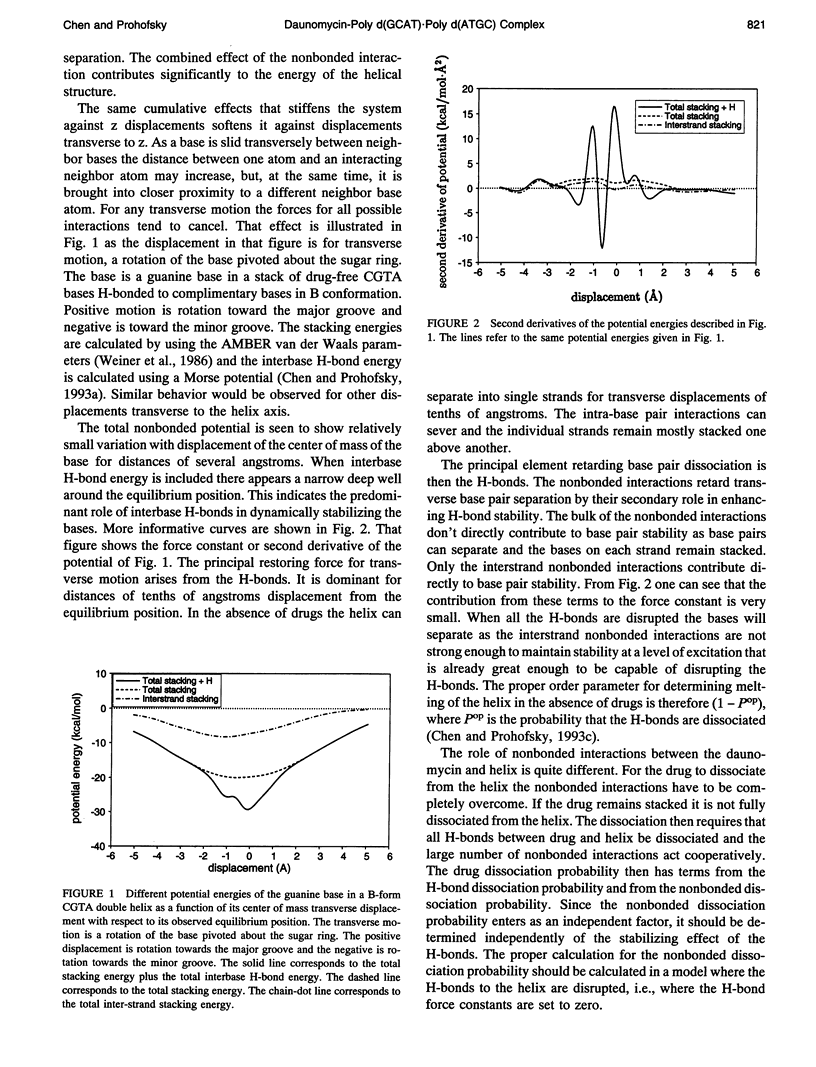
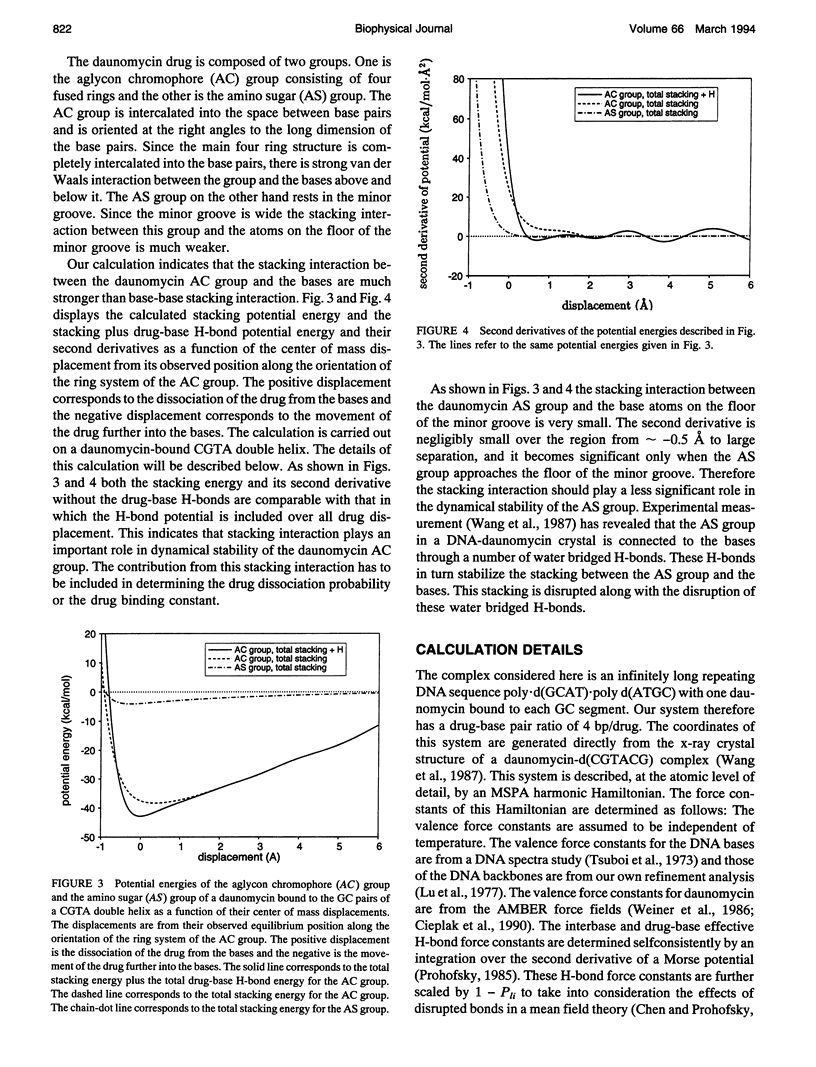




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chen Y. Z., Prohofsky E. W. Synergistic effects in the melting of DNA hydration shell: melting of the minor groove hydration spine in poly(dA).poly(dT) and its effect on base pair stability. Biophys J. 1993 May;64(5):1385–1393. doi: 10.1016/S0006-3495(93)81504-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. Z., Prohofsky E. W. The role of a minor groove spine of hydration in stabilizing poly(dA).poly(dT) against fluctuational interbase H-bond disruption in the premelting temperature regime. Nucleic Acids Res. 1992 Feb 11;20(3):415–419. doi: 10.1093/nar/20.3.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. Z., Zhuang W., Prohofsky E. W. Energy flow considerations and thermal fluctuational opening of DNA base pairs at a replicating fork: unwinding consistent with observed replication rates. J Biomol Struct Dyn. 1992 Oct;10(2):415–427. doi: 10.1080/07391102.1992.10508656. [DOI] [PubMed] [Google Scholar]
- Chen Y. Z., Zhuang W., Prohofsky E. W. Premelting thermal fluctuational interbase hydrogen-bond disrupted states of a B-DNA guanine-cytosine base pair: significance for amino and imino proton exchange. Biopolymers. 1991 Oct;31(11):1273–1281. doi: 10.1002/bip.360311105. [DOI] [PubMed] [Google Scholar]
- Chuprina V. P. Anomalous structure and properties of poly (dA).poly(dT). Computer simulation of the polynucleotide structure with the spine of hydration in the minor groove. Nucleic Acids Res. 1987 Jan 12;15(1):293–311. doi: 10.1093/nar/15.1.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuprina V. P., Heinemann U., Nurislamov A. A., Zielenkiewicz P., Dickerson R. E., Saenger W. Molecular dynamics simulation of the hydration shell of a B-DNA decamer reveals two main types of minor-groove hydration depending on groove width. Proc Natl Acad Sci U S A. 1991 Jan 15;88(2):593–597. doi: 10.1073/pnas.88.2.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cieplak P., Rao S. N., Grootenhuis P. D., Kollman P. A. Free energy calculation on base specificity of drug--DNA interactions: application to daunomycin and acridine intercalation into DNA. Biopolymers. 1990 Mar-Apr;29(4-5):717–727. doi: 10.1002/bip.360290406. [DOI] [PubMed] [Google Scholar]
- Lu K. C., Prohofsky E. W., Van Zandt L. L. Vibrational modes of A-DNA, B-DNA, and A-RNA backbones: an application of a green-function refinement procedure. Biopolymers. 1977 Nov;16(11):2491–2506. doi: 10.1002/bip.1977.360161112. [DOI] [PubMed] [Google Scholar]
- Newlin D. D., Miller K. J., Pilch D. F. Interactions of molecules with nucleic acids. VII. Intercalation and T.A specificity of daunomycin in DNA. Biopolymers. 1984 Jan;23(1):139–158. doi: 10.1002/bip.360230111. [DOI] [PubMed] [Google Scholar]
- Prabhu VV, Young L, Prohofsky EW. Hydrogen-bond melting in B-DNA copolymers in a mean-field self-consistent phonon approach. Phys Rev B Condens Matter. 1989 Mar 15;39(8):5436–5443. doi: 10.1103/physrevb.39.5436. [DOI] [PubMed] [Google Scholar]
- Quintana J. R., Grzeskowiak K., Yanagi K., Dickerson R. E. Structure of a B-DNA decamer with a central T-A step: C-G-A-T-T-A-A-T-C-G. J Mol Biol. 1992 May 20;225(2):379–395. doi: 10.1016/0022-2836(92)90928-d. [DOI] [PubMed] [Google Scholar]
- Remeta D. P., Mudd C. P., Berger R. L., Breslauer K. J. Thermodynamic characterization of daunomycin-DNA interactions: comparison of complete binding profiles for a series of DNA host duplexes. Biochemistry. 1993 May 18;32(19):5064–5073. doi: 10.1021/bi00070a014. [DOI] [PubMed] [Google Scholar]
- Wang A. H., Ughetto G., Quigley G. J., Rich A. Interactions between an anthracycline antibiotic and DNA: molecular structure of daunomycin complexed to d(CpGpTpApCpG) at 1.2-A resolution. Biochemistry. 1987 Feb 24;26(4):1152–1163. doi: 10.1021/bi00378a025. [DOI] [PubMed] [Google Scholar]


