Abstract
Buchnera strains from most aphid subfamilies studied to date have been found to carry the leucine gene cluster (leuA, -B, -C, and -D) on a plasmid, an organization unique among bacteria. Here, however, we demonstrate a classical chromosomal location of the cluster in Buchnera sp. strain PSY from the aphid Pemphigus spyrothecae (subfamily Pemphiginae). The genes that flank leuABCD in Buchnera sp. strain PSY appear to be adjacent in the genome of Buchnera sp. strain APS, a strain carrying a leucine plasmid. We propose that the presence of a leucine plasmid predates the diversification of symbiotic Buchnera and that the chromosomal location observed in Buchnera sp. strain PSY arose by a transfer of the leucine genes from a plasmid to the chromosome.
Aphids are plant-sap-feeding insects that maintain a mutualistic association with bacteria from the genus Buchnera (Proteobacteria) harbored in specialized host cells called bacteriocytes (1, 11). The origin of the aphid-Buchnera association has been estimated at about 200 to 250 million years ago, and host and symbiont lineages have subsequently diverged strictly in parallel (10). The major role of Buchnera in the symbiosis is the provision of amino acids, nutrients that are in short supply in phloem sap (4). The complete genome of Buchnera from the aphid Acyrthosiphon pisum Harris (1776) (Buchnera sp. strain APS), has recently been sequenced (13). It is composed of a chromosome of 641 kb and two small plasmids. One of these plasmids contains genes for tryptophan biosynthesis (7, 12, 18), while the other contains genes for leucine biosynthesis (2, 3, 14, 16, 17, 19). Amplification on plasmids of essential amino acid biosynthesis genes is considered indicative of Buchnera's capacity to overproduce these nutrients to the benefit of its host and hence an adaptation to its symbiotic lifestyle (2).
The first leucine plasmid in Buchnera (pRPE) was described using Rhopalosiphum padi (Linné 1758) (3), a member of the subfamily Aphidinae. It contains genes encoding the key enzymes in the pathway leading to leucine, in the same order as in Escherichia coli (leuABCD), two copies of the replication initiation gene repA, and ORF1, a putative integral membrane protein. Identical plasmids have been shown to be universally present in Buchnera from the subfamily Aphidinae (2, 16), while leucine plasmids with slightly different gene orders and/or gene contents are carried by Buchnera from the subfamilies Pterocommatinae, Thelaxinae, and Lachninae (14, 17, 19). Finally, Buchnera from the subfamily Pemphiginae has been found to carry cryptic plasmids of the same repA1 family that do not encode the structural leucine genes. These are very small plasmids containing only an origin of replication, one or two copies of the repA gene, and one additional gene (ibp, coding for a heat shock protein or ORF1), depending on the tribe within the subfamily (17).
In the present study, we have sequenced the chromosomally encoded leucine genes and flanking regions from Buchnera of the aphid Pemphigus spyrothecae (Passerini 1856) (Buchnera sp. strain PSY), a member of the tribe Pemphigini. This strain carries a small plasmid with the genes repA1 and ibp (19). Why all lineages of Buchnera have not undergone amplification of essential amino acid biosynthesis genes via plasmids is an open and unresolved question but is likely to be related to the quality of phloem sap from different host plants and/or the growth rate of the aphids.
Amplification of the leucine gene cluster and flanking regions
P. spyrothecae was collected from galls of black poplar trees (Populus nigra). DNA was prepared by the cetyltrimethylammonium bromide-NaCl method, after isolation of bacteria from the aphids (5, 19).
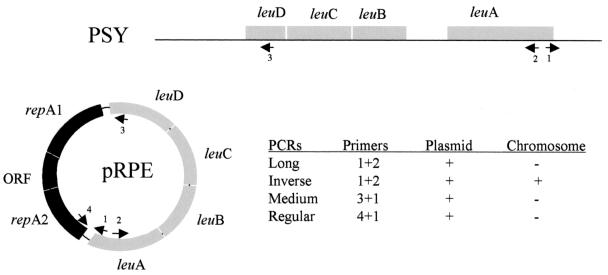
Discrimination between plasmid or chromosomal location of the leucine genes, as well as determination of gene order, followed the procedure of van Ham et al. (17), which is based on a combination of structural PCRs (as shown in Fig. 1) and restriction mapping, followed by Southern blot hybridization. This procedure indicates a chromosomal location of the leucine cluster in Buchnera sp. strain PSY. The resulting restriction map of the leucine cluster chromosomal region showed a 7.4-kb band generated with XbaI that contains the complete cluster and flanking regions (data not shown), selecting this enzyme for the inverse PCR (iPCR) procedure. About 1 μg of Buchnera sp. strain PSY DNA was cut with XbaI and purified on a Qiagen anion-exchange column, and fragments were ligated with T4 DNA ligase (Stratagene). This DNA was subjected to iPCR, using primers leuA_du2 (with a PstI restriction site) and leuA_dl3 (with a SalI restriction site) and the Expand Long Template PCR System (Roche) with a GeneAmp PCR System 2400 thermal cycler (Perkin-Elmer). PCR conditions were as follows: 92°C for 2 min; 10 cycles of 92°C for 10 s, 62°C for 30 s, and 68°C for 10 min; this was followed by 20 more cycles with an autoextension of 20 s/cycle at 68°C and a final extension of 68°C for 7 min.
FIG. 1.
Positions of primers in pRPE plasmid and Buchnera sp. strain PSY chromosome (PSY). The table shows combination of primers and results of structural PCRs according to van Ham et al. (17). +, successful amplification; −, no amplification. Primer names: 1, leuA_du2; 2, leuA_dl3; 3, Leud_up2; 4, repAd2.
Cloning and sequencing
The 7.0-kb fragment obtained by iPCR was purified with the GeneClean II kit (BIO-101), digested with PstI/SalI, and cloned into pUC18, also digested with PstI/SalI. One selected recombinant clone, named ps10, contained two internal EcoRV sites yielding three fragments of 2.6, 3.7, and 0.7 kb plus the vector. The two first fragments were then subcloned into pUC18 EcoRV digested and subjected to nested deletions (Nested Deletion kit; AP Biotech) for complete sequencing. The third one was religated and directly sequenced. Sequencing of all clones was carried out with an ABI 377 automated sequencer using the dRhodamine Dye Terminator Cycle Sequencing kit (Perkin-Elmer) and universal primers T3, T7, UNI17-mer, and UNIrev, as well as specific primers designed to close gaps.
The iPCR with outwardly oriented primers within leuA lack 389 nucleotides (nt) of the original leuA gene. For completion of this sequence, two specific primers were designed and used in a regular PCR to generate a 500-nt fragment containing the missing leuA fragment (PsleuA-F, 5′-CGT GAT GGA CAA GCT TTA ACA AG-3′; PsleuA-R, 5′-TGG AAT TGT AAA ACC GAC AGT ATC-3′). The PCR fragment was purified, cloned into pGEM-T Easy vector (Promega), and sequenced as above.
DNA sequence data were assembled with SEQUENCHER, version 4.0 (GeneCodes). BLAST-X, version 2.2.1, was used for gene assignment of ORF proteins (http://www.ncbi.nlm.nih.gov/BLAST).
Structure of the Buchnera sp. strain PSY leucine gene cluster
The 7.4-kb chromosome region characterized contains seven genes in the following order: rep-leuA-leuB-leuC-leuD-trxA-rho (Fig. 2c), whereas in the chromosome of Buchnera sp. strain APS, the genes rep and trxA are adjacent (13) and the leucine gene cluster is located on a plasmid (16).
FIG. 2.
The three steps of back transfer scenario. (a) ancestral chromosome; (b) intermediate stage; (c) Buchnera sp. strain PSY chromosome. The black line indicates the fragment sequenced; short open arrows indicate tRNA (13).
In Buchnera sp. strain PSY, the intergenic region between leuA and leuB is 441 bp long, suggesting a loss of the leuABCD operon structure found in most other bacteria. No ORF proteins were found in this region. In contrast, the intergenic regions between leuB and leuC and between leuC and leuD are very small (1 and 12 nt, respectively). This suggests that these genes are transcribed as a unique transcription unit, given the lack of space for additional promoters and the absence of putative Buchnera promoters in internal coding regions. Putative −35 and −10 promoter sequences, similar to those found for other Buchnera genes (17) were only found 42 and 22 nt upstream of leuA (TGTTATA and TTAAAAT, respectively). The sequence AGGA, located 7 nt upstream of the initiation codon of leuA, corresponds to the Shine Dalgarno sequence. A search for possible terminators of transcription did not show any inverted repeats downstream of rep and leuD. Such inverted repeats (short inverted repeat type 1) have been found downstream of leuD in various leucine plasmids (14).
Two possible scenarios
The different locations of the leucine gene cluster, either on the chromosome as in Buchnera sp. strain PSY or on a plasmid as in Buchnera sp. strain APS (13), can be explained by two possible scenarios.
(i) The last common ancestor of symbiotic Buchnera spp. carried the leucine operon on the chromosome and contained a cryptic plasmid with, at least, the genes repA1 and ibp. After establishment of the symbiosis, the leucine genes were transferred to plasmids independently in several Buchnera lineages, resulting in leucine plasmids with different gene orders and/or gene contents (17, 19).
(ii) Alternatively, the transfer of the gene cluster to a repA1 plasmid took place only once in the common ancestor of extant Buchnera lineages. Back transfer to the chromosome took place in the lineage leading to Buchnera sp. strain PSY, involving a recombination event between two direct repeat sequences, one in the intergenic region between the chromosomal genes rep and trxA and the other between the genes leuD and leuA of the plasmid (see Fig. 2).
Established aphid phylogenies, which postulate a basal split between the subfamily Pemphiginae and all remaining Buchnera lineages (2, 6), support the first scenario. E. coli, a close free-living relative of Buchnera, carries the leucine gene cluster on the chromosome, in the order leuA, -B, -C, and -D. Although this organization is similar to the one described here for Buchnera sp. strain PSY, it is difficult to explain the great size (441 bp) of the intergenic region between leuA and leuB in the first scenario. Recent studies have estimated the median size of intergenic regions in Buchnera sp. strain APS to be 74 nt (9, 15).
The second scenario seems most plausible in the light of recent molecular phylogenetic studies. These have extended taxonomic sampling relative to previous studies and have questioned traditional aphid phylogenies (8, 17, 19). Most significantly, the subfamily Lachninae was shown to be basal to all other lineages, contrary to such a previous placement for the subfamily Pemphiginae. Buchnera strains from the subfamily Lachninae contain a leucine plasmid. A possible scenario of back transfer of the leucine gene cluster to the chromosome in Buchnera sp. strain PSY is presented in Fig. 2. This back transfer may have been followed by disintegration of some of the genes inserted between leuA and leuB (9, 15), leaving as a remnant of the process the relatively large intergenic region observed between leuA and leuB. Initially, nonrecombinant plasmid copies may have retained the leucine genes, but eventually these too may have disappeared through gene disintegration. Strikingly similar to the leuA-leuB intergenic region, the noncoding region between ibp and repA1, encoded by the plasmid of Buchnera sp. strain PSY , is unusually large, measuring 858 bp (19). This region might correspond to a remnant sequence of the leucine gene cluster's disintegration. Retention of the small, ibp-carrying plasmid could be due to disintegration of the chromosomal copy of this essential gene.
Nucleotide sequence accession number
The nucleotide sequences reported in this paper have been deposited in the GenBank and EMBL databases under accession no. AJ426489.
Acknowledgments
Financial support was provided by grants GR00-27 from Consellería de Cultura y Educació, Comunidad Valenciana, to A.L. and BFM2000-1383 from Ministerio de Ciencia y Tecnología (MCYT) to A.L., F.J.S., and B.S.-M.
We are very grateful to J. M. Michelena for the identification of aphid species and to A. Moya for valuable comments on the manuscript. The facilities of the Servei Central de Suport a la Investigació Experimental of the University of València have been used.
REFERENCES
- 1.Baumann, L., P. Baumann, C. Y. Lai, D. Rouhbakhsh, N. A. Moran, and M. A. Clark. 1995. Genetics, physiology, and evolutionary relationships of the genus Buchnera: intracellular symbionts of aphids. Annu. Rev. Microbiol. 49:55-94. [DOI] [PubMed] [Google Scholar]
- 2.Baumann, L., P. Baumann, N. A. Moran, J. Sandström, and M. L. Thao. 1999. Genetic characterization of plasmids containing genes encoding enzymes of leucine biosynthesis in endosymbionts (Buchnera) of aphids. J. Mol. Evol. 48:77-85. [DOI] [PubMed] [Google Scholar]
- 3.Bracho, A. M., D. Martínez-Torres, A. Moya, and A. Latorre. 1995. Discovery and molecular characterization of a plasmid localized in Buchnera sp. bacterial endosymbiont of the aphid Rhopalosiphum padi. J. Mol. Evol 41:67-73. [DOI] [PubMed] [Google Scholar]
- 4.Douglas, A. E. 1998. Nutritional interactions in insect-microbial symbioses: aphids and their symbiotic bacteria Buchnera. Annu. Rev. Entomol. 43:17-37. [DOI] [PubMed] [Google Scholar]
- 5.Harrison, C. P., A. E. Douglas, and A. F. G. Dixon. 1989. A rapid method to isolate symbiotic bacteria from aphids. J. Invertebr. Pathol. 53:427-428. [Google Scholar]
- 6.Heie, O. E. 1987. Paleontology and phylogeny, p. 367-391. In A. K. Minks and P. Harrewijn (ed.), World Crop Pests, vol. 2A. Aphids: their biology, natural enemies and control. Elsevier, Amsterdam, The Netherlands.
- 7.Lai, C.-Y., L. Baumann, and P. Baumann. 1994. Amplification of trpEG: adaptation of Buchnera aphidicola to an endosymbiotic association with aphids. Proc. Natl. Acad. Sci. USA 91:3819-3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martínez-Torres, D., C. Buades, A. Latorre, and A. Moya. 2001. Molecular systematics of aphids and their primary endosymbionts. Mol. Phylogenet. Evol. 20:437-449. [DOI] [PubMed] [Google Scholar]
- 9.Mira, A., H. Ochman, and N. Moran. 2001. Deletion bias and the evolution of bacterial genomes. Trends Genet. 17:589-596. [DOI] [PubMed] [Google Scholar]
- 10.Moran, N. A., M. A. Munson, P. Baumann, and H. Ishikawa. 1993. A molecular clock in endosymbiotic bacteria is calibrated using the insect host. Proc. R. Soc. Lond. B Biol. Sci. 253:167-171. [Google Scholar]
- 11.Munson, M. A., M. A. Clark, L. Baumann, N. A. Moran, D. J. Voegtlin, and B. C. Campbell. 1991. Evidence for the establishment of aphid-eubacterium endosymbiosis in an ancestor of four aphid families. J. Bacteriol. 173:6321-6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rouhbakhsh, D., C.-Y. Lai, C. von Dohlen, M. A. Clark, L. Baumann, P. Baumann, N. A. Moran, and D. J. Voegtlin. 1996. The tryptophan biosynthesis pathway of aphid endosymbionts (Buchnera): genetics and evolution of plasmid-associated anthranilate synthase (trpEG) within the aphididae. J. Mol. Evol. 42:414-421. [DOI] [PubMed] [Google Scholar]
- 13.Shigenobu, S., H. Watanabe, M. Hatori, Y. Sakaki, and H. Ishikawa. 2000. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature 407:81-86. [DOI] [PubMed] [Google Scholar]
- 14.Silva, F. J., R. C. H. J. van Ham, B. Sabater, and A. Latorre. 1998. Structure and evolution of the leucine plasmids carried by the endosymbiont (Buchnera aphidicola) from aphids of the family Aphididae. FEMS Microbiol. Lett. 168:43-49. [DOI] [PubMed] [Google Scholar]
- 15.Silva, F. J., A. Latorre, and A. Moya. 2001. Genome size reduction through multiple events of gene disintegration in Buchnera APS. Trends Genet. 176:15-618. [DOI] [PubMed] [Google Scholar]
- 16.Soler, T., A. Latorre, B. Sabater, and F. J. Silva. 2000. Molecular characterization of the leucine plasmid from Buchnera aphidicola, primary endosymbiont of the aphid Acyrthosiphon pisum. Curr. Microbiol. 40:264-268. [DOI] [PubMed] [Google Scholar]
- 17.van Ham, R. C. H. J., A. Moya, and A. Latorre. 1997. Putative evolutionary origin of plasmids carrying the genes involved in leucine biosynthesis in Buchnera aphidicola (endosymbiont of aphids). J. Bacteriol. 179:4768-4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Ham, R. C. H. J., D. Martínez-Torres, A. Moya, and A. Latorre. 1999. Plasmid-encoded anthranilate synthase (TrpEG) in Buchnera aphidicola from aphis of the family Pemphigidae. Appl. Environ. Microbiol. 65:117-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Ham, R. C. H. J., F. González-Candelas, F. J. Silva, B. Sabater, A. Moya, and A. Latorre. 2000. Postsymbiotic plasmid acquisition and evolution of the repA1-replicon in Buchnera aphidicola. Proc. Natl. Acad. Sci. USA 97:10855-10860. [DOI] [PMC free article] [PubMed] [Google Scholar]