Abstract
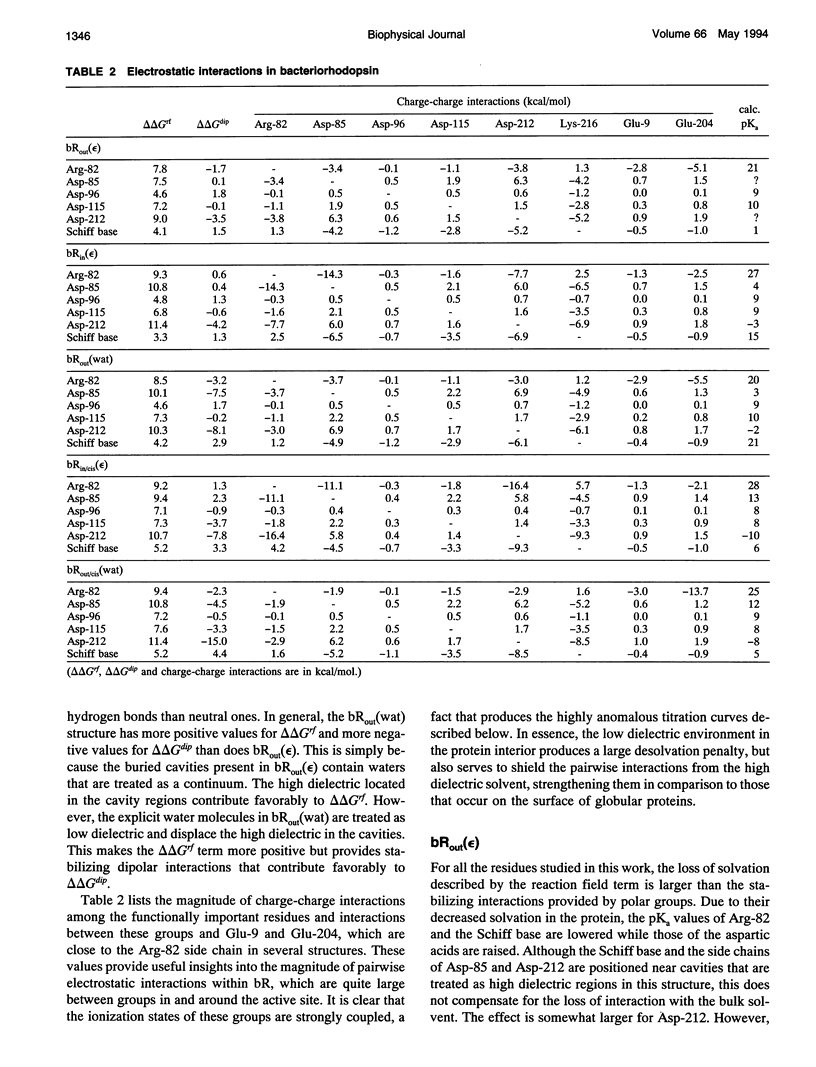
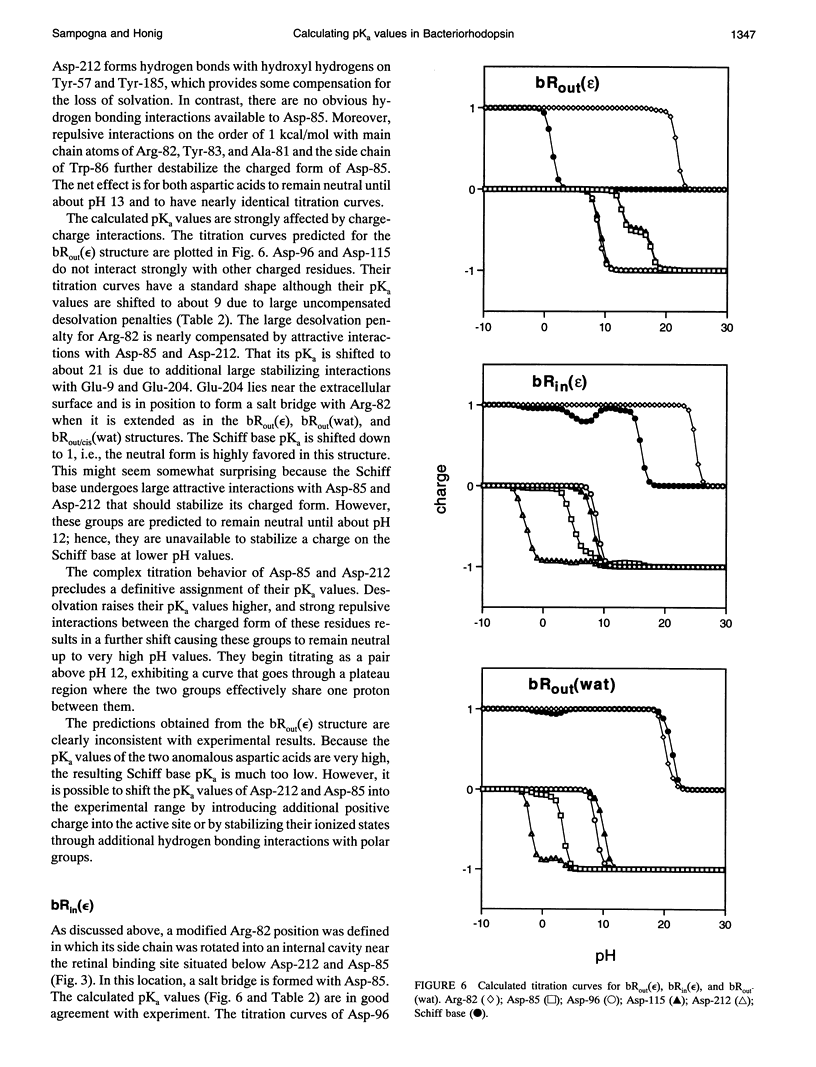
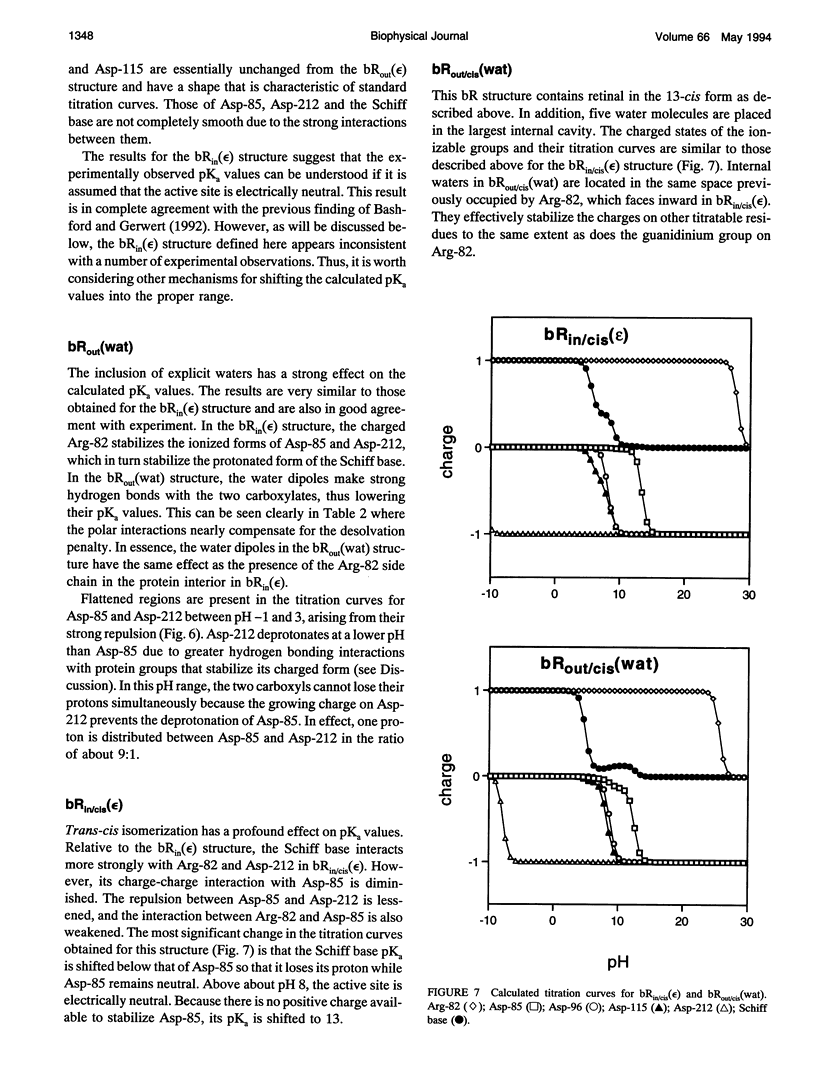
Finite difference solutions of the Poisson-Boltzmann equation are used to calculate the pKa values of the functionally important ionizable groups in bacteriorhodopsin. There are strong charge-charge interactions between the residues in the binding site leading to the possibility of complex titration behavior. Structured water molecules, if they exist in the binding site, can have significant effects on the calculated pKa by strongly stabilizing ionized species. The ionization states of the Schiff base and Asp-85 are found to be strongly coupled. Small environmental changes, which might occur as a consequence of trans-cis isomerization, are capable of causing large shifts in the relative pKa values of these two groups. This provides an explanation for the protonation of Asp-85 and the deprotonation of the Schiff base in the M state of bacteriorhodopsin. The different behavior of Asp-85 and Asp-212 is discussed in this regard.
Full text
PDF

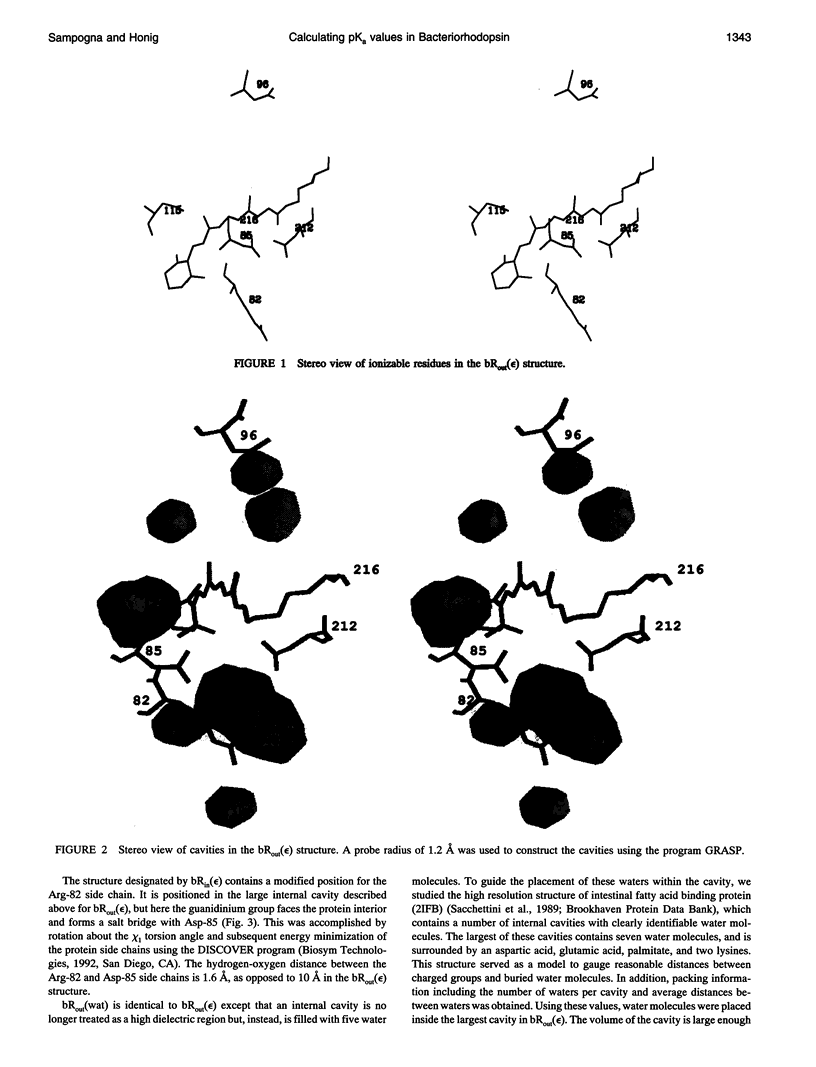
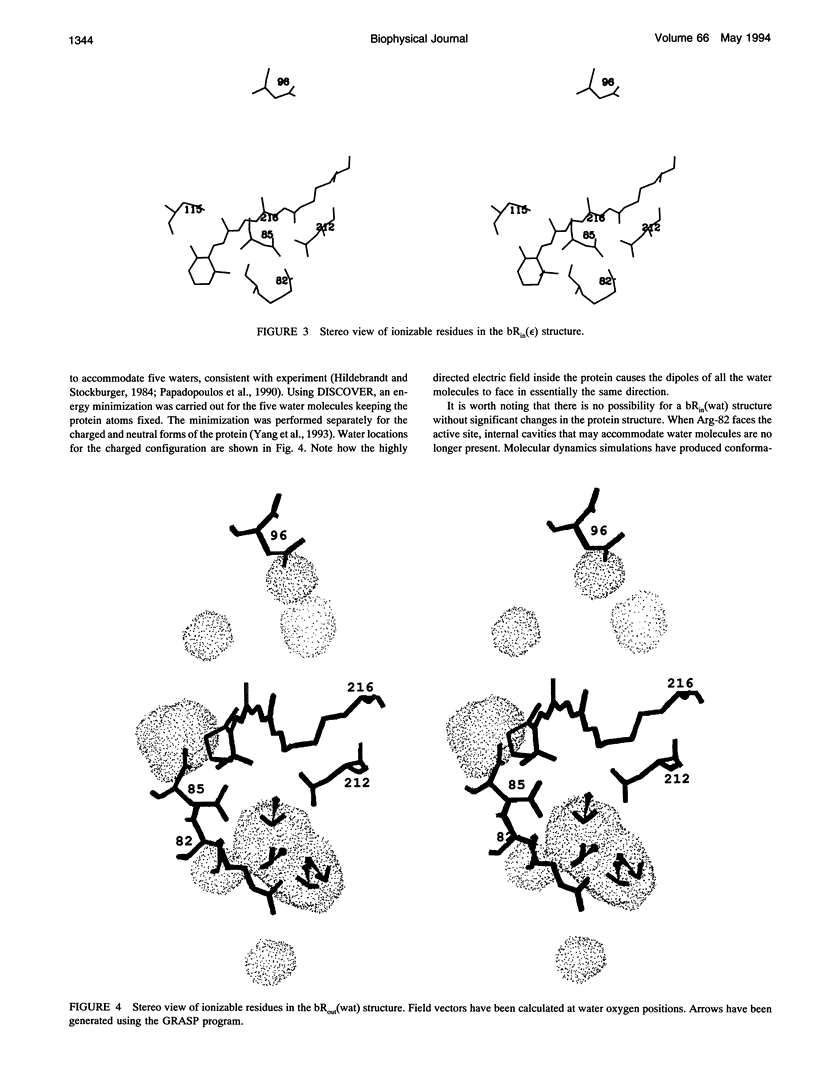
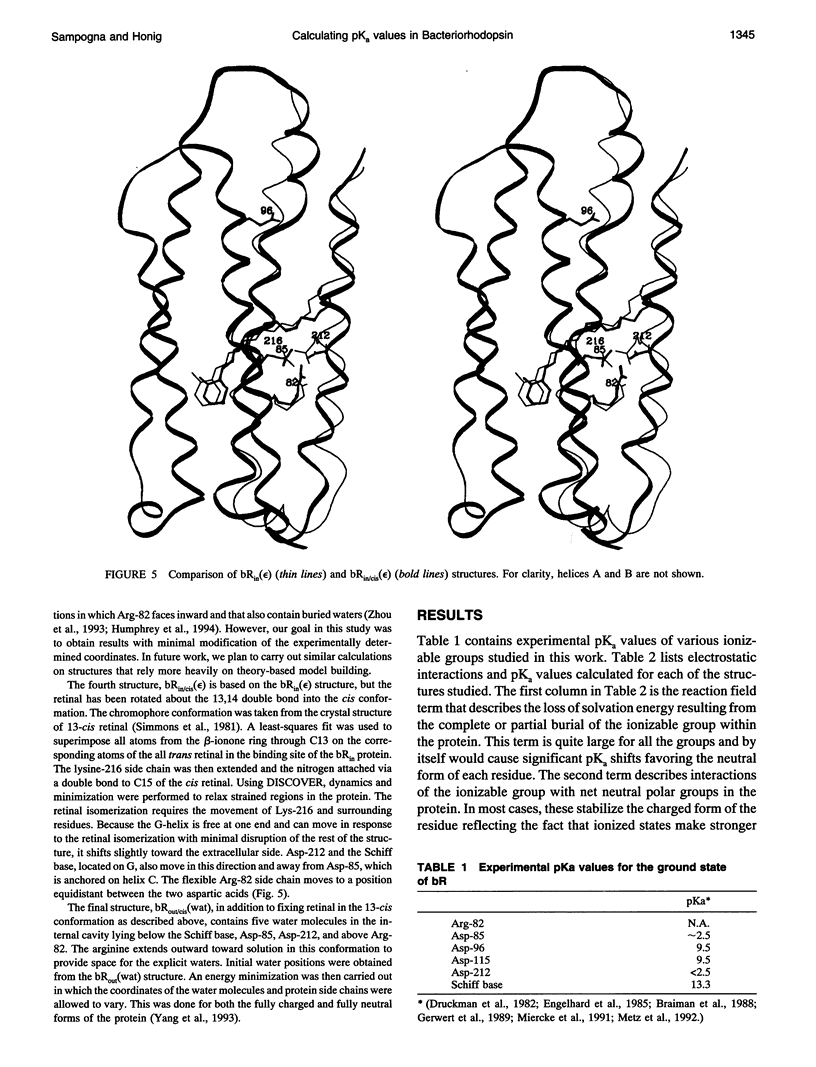




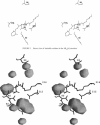
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Balashov S. P., Govindjee R., Kono M., Imasheva E., Lukashev E., Ebrey T. G., Crouch R. K., Menick D. R., Feng Y. Effect of the arginine-82 to alanine mutation in bacteriorhodopsin on dark adaptation, proton release, and the photochemical cycle. Biochemistry. 1993 Oct 5;32(39):10331–10343. doi: 10.1021/bi00090a008. [DOI] [PubMed] [Google Scholar]
- Bashford D., Gerwert K. Electrostatic calculations of the pKa values of ionizable groups in bacteriorhodopsin. J Mol Biol. 1992 Mar 20;224(2):473–486. doi: 10.1016/0022-2836(92)91009-e. [DOI] [PubMed] [Google Scholar]
- Bashford D., Karplus M. pKa's of ionizable groups in proteins: atomic detail from a continuum electrostatic model. Biochemistry. 1990 Nov 6;29(44):10219–10225. doi: 10.1021/bi00496a010. [DOI] [PubMed] [Google Scholar]
- Beroza P., Fredkin D. R., Okamura M. Y., Feher G. Protonation of interacting residues in a protein by a Monte Carlo method: application to lysozyme and the photosynthetic reaction center of Rhodobacter sphaeroides. Proc Natl Acad Sci U S A. 1991 Jul 1;88(13):5804–5808. doi: 10.1073/pnas.88.13.5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braiman M. S., Mogi T., Marti T., Stern L. J., Khorana H. G., Rothschild K. J. Vibrational spectroscopy of bacteriorhodopsin mutants: light-driven proton transport involves protonation changes of aspartic acid residues 85, 96, and 212. Biochemistry. 1988 Nov 15;27(23):8516–8520. doi: 10.1021/bi00423a002. [DOI] [PubMed] [Google Scholar]
- Brown L. S., Bonet L., Needleman R., Lanyi J. K. Estimated acid dissociation constants of the Schiff base, Asp-85, and Arg-82 during the bacteriorhodopsin photocycle. Biophys J. 1993 Jul;65(1):124–130. doi: 10.1016/S0006-3495(93)81064-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Váró G., Klinger A. L., Czajkowsky D. M., Braiman M. S., Needleman R., Lanyi J. K. Proton transfer from Asp-96 to the bacteriorhodopsin Schiff base is caused by a decrease of the pKa of Asp-96 which follows a protein backbone conformational change. Biochemistry. 1993 Mar 2;32(8):1981–1990. doi: 10.1021/bi00059a015. [DOI] [PubMed] [Google Scholar]
- Druckmann S., Ottolenghi M., Pande A., Pande J., Callender R. H. Acid-base equilibrium of the Schiff base in bacteriorhodopsin. Biochemistry. 1982 Sep 28;21(20):4953–4959. doi: 10.1021/bi00263a019. [DOI] [PubMed] [Google Scholar]
- Engelhard M., Gerwert K., Hess B., Kreutz W., Siebert F. Light-driven protonation changes of internal aspartic acids of bacteriorhodopsin: an investigation by static and time-resolved infrared difference spectroscopy using [4-13C]aspartic acid labeled purple membrane. Biochemistry. 1985 Jan 15;24(2):400–407. doi: 10.1021/bi00323a024. [DOI] [PubMed] [Google Scholar]
- Gerwert K., Hess B., Soppa J., Oesterhelt D. Role of aspartate-96 in proton translocation by bacteriorhodopsin. Proc Natl Acad Sci U S A. 1989 Jul;86(13):4943–4947. doi: 10.1073/pnas.86.13.4943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson M. K. Multiple-site titration and molecular modeling: two rapid methods for computing energies and forces for ionizable groups in proteins. Proteins. 1993 Mar;15(3):266–282. doi: 10.1002/prot.340150305. [DOI] [PubMed] [Google Scholar]
- Hagler A. T., Huler E., Lifson S. Energy functions for peptides and proteins. I. Derivation of a consistent force field including the hydrogen bond from amide crystals. J Am Chem Soc. 1974 Aug 21;96(17):5319–5327. doi: 10.1021/ja00824a004. [DOI] [PubMed] [Google Scholar]
- Harvey S. C. Treatment of electrostatic effects in macromolecular modeling. Proteins. 1989;5(1):78–92. doi: 10.1002/prot.340050109. [DOI] [PubMed] [Google Scholar]
- Henderson R., Baldwin J. M., Ceska T. A., Zemlin F., Beckmann E., Downing K. H. Model for the structure of bacteriorhodopsin based on high-resolution electron cryo-microscopy. J Mol Biol. 1990 Jun 20;213(4):899–929. doi: 10.1016/S0022-2836(05)80271-2. [DOI] [PubMed] [Google Scholar]
- Lanyi J. K. Proton transfer and energy coupling in the bacteriorhodopsin photocycle. J Bioenerg Biomembr. 1992 Apr;24(2):169–179. doi: 10.1007/BF00762675. [DOI] [PubMed] [Google Scholar]
- Lee B., Richards F. M. The interpretation of protein structures: estimation of static accessibility. J Mol Biol. 1971 Feb 14;55(3):379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- Mathies R. A., Lin S. W., Ames J. B., Pollard W. T. From femtoseconds to biology: mechanism of bacteriorhodopsin's light-driven proton pump. Annu Rev Biophys Biophys Chem. 1991;20:491–518. doi: 10.1146/annurev.bb.20.060191.002423. [DOI] [PubMed] [Google Scholar]
- McGrath M. E., Vásquez J. R., Craik C. S., Yang A. S., Honig B., Fletterick R. J. Perturbing the polar environment of Asp102 in trypsin: consequences of replacing conserved Ser214. Biochemistry. 1992 Mar 31;31(12):3059–3064. doi: 10.1021/bi00127a005. [DOI] [PubMed] [Google Scholar]
- Metz G., Siebert F., Engelhard M. Asp85 is the only internal aspartic acid that gets protonated in the M intermediate and the purple-to-blue transition of bacteriorhodopsin. A solid-state 13C CP-MAS NMR investigation. FEBS Lett. 1992 Jun 1;303(2-3):237–241. doi: 10.1016/0014-5793(92)80528-o. [DOI] [PubMed] [Google Scholar]
- Miercke L. J., Betlach M. C., Mitra A. K., Shand R. F., Fong S. K., Stroud R. M. Wild-type and mutant bacteriorhodopsins D85N, D96N, and R82Q: purification to homogeneity, pH dependence of pumping, and electron diffraction. Biochemistry. 1991 Mar 26;30(12):3088–3098. doi: 10.1021/bi00226a016. [DOI] [PubMed] [Google Scholar]
- Otto H., Marti T., Holz M., Mogi T., Stern L. J., Engel F., Khorana H. G., Heyn M. P. Substitution of amino acids Asp-85, Asp-212, and Arg-82 in bacteriorhodopsin affects the proton release phase of the pump and the pK of the Schiff base. Proc Natl Acad Sci U S A. 1990 Feb;87(3):1018–1022. doi: 10.1073/pnas.87.3.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos G., Dencher N. A., Zaccai G., Büldt G. Water molecules and exchangeable hydrogen ions at the active centre of bacteriorhodopsin localized by neutron diffraction. Elements of the proton pathway? J Mol Biol. 1990 Jul 5;214(1):15–19. doi: 10.1016/0022-2836(90)90140-h. [DOI] [PubMed] [Google Scholar]
- Sacchettini J. C., Gordon J. I., Banaszak L. J. Crystal structure of rat intestinal fatty-acid-binding protein. Refinement and analysis of the Escherichia coli-derived protein with bound palmitate. J Mol Biol. 1989 Jul 20;208(2):327–339. doi: 10.1016/0022-2836(89)90392-6. [DOI] [PubMed] [Google Scholar]
- Sharp K. A., Honig B. Electrostatic interactions in macromolecules: theory and applications. Annu Rev Biophys Biophys Chem. 1990;19:301–332. doi: 10.1146/annurev.bb.19.060190.001505. [DOI] [PubMed] [Google Scholar]
- Tavan P., Schulten K., Oesterhelt D. The effect of protonation and electrical interactions on the stereochemistry of retinal schiff bases. Biophys J. 1985 Mar;47(3):415–430. doi: 10.1016/S0006-3495(85)83933-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Váró G., Lanyi J. K. Thermodynamics and energy coupling in the bacteriorhodopsin photocycle. Biochemistry. 1991 May 21;30(20):5016–5022. doi: 10.1021/bi00234a025. [DOI] [PubMed] [Google Scholar]
- Warshel A., Aqvist J. Electrostatic energy and macromolecular function. Annu Rev Biophys Biophys Chem. 1991;20:267–298. doi: 10.1146/annurev.bb.20.060191.001411. [DOI] [PubMed] [Google Scholar]
- Warshel A. Calculations of enzymatic reactions: calculations of pKa, proton transfer reactions, and general acid catalysis reactions in enzymes. Biochemistry. 1981 May 26;20(11):3167–3177. doi: 10.1021/bi00514a028. [DOI] [PubMed] [Google Scholar]
- Yang A. S., Gunner M. R., Sampogna R., Sharp K., Honig B. On the calculation of pKas in proteins. Proteins. 1993 Mar;15(3):252–265. doi: 10.1002/prot.340150304. [DOI] [PubMed] [Google Scholar]
- Zhou F., Windemuth A., Schulten K. Molecular dynamics study of the proton pump cycle of bacteriorhodopsin. Biochemistry. 1993 Mar 9;32(9):2291–2306. doi: 10.1021/bi00060a022. [DOI] [PubMed] [Google Scholar]