Abstract
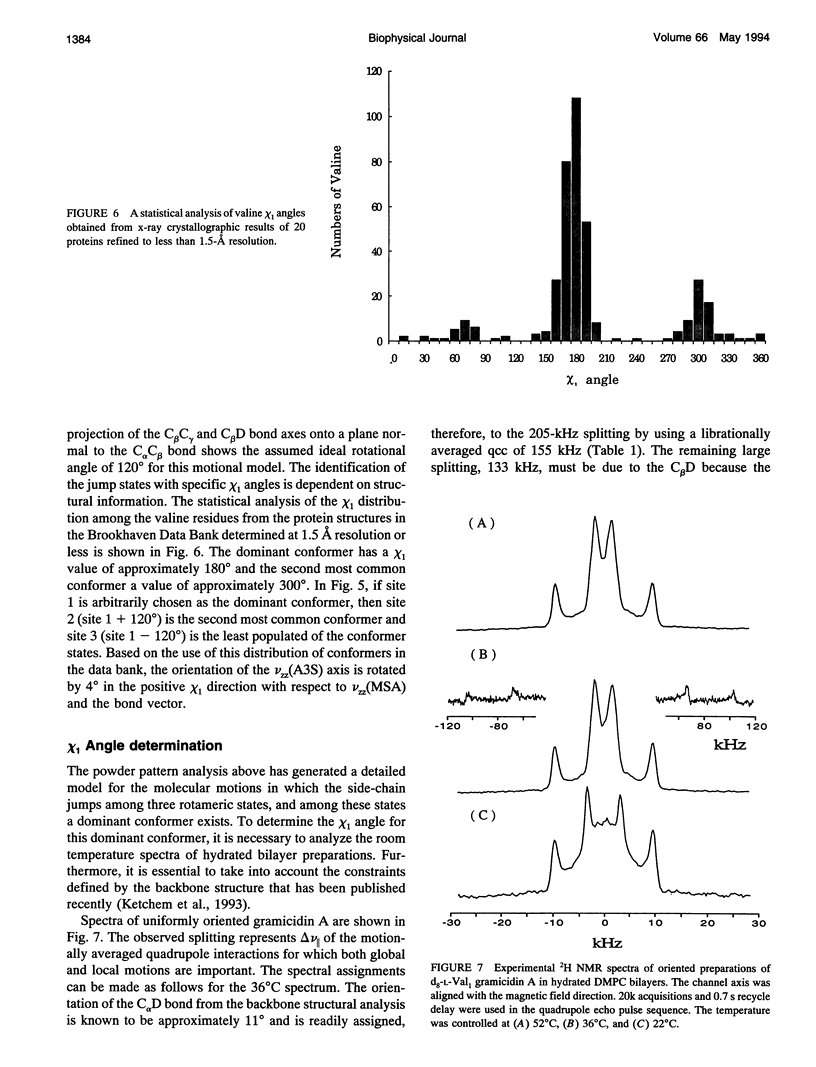
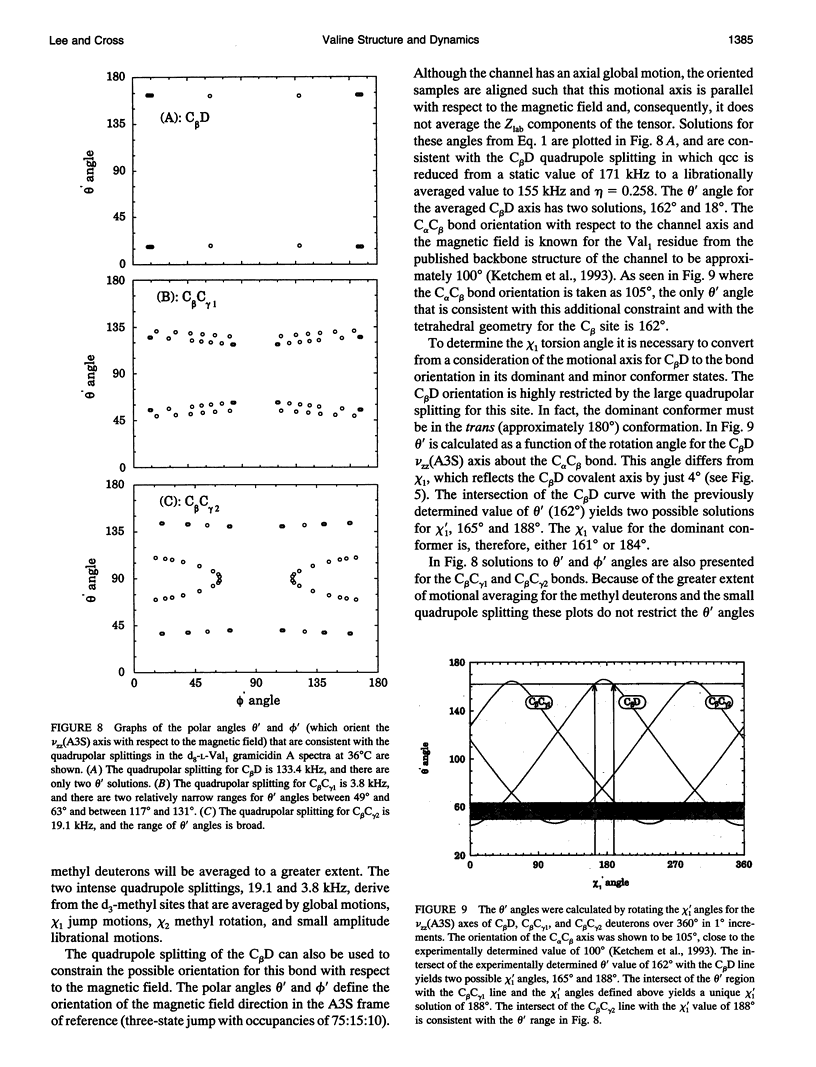
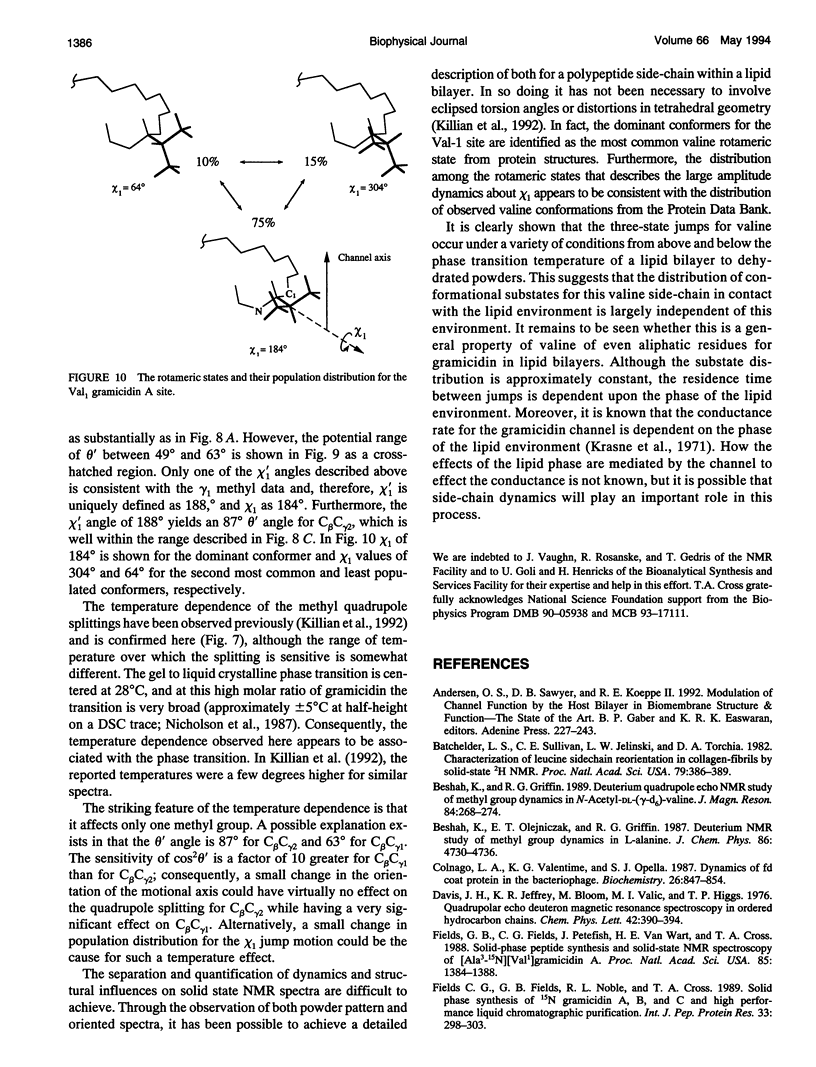
High resolution dynamics and structural information has been resolved from 2H solid-state NMR spectra of the Val-1 side-chain of the gramicidin channel in a lipid bilayer. Both powder pattern lineshapes and spectra from uniformly aligned samples of gramicidin in lipid bilayers have been analyzed to achieve a fully consistant interpretation of the data. Torsional motions about the C alpha C beta axis (chi 1) are shown to be three-state jumps in which the occupancy of the states is given by the ratio, 75:15:10 for the chi 1 angles of 184 degrees:304 degrees:64 degrees. The dominant conformer is also the most common conformation observed for valines in well defined protein structures. The distribution of conformational substates that represents the chi 1 dynamics appears to be largely independent of the lipid phase transition and the hydration of the sample. However, there is evidence that the residence time between jumps is dependent on the lipid phase transition. Although this time is shown to be approximately 1 microseconds below the phase transition temperature, it is in the fast exchange limit above the transition temperature.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Batchelder L. S., Sullivan C. E., Jelinski L. W., Torchia D. A. Characterization of leucine side-chain reorientation in collagen-fibrils by solid-state 2H NMR. Proc Natl Acad Sci U S A. 1982 Jan;79(2):386–389. doi: 10.1073/pnas.79.2.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colnago L. A., Valentine K. G., Opella S. J. Dynamics of fd coat protein in the bacteriophage. Biochemistry. 1987 Feb 10;26(3):847–854. doi: 10.1021/bi00377a028. [DOI] [PubMed] [Google Scholar]
- Fields C. G., Fields G. B., Noble R. L., Cross T. A. Solid phase peptide synthesis of 15N-gramicidins A, B, and C and high performance liquid chromatographic purification. Int J Pept Protein Res. 1989 Apr;33(4):298–303. doi: 10.1111/j.1399-3011.1989.tb01285.x. [DOI] [PubMed] [Google Scholar]
- Fields G. B., Fields C. G., Petefish J., Van Wart H. E., Cross T. A. Solid-phase peptide synthesis and solid-state NMR spectroscopy of [Ala3-15N][Val1]gramicidin A. Proc Natl Acad Sci U S A. 1988 Mar;85(5):1384–1388. doi: 10.1073/pnas.85.5.1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keniry M. A., Kintanar A., Smith R. L., Gutowsky H. S., Oldfield E. Nuclear magnetic resonance studies of amino acids and proteins. Deuterium nuclear magnetic resonance relaxation of deuteriomethyl-labeled amino acids in crystals and in Halobacterium halobium and Escherichia coli cell membranes. Biochemistry. 1984 Jan 17;23(2):288–298. doi: 10.1021/bi00297a018. [DOI] [PubMed] [Google Scholar]
- Keniry M. A., Rothgeb T. M., Smith R. L., Gutowsky H. S., Oldfield E. Nuclear magnetic resonance studies of amino acids and proteins. Side-chain mobility of methionine in the crystalline amino acid and in crystalline sperm whale (Physeter catodon) myoglobin. Biochemistry. 1983 Apr 12;22(8):1917–1926. doi: 10.1021/bi00277a028. [DOI] [PubMed] [Google Scholar]
- Ketchem R. R., Hu W., Cross T. A. High-resolution conformation of gramicidin A in a lipid bilayer by solid-state NMR. Science. 1993 Sep 10;261(5127):1457–1460. doi: 10.1126/science.7690158. [DOI] [PubMed] [Google Scholar]
- Killian J. A. Gramicidin and gramicidin-lipid interactions. Biochim Biophys Acta. 1992 Dec 11;1113(3-4):391–425. doi: 10.1016/0304-4157(92)90008-x. [DOI] [PubMed] [Google Scholar]
- Killian J. A., Taylor M. J., Koeppe R. E., 2nd Orientation of the valine-1 side chain of the gramicidin transmembrane channel and implications for channel functioning. A 2H NMR study. Biochemistry. 1992 Nov 24;31(46):11283–11290. doi: 10.1021/bi00161a004. [DOI] [PubMed] [Google Scholar]
- Kinsey R. A., Kintanar A., Tsai M. D., Smith R. L., Janes N., Oldfield E. First observation of amino acid side chain dynamics in membrane proteins using high field deuterium nuclear magnetic resonance spectroscopy. J Biol Chem. 1981 May 10;256(9):4146–4149. [PubMed] [Google Scholar]
- Krasne S., Eisenman G., Szabo G. Freezing and melting of lipid bilayers and the mode of action of nonactin, valinomycin, and gramicidin. Science. 1971 Oct 22;174(4007):412–415. doi: 10.1126/science.174.4007.412. [DOI] [PubMed] [Google Scholar]
- Lee C. W., Waugh J. S., Griffin R. G. Solid-state NMR study of trehalose/1,2-dipalmitoyl-sn-phosphatidylcholine interactions. Biochemistry. 1986 Jul 1;25(13):3737–3742. doi: 10.1021/bi00361a001. [DOI] [PubMed] [Google Scholar]
- Lee K. C., Hu W., Cross T. A. 2H NMR determination of the global correlation time of the gramicidin channel in a lipid bilayer. Biophys J. 1993 Sep;65(3):1162–1167. doi: 10.1016/S0006-3495(93)81150-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leo G. C., Colnago L. A., Valentine K. G., Opella S. J. Dynamics of fd coat protein in lipid bilayers. Biochemistry. 1987 Feb 10;26(3):854–862. doi: 10.1021/bi00377a029. [DOI] [PubMed] [Google Scholar]
- Mai W., Hu W., Wang C., Cross T. A. Orientational constraints as three-dimensional structural constraints from chemical shift anisotropy: the polypeptide backbone of gramicidin A in a lipid bilayer. Protein Sci. 1993 Apr;2(4):532–542. doi: 10.1002/pro.5560020405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson L. K., Moll F., Mixon T. E., LoGrasso P. V., Lay J. C., Cross T. A. Solid-state 15N NMR of oriented lipid bilayer bound gramicidin A'. Biochemistry. 1987 Oct 20;26(21):6621–6626. doi: 10.1021/bi00395a009. [DOI] [PubMed] [Google Scholar]
- Nicholson L. K., Teng Q., Cross T. A. Solid-state nuclear magnetic resonance derived model for dynamics in the polypeptide backbone of the gramicidin A channel. J Mol Biol. 1991 Apr 5;218(3):621–637. doi: 10.1016/0022-2836(91)90706-c. [DOI] [PubMed] [Google Scholar]
- Prosser R. S., Davis J. H., Dahlquist F. W., Lindorfer M. A. 2H nuclear magnetic resonance of the gramicidin A backbone in a phospholipid bilayer. Biochemistry. 1991 May 14;30(19):4687–4696. doi: 10.1021/bi00233a008. [DOI] [PubMed] [Google Scholar]
- Wallace B. A. Gramicidin channels and pores. Annu Rev Biophys Biophys Chem. 1990;19:127–157. doi: 10.1146/annurev.bb.19.060190.001015. [DOI] [PubMed] [Google Scholar]