Abstract
An efficient transformation protocol for Gluconobacter oxydans and Acetobacter liquefaciens strains was developed by preparation of electrocompetent cells grown on yeast extract-ethanol medium. Plasmid pBBR122 was used as broad-host-range vector to clone the Escherichia coli lacZY genes in G. oxydans and A. liquefaciens. Although both lac genes were functionally expressed in both acetic acid bacteria, only a few transformants were able to grow on lactose. However, this ability strictly depended on the presence of a plasmid expressing both lac genes. Mutations in the plasmids and/or in the chromosome were excluded as the cause of growth ability on lactose.
Gluconobacter and Acetobacter are industrially important acetic acid bacteria. They oxidize many sugars and sugar alcohols to yield valuable products such as sorbose and gluconic and ketogluconic acids. So far, only a few genetic tools have been developed for and applied to these bacteria. Transposon Tn5 mutagenesis in the direct glucose oxidation pathway of Gluconobacter oxydans ATCC 9937 has been carried out (10). Transfer of RP4::Mu from Escherichia coli into G. oxydans has been shown, but with extremely low transfer frequencies (14). Some shuttle vector transformation protocols for G. oxydans have been developed (7, 8, 9, 21, 24) and used for the cloning of genes from Acetobacter liquefaciens and G. oxydans into G. oxydans strains (20, 22).
Gluconobacter is metabolically active even at rather low pH. This would make it a promising candidate, e.g., for conversion of residual lactose in whey, if it were able to metabolize lactose (4). However, conversion of lactose to gluconic acid in whey by Gluconobacter currently depends on a costly hydrolysis process using externally added β-galactosidase (5, 25). So far, only one attempt to introduce the genes of the lac operon of E. coli into G. oxydans has been reported. Transposon Tn951, carrying the lacIZY genes (3), was conjugally transferred from E. coli into G. oxydans. The Tn951-encoded β-galactosidase was weakly expressed; however, the transconjugants were unable to grow on lactose as the sole carbon source (2).
In order to test whether lactose metabolism could be established in G. oxydans, the following steps were undertaken: (i) establishment of an efficient transformation system in G. oxydans, (ii) cloning of the E. coli lacZY genes in a shuttle vector and transformation into G. oxydans, (iii) determination of functional gene expression of the transgenes, and (iv) characterization of recombinant G. oxydans strains obtained.
Electrotransformation of Gluconobacter and Acetobacter.
Plasmid pBBR122 (mob Cmr Kmr, 5304 bp), derived from Bordetella bronchiseptica plasmid pBBR1 (1, 6, 11) (purchased from MoBiTec, Göttingen, Germany), stably replicates in many gram-negative bacteria. However, transformation of the plasmid into different G. oxydans strains grown in glucose-yeast extract-peptone medium (YPG) (15) proved inefficient. Since growth on glucose is known to favor slime production, we used YE (4% ethanol, 1% yeast extract, pH 6.0) as growth medium instead.
G. oxydans and A. liquefaciens strains were grown overnight in YE. Cultures were diluted 1:10 into 25 ml of prewarmed YE and incubated with aeration until the cells reached early log phase (1 × 108 to 2 × 108 cells/ml; optical density at 550 nm, 0.5 to 0.8). They were transferred to centrifuge tubes, incubated on ice for 15 min, and kept cold through the rest of the procedure. The cells were sedimented at 2,700 × g for 10 min at 4°C and washed with 10 ml of cold 1 mM HEPES (pH 7.0). The sedimenting and washing procedure was repeated once. The cells were suspended in 5 ml of cold 10% glycerol and sedimented again. Finally, the cells were suspended in 0.5 ml of cold 10% glycerol. Aliquots of 65 μl were shock-frozen in liquid nitrogen and stored at −80°C. After thawing on ice, 65 μl of competent Gluconobacter or Acetobacter cells was mixed with DNA (0.25 to 0.5 μg/ml) in a chilled microcentrifuge tube. The cell-DNA mixture was transferred to a cold 0.1-cm-diameter cuvette. One pulse with the gene pulser (Bio-Rad Laboratories, Richmond, Calif.) was set at 2.0 kV and 25 μF, with the pulse controller set at 200 Ω. The cells were immediately diluted with 0.9 ml of YE, transferred to a tube, and incubated at 30°C for 1 to 4 h. Thereafter, the cells were plated on agar medium selective for transformants and incubated at 30°C.
As a control, E. coli HB101 (17) was electroporated as described by Provence and Curtiss (16).
Competent cells prepared from ethanol-grown G. oxydans or Acetobacter strains were efficiently transformed with pBBRI22 by our electroporation protocol. Transformation rates of up to 1.7 × 105 transformed cells/μg of DNA were obtained, depending on the strain and on the duration of phenotypic expression (pe) (Table 1).
TABLE 1.
Transformation of pBBR122
| Strain | No. of transformants/μg of DNA with pe lasting:
|
|||
|---|---|---|---|---|
| 1 h | 2 h | 3 h | 4 h | |
| E. coli HB101 | 3.8 × 103 | —c | — | — |
| G. oxydans DSM 3504a | 2.1 × 103 | 2.1 × 103 | 2.6 × 103 | 2.1 × 103 |
| G. oxydans DSM 3503a | 9.9 × 104 | 1.4 × 105 | 1.6 × 105 | 1.7 × 105 |
| G. oxydans DSM 2343a | 1.9 × 104 | 4.1 × 104 | 3.2 × 104 | 4.9 × 104 |
| A. liquefaciens LMG1382b | 9.3 × 104 | 1.2 × 105 | 1.3 × 105 | 1.6 × 105 |
From the German Collection of Microorganisms and Cell Cultures.
From the Laboratory of Microbiology, University of Ghent.
—, not determined.
Construction of pBBR122 derivatives bearing the E. coli lacZY genes.
E. coli XL1-Blue (17) was used as host for the construction of recombinant plasmids. All constructions, as shown in Fig. 1, were verified by restriction analyses (not shown).
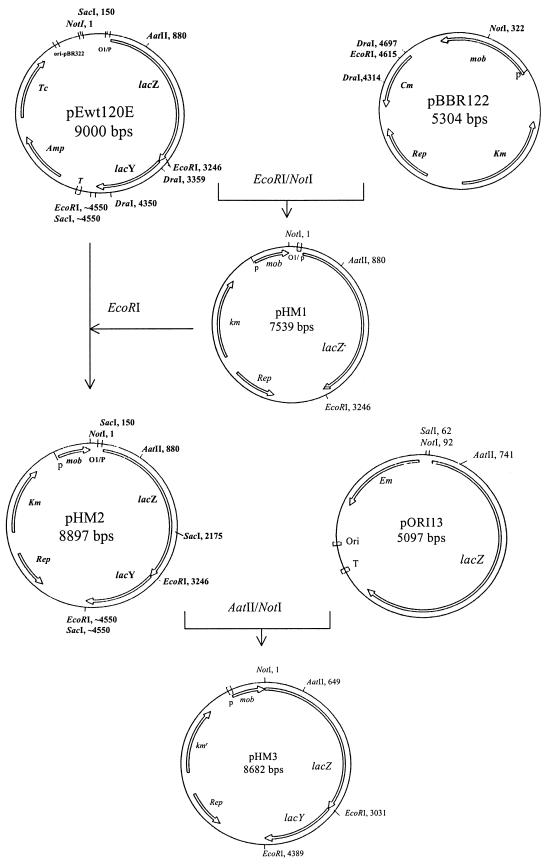
FIG. 1.
Construction scheme for the recombinant plasmids pHM1, pHM2, and pHM3 used for the heterologous expression of the E. coli β-galactosidase and lactose permease genes in G. oxydans and A. liquefaciens.
Plasmid pEwt120E harboring part of the E. coli lac operon (O1/P lacZ lacY; kindly provided by H. Assmann, University of Cologne) was digested with EcoRI and NotI. The 3.25-kb fragment consisting of the lac operator/promoter sequence and a 3′-end-truncated lacZ gene was ligated with a 4.3-kb EcoRI-NotI fragment of the vector plasmid pBBR122 containing the kanamycin resistance (Kmr) marker and replication functions, resulting in plasmid pHM1 (7,539 bp). This plasmid was digested with EcoRI and ligated with a 1.3-kb EcoRI fragment of pEwt120E carrying the 3′ end of lacZ and the complete lacY gene. The resulting plasmid, pHM2 (8,897 bp), contains the lacZ and lacY genes in proper orientation.
To remove the original E. coli promoter sequence from pHM2, an 880-bp AatII-NotI fragment was replaced by a 649-bp AatII-NotI fragment of plasmid pORI13 (18). This fragment restored the lacZ gene but did not contain a promoter sequence. In the resulting plasmid, pHM3, the lacZY genes were fused out of frame with the mob gene of the vector and, thus, were set under the control of the mob promoter located about 600 bp upstream of the start of the lacZ gene.
All constructed plasmids were successfully transformed into the Gluconobacter and Acetobacter strains listed in Table 1. Maximal transformation rates per microgram of DNA were as follows: for pHM1, 5.5 × 104 in G. oxydans DSM 3503, with 2 h of pe; pHM2, 1.1 × 104 in A. liquefaciens LMG 1382, with 1 h of pe; pHM3, 2.5 × 104 in A. liquefaciens LMG 1382, with 3 h of pe.
Functional expression of the cloned lacZ and lacY genes in E. coli.
Functional expression of the cloned genes was tested by complementation in lac-deficient mutants of E. coli. Plasmids pHM1 through pHM3 were transformed into E. coli XL1-Blue (lacZ) (17) and HB101 (lacY) and tested for Lac+ phenotypes on 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal)-Luria-Bertani-eosin-methylene blue-lactose-agar and for β-galactosidase activity in permeabilized cells and in cell extracts (13). Plasmids pHM2 and pHM3 restored β-galactosidase activity in XL1-Blue and complemented the lactose permease defect in HB101.
Expression of lacZ and lacZ in G. oxydans and A. liquefaciens.
Plasmids pHM2 and pHM3 were successfully transferred by electroporation into five G. oxydans strains (DSM 2003, DSM 2343, DSM 3503, DSM 3504, and DSM 50049) and A. liquefaciens LMG 1832. Deep- and light-blue colonies were obtained from YPG-X-Gal-kanamycin plates. Deep-blue ones were picked, grown in YPG-kanamycin medium, and tested for β-galactosidase activity. Enzyme activities of the acetic acid bacteria transformed with pHM2 or pHM3 were comparable to the activities of isopropyl-β-d-thiogalactopyranoside (IPTG)-induced E. coli XL1-Blue cells transformed with the same plasmids and grown in lactose medium. In cell extracts of XL1-Blue and strains DSM 2343, 3503, and 3504, β-galactosidase activities ranged between 470 and 690 U/ml, whereas activities between 100 and 350 U/ml were recorded for cell extracts of strains DSM 50049 and LMG 1832.
Cell lysates of G. oxydans 3503 and G. oxydans 3503(pHM2) were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (12). A protein band with an apparent molecular mass of 116 kDa was detected only in the recombinant strain (not shown). Activity staining with o-nitrophenyl-β-d-galactopyranoside (ONPG) on nondenaturing polyacrylamide gels revealed single bands with identical mobilities for cell extracts of strain 3503(pHM2) (not shown). These results demonstrated that—in contrast to the data of Condon et al. (2)—active β-galactosidase can be produced in G. oxydans at a high level.
The presence of intact lacY genes on the plasmids was further verified by DNA sequencing using the dideoxy-chain termination procedure (19) with synthetic primers binding upstream and downstream of lacY. The lacY sequences of pHM2 and pHM3 were identical to the lacY sequence of pEwt120E and to the published lacY sequence of E. coli (23). However, since improper integration into or orientation within the membrane of the lactose permease may result in impaired lactose transport, G. oxydans 3503(pHM3) was tested for lactose transport by measuring ONPG hydrolysis by intact cells (26). Transport rates comparable to the transport rate of E. coli DH1 (lac+) (17) were observed (data not shown). All together, our results demonstrated that the recombinant G. oxydans strains, harboring plasmid pHM2 or pHM3, expressed active lactose transport and β-galactosidase systems.
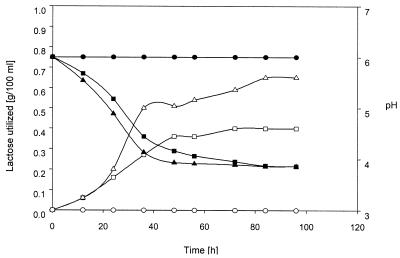
G. oxydans cells functionally expressing lacZY should be able to grow on lactose, because the lactose taken up is cleaved into galactose and glucose, the latter of which is a sugar that is readily metabolized intracellularly. Galactose, on the other hand, is metabolized only after exhaustion of glucose and after a rather long adaption phase. Only about 1% of the glucose-starved cells become able to metabolize galactose (25). Two hundred deep-blue colonies of transformants carrying either pHM2 or pHM3 were transferred into yeast extract-peptone-lactose medium (YPL), containing lactose as the sole carbon source. Despite the fact that all tested transformants showed Lac+ phenotypes on X-Gal plates and expressed lacZ efficiently, only four transformants were able to grow in lactose medium after a long (48- to 96-h) adaption phase. In contrast to transformants grown in YPG, those pregrown in YPL started growing immediately after being subcultured in fresh YPL (Fig. 2). Metabolism of lactose was clearly indicated by reduction of the lactose content in the growth medium and concomitant pH reduction indicating acid production (Fig. 2). Growth on lactose was dependent on the presence of pHM2 or pHM3, since plasmid-cured purified derivatives were unable to grow on lactose (not shown).
FIG. 2.
Lactose utilization and pH changes during growth of G. oxydans in YPL. Shown are G. oxydans 3305 utilization of lactose (○) and change of pH (•), G. oxydans 3305 (pHM2) utilization of lactose (□) and change of pH (▪), and G. oxydans 3305 (pHM3) utilization of lactose (▵) and change of pH (▴).
Since only a few transformants were able to grow on lactose, the possibility of whether mutations in the plasmid and/or in the chromosome were responsible for that growth phenotype had to be tested. First, plasmids were isolated from transformants growing in lactose (lactose-growing transformants) and from those not growing in lactose (non-lactose-growing transformants), and they were tested for functional lac genes in complementation assays. Independently of their origins, all plasmids complemented lacZ in XL1-Blue and the defective lacY gene in E. coli HB101. When they were determined, the lacY sequences of two (one carrying pHM2 and one carrying pHM3) lactose-growing isolates and two non-lactose-growing isolates were found to be identical to each other and to the published lacY sequence of E. coli (23). Furthermore, similar lactose transport activities were observed for a lactose-growing strain and a non-lactose-growing strain (not shown). Second, plasmid-cured strains of lactose-growing cells were transformed with pHM3 isolated from lactose-growing cells. If the lactose-growing phenotype was due to mutations in the plasmid and/or the chromosome, all transformants should be able to immediately grow on lactose after transformation. Plasmid curing was performed by growing strains in medium without antibiotic. Table 2 shows that plasmids pHM2 and pHM3 were more easily lost in glucose medium than in lactose medium and that pHM2 appeared to be segregationally more stable than pHM3. Plasmid-cured cells of G. oxydans 3503 (pHM3) isolated from the culture grown on lactose were retransformed with pHM3 purified from the lactose-grown parent strain. Dark-blue colonies appearing on YPG-X-Gal-kanamycin plates were transferred into YPL. None of about 200 colonies tested was able to grow on lactose. This clearly indicated that neither mutations on the chromosome nor those on the plasmid nor those on both DNA molecules were responsible for growth on lactose of the lacZ+Y+ G. oxydans strains. However, formally we cannot exclude the possibility that a tiny fraction of mutated plasmids gave rise to cells growing on lactose. In line with this argument could be the observation from the experiments on segregational stability that apparently a rather small fraction of cells able to grow on lactose (11% of cells carrying pHM3 after eight growth cycles on YPL medium) sustained those cells which had lost the plasmid and were thus unable to grow on lactose (Table 2). It should be noted that yeast extract as the cause for growth of the latter cells had been ruled out.
TABLE 2.
Segregational stabilities of plasmids pHM2 and pHM3 in G. oxydans 3503
| No. of cyclesb | Relative number of cells (%) bearing plasmid pHM2 or pHM3a
|
|||
|---|---|---|---|---|
|
G. oxydans DSM 3503 (pHM2)
|
G. oxydans DSM 3503 (pHM3)
|
|||
| YPG | YPL | YPG | YPL | |
| 1 | 96 | 98 | 85 | 86 |
| 2 | 93 | 93 | 45 | 84 |
| 3 | 85 | 91 | 28 | 79 |
| 4 | 74 | 82 | 24 | 52 |
| 5 | 67 | 67 | 3 | 38 |
| 6 | 51 | 53 | 0 | 22 |
| 7 | 43 | 46 | 0 | 15 |
| 8 | 19 | 31 | 0 | 11 |
Calculated by testing Km resistance of 100 colonies obtained by plating suitable dilutions of the cultures on YPG-agar plates.
One growth cycle (30°C for 48 h) represents about six generations.
So far, the findings regarding growth on lactose remain a puzzle, and certainly more investigations are needed to gain insight into this problem. One aspect that may be relevant is that when lactose is provided as the carbon source to lacZ+Y+ Gluconobacter cells, cleavage by β-galactosidase results in the simultaneous accumulation of glucose and galactose inside the cell. This is a very unusual situation for Gluconobacter cells, since glucose and galactose, even if provided simultaneously as carbon sources, will usually never be present together inside the cells. Galactose is metabolized slowly only after exhaustion of glucose, and only about 1% of the starved cells become able to metabolize galactose (25). The inability to grow on lactose of most of the recombinant G. oxydans cells bearing functional lacZ and lacY genes thus may be due to an unknown inhibitory or regulatory effect of galactose present intracellularly during glucose metabolism. Since such an effect would be of importance for the biotechnological exploitation of Gluconobacter in whey fermentations, we will further analyze this possible effect by testing in cell extracts inhibition by galactose of glycolytic enzymes. A first attempt to approach the problem by analyzing lacZ+Y+ G. oxidans strains grown on lactose, glucose, or galactose for growth on each of the three sugars has failed so far, since the growth patterns observed did not provide any clues to a solution of the problem.
Acknowledgments
The help of M. Menzel and V. Wind with DNA sequencing and useful advice on molecular techniques from S. Lick are gratefully acknowledged.
H.E.M. was supported by a scholarship from the Egyptian Government and DAAD (Channel Program).
REFERENCES
- 1.Antoine, R., and C. Locht. 1992. Isolation and molecular characterization of a novel broad-host-range plasmid from Bordetella bronchiseptica with sequence similarities to plasmids from gram-positive organisms. Mol. Microbiol. 6:1785-1799. [DOI] [PubMed] [Google Scholar]
- 2.Condon, C., R. J. Fitz Gerald, and F. O'Gara. 1991. Conjugation and heterologous gene expression in Gluconobacter oxydans ssp. suboxydans. FEMS Microbiol. Lett. 80:173-178. [Google Scholar]
- 3.Cornelis, G., D. Ghosal, and H. Saedler. 1978. Tn 951: a new transposon carrying a lactose operon. Mol. Gen. Genet. 160:215-224. [DOI] [PubMed] [Google Scholar]
- 4.De Ley, J., and J. Swings. 1984. Gram negative aerobic rods and cocci, p. 140-406. In N. R. Krieg and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 1. Williams and Wilkins, Baltimore, Md. [Google Scholar]
- 5.El-Sayed, M. M., S. A. El-Deeb, N. Z. Abdel-Rehim, H. E. Mostafa, and M. A. Khorshid. 1996. Utilization of cheese whey for producing gluconic acid as a means for pollution control. Milchwissenschaft 51:266-268. [Google Scholar]
- 6.Elzer, P., M. E. Kovach, R. W. Phillips, G. T. Robertson, K. M. Peterson, and M. R. Roop. 1995. In vivo and in vitro stability of the broad-host-range cloning vector pBBR1MCS in six Brucella species. Plasmid 33:51-57. [DOI] [PubMed] [Google Scholar]
- 7.Fukaya, M., T. Iwata, E. Entani, H. Masai, and T. Uozmi. 1985. Distribution and characterization of plasmids in acetic acid bacteria. Agric. Biol. Chem. 49:1349-1355. [Google Scholar]
- 8.Fukaya, M., H. Okumura, and H. Masai. 1985. Development of a host-vector system for Gluconobacter suboxydans. Agric. Biol. Chem. 49:2407-2411. [Google Scholar]
- 9.Fukaya, M., J. Tayama, H. Okumura, H. Masai, T. Uozmi, and T. Beppu. 1985. Improved transformation method for Acetobacter with plasmid DNA. Agric. Biol. Chem. 49:2091-2097. [Google Scholar]
- 10.Gupta, A., V. Verma, and G. N. Qazi. 1997. Transposon induced mutation in Gluconobacter oxydans with social reference to its direct-glucose oxidation metabolism. FEMS Microbiol. Lett. 147:181-188. [DOI] [PubMed] [Google Scholar]
- 11.Kovach, M. E., R. W. Phillips, P. Elzer, M. R. Roop II, and K. M. Peterson. 1994. pBBR1MCS: a broad-host-range cloning vector. BioTechniques 16:800-802. [PubMed] [Google Scholar]
- 12.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 13.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 14.Murooka, Y., N. Takizawa, and T. Harada. 1981. Introduction of bacteriophage Mu into bacteria of various genera and intergenetic gene transfer by RP4::Mu. J. Bacteriol. 145:358-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okumura, H., T. Uozumi, and T. Beppu. 1985. Construction of plasmid vectors and a genetic transformation system for Acetobacter aceti. Agric. Biol. Chem. 49:1011-1017. [Google Scholar]
- 16.Provence, D. L., and R. Curtiss. 1994. Gene transfer in gram-negative bacteria, p. 319-347. In P. Gerhardt, R. G. E. Murray, W. A. Wood, and N. R. Krieg (ed.), Methods for general and molecular bacteriology. American Society for Microbiology, Washington, D.C.
- 17.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 18.Sanders, J. W., K. J. Leenhouts, A. J. Haandrikman, G. Venema, and J. Kok. 1995. Stress response in Lactococcus lactis: cloning, expression analysis, and mutation of the lactococcal superoxide dismutase gene. J. Bacteriol. 177:5254-5260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain- terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Satio, Y., Y. Ishii, H. Hayashi, Y. Imao, T. Akashi, K. Yoshikawa, Y. Noguchi, S. Soeda, M. Yoshida, M. Niwa, J. Hosoda, and K. Shimomura. 1997. Cloning of genes coding for l-sorbose and l-sorbosone dehydrogenases from Gluconobacter oxydans and microbial production of 2-keto-l-gulonate, a precursor of l-ascorbic acid, in a recombinant G. oxydans strain. Appl. Environ. Microbiol. 63:454-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shinyoh, M., and T. Hoshino. 1995. Development of a stable shuttle vector and a conjugative transfer system for Gluconobacter oxydans. J. Ferment. Bioeng. 79:95-99. [Google Scholar]
- 22.Shinyoh, M., N. Tomiyama, A. Asakura, and T. Hoshino. 1995. Cloning and nucleotide sequencing of the membrane-bound l-sorbosone dehydrogenase of Acetobacter liquefaciens IFO 12258 and its expression in Gluconobacter oxydans. Appl. Environ. Microbiol. 61:413-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Silhavy, T. J., L. Berman, and L. W. Enquist. 1984. Experiments with gene fusions. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 24.Tayama, K., M. Fukaya, H. Okumura, Y. Kawamura, S. Horinouchi, and T. Beppu. 1994. Transformation of Acetobacter polyoxogenes with plasmid DNA by electroporation. Biosci. Biotech. Biochem. 58:974-975. [Google Scholar]
- 25.Vanhuynh, N., M. Decleire, J. C. Voets, J. C. Motte, and X. Monseur. 1986. Production of gluconic acid from whey hydrolysate by Gluconobacter oxydans. Process Biochem. 2:31-32. [Google Scholar]
- 26.Weisberg, L. J., J. E. Cronan, and W. D. Nunn. 1975. Induction of lactose transport in Escherichia coli during the absence of phospholipid synthesis. J. Bacteriol. 123:492-496. [DOI] [PMC free article] [PubMed] [Google Scholar]