Abstract
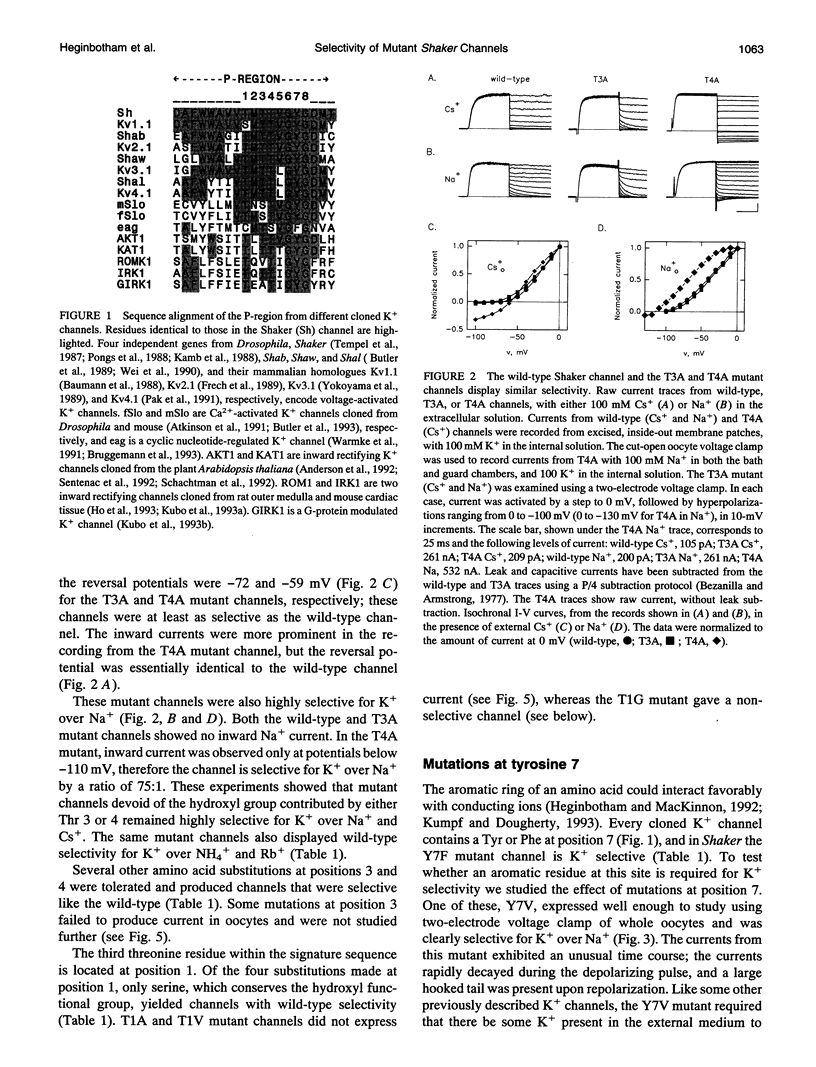
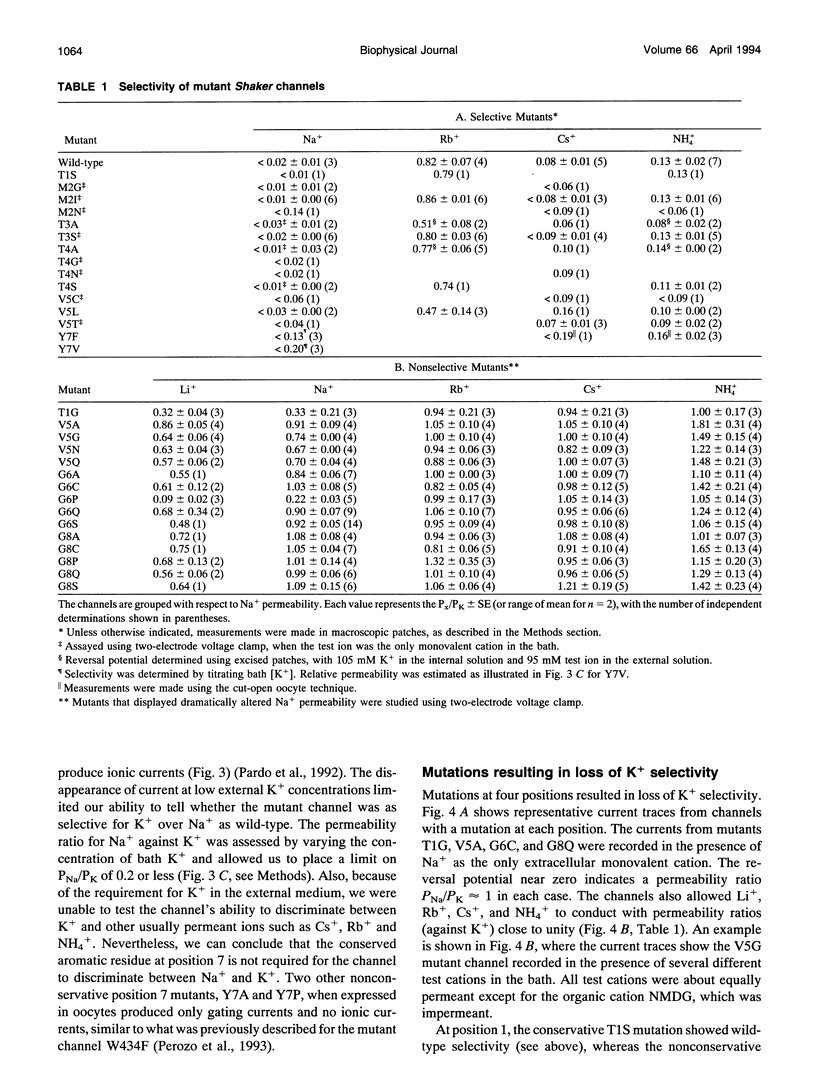
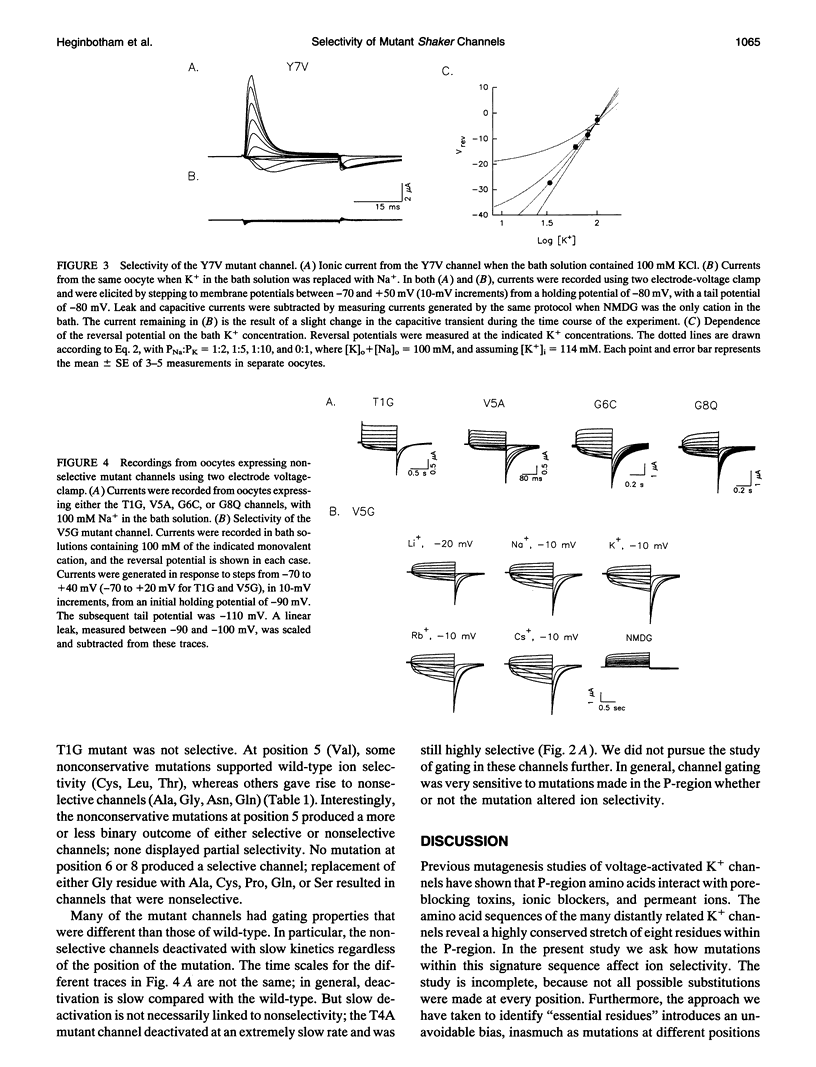
Potassium channels share a highly conserved stretch of eight amino acids, a K+ channel signature sequence. The conserved sequence falls within the previously defined P-region of voltage-activated K+ channels. In this study we investigate the effect of mutations in the signature sequence of the Shaker channel on K+ selectivity determined under bi-ionic conditions. Nonconservative substitutions of two threonine residues and the tyrosine residue leave selectivity intact. In contrast, mutations at some positions render the channel nonselective among monovalent cations. These findings are consistent with a proposal that the signature sequence contributes to a selectivity filter. Furthermore, the results illustrate that the hydroxyl groups at the third and fourth positions, and the aromatic group at position seven, are not essential in determining K+ selectivity.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Acharya K. R., Stuart D. I., Walker N. P., Lewis M., Phillips D. C. Refined structure of baboon alpha-lactalbumin at 1.7 A resolution. Comparison with C-type lysozyme. J Mol Biol. 1989 Jul 5;208(1):99–127. doi: 10.1016/0022-2836(89)90091-0. [DOI] [PubMed] [Google Scholar]
- Anderson J. A., Huprikar S. S., Kochian L. V., Lucas W. J., Gaber R. F. Functional expression of a probable Arabidopsis thaliana potassium channel in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1992 May 1;89(9):3736–3740. doi: 10.1073/pnas.89.9.3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson N. S., Robertson G. A., Ganetzky B. A component of calcium-activated potassium channels encoded by the Drosophila slo locus. Science. 1991 Aug 2;253(5019):551–555. doi: 10.1126/science.1857984. [DOI] [PubMed] [Google Scholar]
- Baumann A., Grupe A., Ackermann A., Pongs O. Structure of the voltage-dependent potassium channel is highly conserved from Drosophila to vertebrate central nervous systems. EMBO J. 1988 Aug;7(8):2457–2463. doi: 10.1002/j.1460-2075.1988.tb03092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezanilla F., Armstrong C. M. Inactivation of the sodium channel. I. Sodium current experiments. J Gen Physiol. 1977 Nov;70(5):549–566. doi: 10.1085/jgp.70.5.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezanilla F., Armstrong C. M. Negative conductance caused by entry of sodium and cesium ions into the potassium channels of squid axons. J Gen Physiol. 1972 Nov;60(5):588–608. doi: 10.1085/jgp.60.5.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brüggemann A., Pardo L. A., Stühmer W., Pongs O. Ether-à-go-go encodes a voltage-gated channel permeable to K+ and Ca2+ and modulated by cAMP. Nature. 1993 Sep 30;365(6445):445–448. doi: 10.1038/365445a0. [DOI] [PubMed] [Google Scholar]
- Butler A., Tsunoda S., McCobb D. P., Wei A., Salkoff L. mSlo, a complex mouse gene encoding "maxi" calcium-activated potassium channels. Science. 1993 Jul 9;261(5118):221–224. doi: 10.1126/science.7687074. [DOI] [PubMed] [Google Scholar]
- Butler A., Wei A. G., Baker K., Salkoff L. A family of putative potassium channel genes in Drosophila. Science. 1989 Feb 17;243(4893):943–947. doi: 10.1126/science.2493160. [DOI] [PubMed] [Google Scholar]
- Choi K. L., Mossman C., Aubé J., Yellen G. The internal quaternary ammonium receptor site of Shaker potassium channels. Neuron. 1993 Mar;10(3):533–541. doi: 10.1016/0896-6273(93)90340-w. [DOI] [PubMed] [Google Scholar]
- Frech G. C., VanDongen A. M., Schuster G., Brown A. M., Joho R. H. A novel potassium channel with delayed rectifier properties isolated from rat brain by expression cloning. Nature. 1989 Aug 24;340(6235):642–645. doi: 10.1038/340642a0. [DOI] [PubMed] [Google Scholar]
- Goldman D. E. POTENTIAL, IMPEDANCE, AND RECTIFICATION IN MEMBRANES. J Gen Physiol. 1943 Sep 20;27(1):37–60. doi: 10.1085/jgp.27.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HODGKIN A. L., KATZ B. The effect of sodium ions on the electrical activity of giant axon of the squid. J Physiol. 1949 Mar 1;108(1):37–77. doi: 10.1113/jphysiol.1949.sp004310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamill O. P., Marty A., Neher E., Sakmann B., Sigworth F. J. Improved patch-clamp techniques for high-resolution current recording from cells and cell-free membrane patches. Pflugers Arch. 1981 Aug;391(2):85–100. doi: 10.1007/BF00656997. [DOI] [PubMed] [Google Scholar]
- Hartmann H. A., Kirsch G. E., Drewe J. A., Taglialatela M., Joho R. H., Brown A. M. Exchange of conduction pathways between two related K+ channels. Science. 1991 Feb 22;251(4996):942–944. doi: 10.1126/science.2000495. [DOI] [PubMed] [Google Scholar]
- Heginbotham L., Abramson T., MacKinnon R. A functional connection between the pores of distantly related ion channels as revealed by mutant K+ channels. Science. 1992 Nov 13;258(5085):1152–1155. doi: 10.1126/science.1279807. [DOI] [PubMed] [Google Scholar]
- Heginbotham L., MacKinnon R. The aromatic binding site for tetraethylammonium ion on potassium channels. Neuron. 1992 Mar;8(3):483–491. doi: 10.1016/0896-6273(92)90276-j. [DOI] [PubMed] [Google Scholar]
- Hille B. Potassium channels in myelinated nerve. Selective permeability to small cations. J Gen Physiol. 1973 Jun;61(6):669–686. doi: 10.1085/jgp.61.6.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho K., Nichols C. G., Lederer W. J., Lytton J., Vassilev P. M., Kanazirska M. V., Hebert S. C. Cloning and expression of an inwardly rectifying ATP-regulated potassium channel. Nature. 1993 Mar 4;362(6415):31–38. doi: 10.1038/362031a0. [DOI] [PubMed] [Google Scholar]
- Hoshi T., Zagotta W. N., Aldrich R. W. Biophysical and molecular mechanisms of Shaker potassium channel inactivation. Science. 1990 Oct 26;250(4980):533–538. doi: 10.1126/science.2122519. [DOI] [PubMed] [Google Scholar]
- Kamb A., Iverson L. E., Tanouye M. A. Molecular characterization of Shaker, a Drosophila gene that encodes a potassium channel. Cell. 1987 Jul 31;50(3):405–413. doi: 10.1016/0092-8674(87)90494-6. [DOI] [PubMed] [Google Scholar]
- Kamb A., Tseng-Crank J., Tanouye M. A. Multiple products of the Drosophila Shaker gene may contribute to potassium channel diversity. Neuron. 1988 Jul;1(5):421–430. doi: 10.1016/0896-6273(88)90192-4. [DOI] [PubMed] [Google Scholar]
- Kavanaugh M. P., Hurst R. S., Yakel J., Varnum M. D., Adelman J. P., North R. A. Multiple subunits of a voltage-dependent potassium channel contribute to the binding site for tetraethylammonium. Neuron. 1992 Mar;8(3):493–497. doi: 10.1016/0896-6273(92)90277-k. [DOI] [PubMed] [Google Scholar]
- Kavanaugh M. P., Varnum M. D., Osborne P. B., Christie M. J., Busch A. E., Adelman J. P., North R. A. Interaction between tetraethylammonium and amino acid residues in the pore of cloned voltage-dependent potassium channels. J Biol Chem. 1991 Apr 25;266(12):7583–7587. [PubMed] [Google Scholar]
- Kim S. S., Smith T. J., Chapman M. S., Rossmann M. C., Pevear D. C., Dutko F. J., Felock P. J., Diana G. D., McKinlay M. A. Crystal structure of human rhinovirus serotype 1A (HRV1A). J Mol Biol. 1989 Nov 5;210(1):91–111. doi: 10.1016/0022-2836(89)90293-3. [DOI] [PubMed] [Google Scholar]
- Kirsch G. E., Drewe J. A., Hartmann H. A., Taglialatela M., de Biasi M., Brown A. M., Joho R. H. Differences between the deep pores of K+ channels determined by an interacting pair of nonpolar amino acids. Neuron. 1992 Mar;8(3):499–505. doi: 10.1016/0896-6273(92)90278-l. [DOI] [PubMed] [Google Scholar]
- Kubo Y., Baldwin T. J., Jan Y. N., Jan L. Y. Primary structure and functional expression of a mouse inward rectifier potassium channel. Nature. 1993 Mar 11;362(6416):127–133. doi: 10.1038/362127a0. [DOI] [PubMed] [Google Scholar]
- Kubo Y., Reuveny E., Slesinger P. A., Jan Y. N., Jan L. Y. Primary structure and functional expression of a rat G-protein-coupled muscarinic potassium channel. Nature. 1993 Aug 26;364(6440):802–806. doi: 10.1038/364802a0. [DOI] [PubMed] [Google Scholar]
- Kumpf R. A., Dougherty D. A. A mechanism for ion selectivity in potassium channels: computational studies of cation-pi interactions. Science. 1993 Sep 24;261(5129):1708–1710. doi: 10.1126/science.8378771. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez G. A., Jan Y. N., Jan L. Y. Evidence that the S6 segment of the Shaker voltage-gated K+ channel comprises part of the pore. Nature. 1994 Jan 13;367(6459):179–182. doi: 10.1038/367179a0. [DOI] [PubMed] [Google Scholar]
- MacKinnon R., Heginbotham L., Abramson T. Mapping the receptor site for charybdotoxin, a pore-blocking potassium channel inhibitor. Neuron. 1990 Dec;5(6):767–771. doi: 10.1016/0896-6273(90)90335-d. [DOI] [PubMed] [Google Scholar]
- MacKinnon R., Miller C. Mutant potassium channels with altered binding of charybdotoxin, a pore-blocking peptide inhibitor. Science. 1989 Sep 22;245(4924):1382–1385. doi: 10.1126/science.2476850. [DOI] [PubMed] [Google Scholar]
- MacKinnon R., Reinhart P. H., White M. M. Charybdotoxin block of Shaker K+ channels suggests that different types of K+ channels share common structural features. Neuron. 1988 Dec;1(10):997–1001. doi: 10.1016/0896-6273(88)90156-0. [DOI] [PubMed] [Google Scholar]
- MacKinnon R., Yellen G. Mutations affecting TEA blockade and ion permeation in voltage-activated K+ channels. Science. 1990 Oct 12;250(4978):276–279. doi: 10.1126/science.2218530. [DOI] [PubMed] [Google Scholar]
- Pak M. D., Baker K., Covarrubias M., Butler A., Ratcliffe A., Salkoff L. mShal, a subfamily of A-type K+ channel cloned from mammalian brain. Proc Natl Acad Sci U S A. 1991 May 15;88(10):4386–4390. doi: 10.1073/pnas.88.10.4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo L. A., Heinemann S. H., Terlau H., Ludewig U., Lorra C., Pongs O., Stühmer W. Extracellular K+ specifically modulates a rat brain K+ channel. Proc Natl Acad Sci U S A. 1992 Mar 15;89(6):2466–2470. doi: 10.1073/pnas.89.6.2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perozo E., MacKinnon R., Bezanilla F., Stefani E. Gating currents from a nonconducting mutant reveal open-closed conformations in Shaker K+ channels. Neuron. 1993 Aug;11(2):353–358. doi: 10.1016/0896-6273(93)90190-3. [DOI] [PubMed] [Google Scholar]
- Pongs O., Kecskemethy N., Müller R., Krah-Jentgens I., Baumann A., Kiltz H. H., Canal I., Llamazares S., Ferrus A. Shaker encodes a family of putative potassium channel proteins in the nervous system of Drosophila. EMBO J. 1988 Apr;7(4):1087–1096. doi: 10.1002/j.1460-2075.1988.tb02917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachtman D. P., Schroeder J. I., Lucas W. J., Anderson J. A., Gaber R. F. Expression of an inward-rectifying potassium channel by the Arabidopsis KAT1 cDNA. Science. 1992 Dec 4;258(5088):1654–1658. doi: 10.1126/science.8966547. [DOI] [PubMed] [Google Scholar]
- Sentenac H., Bonneaud N., Minet M., Lacroute F., Salmon J. M., Gaymard F., Grignon C. Cloning and expression in yeast of a plant potassium ion transport system. Science. 1992 May 1;256(5057):663–665. doi: 10.1126/science.1585180. [DOI] [PubMed] [Google Scholar]
- Slesinger P. A., Jan Y. N., Jan L. Y. The S4-S5 loop contributes to the ion-selective pore of potassium channels. Neuron. 1993 Oct;11(4):739–749. doi: 10.1016/0896-6273(93)90083-4. [DOI] [PubMed] [Google Scholar]
- Taglialatela M., Toro L., Stefani E. Novel voltage clamp to record small, fast currents from ion channels expressed in Xenopus oocytes. Biophys J. 1992 Jan;61(1):78–82. doi: 10.1016/S0006-3495(92)81817-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tempel B. L., Papazian D. M., Schwarz T. L., Jan Y. N., Jan L. Y. Sequence of a probable potassium channel component encoded at Shaker locus of Drosophila. Science. 1987 Aug 14;237(4816):770–775. doi: 10.1126/science.2441471. [DOI] [PubMed] [Google Scholar]
- Toney M. D., Hohenester E., Cowan S. W., Jansonius J. N. Dialkylglycine decarboxylase structure: bifunctional active site and alkali metal sites. Science. 1993 Aug 6;261(5122):756–759. doi: 10.1126/science.8342040. [DOI] [PubMed] [Google Scholar]
- Warmke J., Drysdale R., Ganetzky B. A distinct potassium channel polypeptide encoded by the Drosophila eag locus. Science. 1991 Jun 14;252(5012):1560–1562. doi: 10.1126/science.1840699. [DOI] [PubMed] [Google Scholar]
- Wei A., Covarrubias M., Butler A., Baker K., Pak M., Salkoff L. K+ current diversity is produced by an extended gene family conserved in Drosophila and mouse. Science. 1990 May 4;248(4955):599–603. doi: 10.1126/science.2333511. [DOI] [PubMed] [Google Scholar]
- Yellen G., Jurman M. E., Abramson T., MacKinnon R. Mutations affecting internal TEA blockade identify the probable pore-forming region of a K+ channel. Science. 1991 Feb 22;251(4996):939–942. doi: 10.1126/science.2000494. [DOI] [PubMed] [Google Scholar]
- Yokoyama S., Imoto K., Kawamura T., Higashida H., Iwabe N., Miyata T., Numa S. Potassium channels from NG108-15 neuroblastoma-glioma hybrid cells. Primary structure and functional expression from cDNAs. FEBS Lett. 1989 Dec 18;259(1):37–42. doi: 10.1016/0014-5793(89)81488-7. [DOI] [PubMed] [Google Scholar]
- Yool A. J., Schwarz T. L. Alteration of ionic selectivity of a K+ channel by mutation of the H5 region. Nature. 1991 Feb 21;349(6311):700–704. doi: 10.1038/349700a0. [DOI] [PubMed] [Google Scholar]




