Abstract
We discuss the requirement of type II DNA topoisomerase in the process of mitotic chromosome condensation. Using a known model describing the collapse of homopolymers, we propose that the compaction process necessitates a change in the topological state (i.e., a self-knotting) of the chromosomal chain. We argue that the enzymes are necessary to reach the compact metaphase state in a time interval that is much smaller than the time expected in the uncatalyzed process. The folding process is such that the potential entanglement points are localized at particular regions of the chromosome known as the scaffold-associated regions. The concentration of entanglements in the metaphase chromosome is related to the average size of the radial loops. A phantom chain model for the condensation process, in which each potential entanglement point is dealt with by a topoisomerase II molecule, is proposed.
Full text
PDF

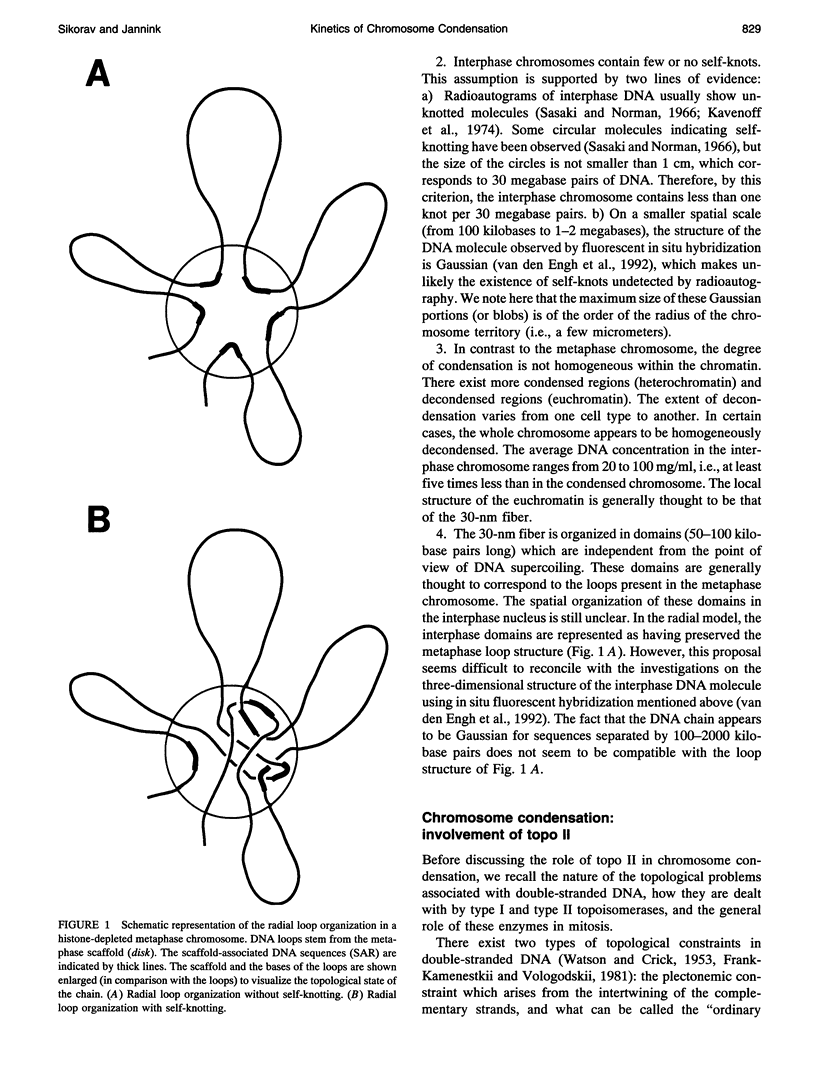








Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adachi Y., Käs E., Laemmli U. K. Preferential, cooperative binding of DNA topoisomerase II to scaffold-associated regions. EMBO J. 1989 Dec 20;8(13):3997–4006. doi: 10.1002/j.1460-2075.1989.tb08582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adachi Y., Luke M., Laemmli U. K. Chromosome assembly in vitro: topoisomerase II is required for condensation. Cell. 1991 Jan 11;64(1):137–148. doi: 10.1016/0092-8674(91)90215-k. [DOI] [PubMed] [Google Scholar]
- Boles T. C., White J. H., Cozzarelli N. R. Structure of plectonemically supercoiled DNA. J Mol Biol. 1990 Jun 20;213(4):931–951. doi: 10.1016/S0022-2836(05)80272-4. [DOI] [PubMed] [Google Scholar]
- Boy de la Tour E., Laemmli U. K. The metaphase scaffold is helically folded: sister chromatids have predominantly opposite helical handedness. Cell. 1988 Dec 23;55(6):937–944. doi: 10.1016/0092-8674(88)90239-5. [DOI] [PubMed] [Google Scholar]
- DiNardo S., Voelkel K., Sternglanz R. DNA topoisomerase II mutant of Saccharomyces cerevisiae: topoisomerase II is required for segregation of daughter molecules at the termination of DNA replication. Proc Natl Acad Sci U S A. 1984 May;81(9):2616–2620. doi: 10.1073/pnas.81.9.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw W. C., Heck M. M. Localization of topoisomerase II in mitotic chromosomes. J Cell Biol. 1985 May;100(5):1716–1725. doi: 10.1083/jcb.100.5.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried M. G., Bloomfield V. A. DNA gelation in concentrated solutions. Biopolymers. 1984 Nov;23(11 Pt 1):2141–2155. doi: 10.1002/bip.360231104. [DOI] [PubMed] [Google Scholar]
- Gautier T., Robert-Nicoud M., Guilly M. N., Hernandez-Verdun D. Relocation of nucleolar proteins around chromosomes at mitosis. A study by confocal laser scanning microscopy. J Cell Sci. 1992 Aug;102(Pt 4):729–737. doi: 10.1242/jcs.102.4.729. [DOI] [PubMed] [Google Scholar]
- Haaf T., Schmid M. Chromosome topology in mammalian interphase nuclei. Exp Cell Res. 1991 Feb;192(2):325–332. doi: 10.1016/0014-4827(91)90048-y. [DOI] [PubMed] [Google Scholar]
- Heck M. M., Hittelman W. N., Earnshaw W. C. Differential expression of DNA topoisomerases I and II during the eukaryotic cell cycle. Proc Natl Acad Sci U S A. 1988 Feb;85(4):1086–1090. doi: 10.1073/pnas.85.4.1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T., Mitchison T. J. Topoisomerase II does not play a scaffolding role in the organization of mitotic chromosomes assembled in Xenopus egg extracts. J Cell Biol. 1993 Feb;120(3):601–612. doi: 10.1083/jcb.120.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraoka Y., Minden J. S., Swedlow J. R., Sedat J. W., Agard D. A. Focal points for chromosome condensation and decondensation revealed by three-dimensional in vivo time-lapse microscopy. Nature. 1989 Nov 16;342(6247):293–296. doi: 10.1038/342293a0. [DOI] [PubMed] [Google Scholar]
- Holm C., Goto T., Wang J. C., Botstein D. DNA topoisomerase II is required at the time of mitosis in yeast. Cell. 1985 Jun;41(2):553–563. doi: 10.1016/s0092-8674(85)80028-3. [DOI] [PubMed] [Google Scholar]
- Jencks W. P. Binding energy, specificity, and enzymic catalysis: the circe effect. Adv Enzymol Relat Areas Mol Biol. 1975;43:219–410. doi: 10.1002/9780470122884.ch4. [DOI] [PubMed] [Google Scholar]
- Kavenoff R., Klotz L. C., Zimm B. H. One the nature of chromosome-sized DNA molecules. Cold Spring Harb Symp Quant Biol. 1974;38:1–8. doi: 10.1101/sqb.1974.038.01.003. [DOI] [PubMed] [Google Scholar]
- Kavenoff R., Zimm B. H. Chromosome-sized DNA molecules from Drosophila. Chromosoma. 1973;41(1):1–27. doi: 10.1007/BF00284071. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K., Käs E., Poljak L., Adachi Y. Scaffold-associated regions: cis-acting determinants of chromatin structural loops and functional domains. Curr Opin Genet Dev. 1992 Apr;2(2):275–285. doi: 10.1016/s0959-437x(05)80285-0. [DOI] [PubMed] [Google Scholar]
- Lindsley J. E., Wang J. C. On the coupling between ATP usage and DNA transport by yeast DNA topoisomerase II. J Biol Chem. 1993 Apr 15;268(11):8096–8104. [PubMed] [Google Scholar]
- Liu L. F., Liu C. C., Alberts B. M. Type II DNA topoisomerases: enzymes that can unknot a topologically knotted DNA molecule via a reversible double-strand break. Cell. 1980 Mar;19(3):697–707. doi: 10.1016/s0092-8674(80)80046-8. [DOI] [PubMed] [Google Scholar]
- Manuelidis L. A view of interphase chromosomes. Science. 1990 Dec 14;250(4987):1533–1540. doi: 10.1126/science.2274784. [DOI] [PubMed] [Google Scholar]
- McDowall A. W., Smith J. M., Dubochet J. Cryo-electron microscopy of vitrified chromosomes in situ. EMBO J. 1986 Jun;5(6):1395–1402. doi: 10.1002/j.1460-2075.1986.tb04373.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeon F. D., Tuffanelli D. L., Kobayashi S., Kirschner M. W. The redistribution of a conserved nuclear envelope protein during the cell cycle suggests a pathway for chromosome condensation. Cell. 1984 Jan;36(1):83–92. doi: 10.1016/0092-8674(84)90076-x. [DOI] [PubMed] [Google Scholar]
- Newport J., Spann T. Disassembly of the nucleus in mitotic extracts: membrane vesicularization, lamin disassembly, and chromosome condensation are independent processes. Cell. 1987 Jan 30;48(2):219–230. doi: 10.1016/0092-8674(87)90425-9. [DOI] [PubMed] [Google Scholar]
- Ott G. S., Bastia D., Bauer W. A spectroscopic and electron microscopic examination of the highly condensed DNA structures formed by denaturation in Mg(ClO4)2. Biochim Biophys Acta. 1978 Apr 27;518(2):216–232. doi: 10.1016/0005-2787(78)90179-x. [DOI] [PubMed] [Google Scholar]
- Porschke D. Dynamics of DNA condensation. Biochemistry. 1984 Oct 9;23(21):4821–4828. doi: 10.1021/bi00316a002. [DOI] [PubMed] [Google Scholar]
- Post C. B., Zimm B. H. Light-scattering study of DNA condensation: competition between collapse and aggregation. Biopolymers. 1982 Nov;21(11):2139–2160. doi: 10.1002/bip.360211105. [DOI] [PubMed] [Google Scholar]
- Rattner J. B., Lin C. C. Radial loops and helical coils coexist in metaphase chromosomes. Cell. 1985 Aug;42(1):291–296. doi: 10.1016/s0092-8674(85)80124-0. [DOI] [PubMed] [Google Scholar]
- Rika J, Meewes M, Nyffenegger R, Binkert T. Intermolecular and intramolecular solubilization: Collapse and expansion of a polymer chain in surfactant solutions. Phys Rev Lett. 1990 Jul 30;65(5):657–660. doi: 10.1103/PhysRevLett.65.657. [DOI] [PubMed] [Google Scholar]
- Sasaki M. S., Norman A. DNA fibres from human lymphocyte nuclei. Exp Cell Res. 1966 Nov-Dec;44(2):642–645. doi: 10.1016/0014-4827(66)90474-5. [DOI] [PubMed] [Google Scholar]
- Schwartz D. C., Cantor C. R. Separation of yeast chromosome-sized DNAs by pulsed field gradient gel electrophoresis. Cell. 1984 May;37(1):67–75. doi: 10.1016/0092-8674(84)90301-5. [DOI] [PubMed] [Google Scholar]
- Shamu C. E., Murray A. W. Sister chromatid separation in frog egg extracts requires DNA topoisomerase II activity during anaphase. J Cell Biol. 1992 Jun;117(5):921–934. doi: 10.1083/jcb.117.5.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundin O., Varshavsky A. Terminal stages of SV40 DNA replication proceed via multiply intertwined catenated dimers. Cell. 1980 Aug;21(1):103–114. doi: 10.1016/0092-8674(80)90118-x. [DOI] [PubMed] [Google Scholar]
- Swedlow J. R., Sedat J. W., Agard D. A. Multiple chromosomal populations of topoisomerase II detected in vivo by time-lapse, three-dimensional wide-field microscopy. Cell. 1993 Apr 9;73(1):97–108. doi: 10.1016/0092-8674(93)90163-k. [DOI] [PubMed] [Google Scholar]
- Uemura T., Ohkura H., Adachi Y., Morino K., Shiozaki K., Yanagida M. DNA topoisomerase II is required for condensation and separation of mitotic chromosomes in S. pombe. Cell. 1987 Sep 11;50(6):917–925. doi: 10.1016/0092-8674(87)90518-6. [DOI] [PubMed] [Google Scholar]
- Uemura T., Tanagida M. Mitotic spindle pulls but fails to separate chromosomes in type II DNA topoisomerase mutants: uncoordinated mitosis. EMBO J. 1986 May;5(5):1003–1010. doi: 10.1002/j.1460-2075.1986.tb04315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WATSON J. D., CRICK F. H. The structure of DNA. Cold Spring Harb Symp Quant Biol. 1953;18:123–131. doi: 10.1101/sqb.1953.018.01.020. [DOI] [PubMed] [Google Scholar]
- Wang J. C. DNA topoisomerases: why so many? J Biol Chem. 1991 Apr 15;266(11):6659–6662. [PubMed] [Google Scholar]
- van den Engh G., Sachs R., Trask B. J. Estimating genomic distance from DNA sequence location in cell nuclei by a random walk model. Science. 1992 Sep 4;257(5075):1410–1412. doi: 10.1126/science.1388286. [DOI] [PubMed] [Google Scholar]


