Abstract
Gibberella zeae, a major cause of cereal scab, can be divided into two chemotypes based on production of the 8-ketotrichothecenes deoxynivalenol (DON) and nivalenol (NIV). We cloned and sequenced a Tri13 homolog from each chemotype. The Tri13 from a NIV chemotype strain (88-1) is located in the trichothecene gene cluster and carries an open reading frame similar to that of Fusarium sporotrichioides, whereas the Tri13 from a DON chemotype strain (H-11) carries several mutations. To confirm the roles of the Tri13 and Tri7 genes in trichothecene production by G. zeae, we genetically altered toxin production in 88-1 and H-11. In transgenic strains, the targeted deletion of Tri13 from the genome of 88-1 caused production of DON rather than NIV. Heterologous expression of the 88-1 Tri13 gene alone or in combination with the 88-1 Tri7 gene conferred on H-11 the ability to synthesize NIV; in the latter case, 4-acetylnivalenol (4-ANIV) also was produced. These results suggest that Tri13 and Tri7 are required for oxygenation and acetylation of the oxygen at C-4 during synthesis of NIV and 4-ANIV in G. zeae. These functional analyses of the Tri13 and Tri7 genes provide the first clear evidence for the genetic basis of the DON and NIV chemotypes in G. zeae.
Gibberella zeae (Schwein.) Petch (anamorph: Fusarium graminearum Schwabe) is an important pathogen of cereal crops such as maize, wheat, and barley. G. zeae causes root rot and seedling diseases (6, 25), head blight of wheat and barley, and stalk and ear rot of maize (7, 21). Head blight and ear rot reduce grain yield, and the harvested grain often is contaminated with mycotoxins, such as trichothecenes and zearalenone. Cereals contaminated with trichothecenes are associated with feed refusal, vomiting, diarrhea, dermatitis, and hemorrhages in farm animals (26). Trichothecenes also appear to contribute to the virulence of G. zeae on host plants (29, 33).
G. zeae may be divided into two chemotaxonomic groups based on production of 8-ketotrichothecenes (16). The deoxynivalenol (DON) chemotype produces DON and acetyl-DONs such as 3-acetyl-DON (3-ADON) and 15-acetyldeoxynivalenol (15-ADON). The nivalenol (NIV) chemotype produces NIV and 4-acetyl-NIV (4-ANIV) (also known as fusarenon-X). The two chemotypes appear to differ in geographic distribution, with both DON and NIV chemotypes reported in several countries of Africa, Asia, and Europe (10, 15, 24, 36-38) but only the DON chemotype reported in North America (1, 30). DON and NIV are frequently found in cereals harvested in some Asian countries, e.g., Korea and Japan (18, 40, 42). NIV is present at higher levels than DON in cereals from these countries.
Trichothecenes are biosynthesized in a complex pathway involving a series of oxygenation, isomerization, and esterification steps, and the molecular genetics of T-2 toxin production by Fusarium sporotrichioides have been studied intensively (9, 14). Many of the trichothecene biosynthesis genes are localized in a gene cluster of at least 10 genes. The genes in this cluster include those for trichodiene synthetase (Tri5) (12), P450 oxygenase (Tri4 and Tri11) (2, 13), acetyltransferase (Tri3 and Tri7) (5, 28), transcription factors (Tri6 and Tri10) (34, 39), a toxin efflux pump (Tri12) (3), and two unidentified hypothetical proteins (Tri8 and Tri9) (14, 29). Another acetyltransferase gene (Tri101) (20) is unlinked to the cluster. Recently, two F. sporotrichioides genes, Tri13 and Tri14, were found to be under the control of Tri10 (39), but the functions of these genes are not known. Homologs of Tri genes have been reported for G. zeae (5, 19, 22, 27, 34, 39, 41).
In a previous study (22), we analyzed the sequences of Tri genes from G. zeae DON and NIV chemotypes. Of the 10 Tri gene homologs in the Tri gene cluster, all except Tri7 were conserved; the Tri7 open reading frame (ORF) is intact in NIV chemotypes, whereas it is defective in DON chemotypes. However, there has been no clear evidence for the genetic determinants of DON and NIV in G. zeae. Brown et al. (5) compared the sequences of the Tri gene clusters from F. sporotrichioides and F. graminearum. They reported that Tri7 is required for acetylation of the oxygen on C-4 of the T-2 toxin in F. sporotrichioides and is nonfunctional in F. graminearum.
The general objective of this study was to better understand trichothecene biosynthesis by the two chemotypes of G. zeae. We reasoned that comparison of sequences within each trichothecene gene cluster would identify the determinants of DON and NIV production. The specific objectives of this study were (i) to sequence Tri13 from DON and NIV producers, (ii) to study the functions of Tri13 and Tri7, and (iii) to correlate differences in gene structure with differences in production of 8-ketotrichothecenes by the two chemotypes of G. zeae.
MATERIALS AND METHODS
Strains, media, and culture conditions.
G. zeae strains H-11 (a DON producer) and 88-1 (a NIV producer) were described previously (22) and were used for sequence analysis and functional studies. Fungal strains from 25% glycerol stock cultures stored at −80°C were maintained on potato dextrose agar (Difco Laboratories, Detroit, Mich.). To isolate genomic DNA, fungal conidia were inoculated into 100 ml of complete liquid medium (8) at 106 per ml. Cultures were incubated at 25°C for 48 h on a rotary shaker (200 rpm), after which mycelia were harvested and lyophilized. Recombinant Escherichia coli strains were grown on Luria-Bertani agar (35) or liquid medium supplemented with 75 μg of ampicillin per ml.
DNA manipulation and PCR conditions.
Fungal genomic DNA was prepared as previously described (17). E. coli colonies carrying recombinant plasmids were screened by a single-tube miniprep method (23). For sequencing, plasmids were purified from 5-ml E. coli cultures by using a kit from Qiagen Inc. (Valencia, Calif.). Standard procedures were used for restriction endonuclease digestions, ligations, gel blot analysis, and agarose gel electrophoresis (35). The PCR primers used in this study (Table 1) were obtained from the Bioneer oligonucleotide synthesis facility (Bioneer Corporation, Chungwon, Korea), dissolved at 100 μM in sterilized water, and stored at −20°C. PCRs were performed as described previously (44).
TABLE 1.
PCR primers and plasmids used in this study
| Primer or plasmid | Sequence (5′ to 3′) or characteristicsa | GenBank accession no. (bpb) or reference |
|---|---|---|
| Primers | ||
| Ntri7/p1 | ACAGAACAGCGCGAATTGAGTCCA | AF336365 (3362-3385) |
| Ntri7/p2 | AAAGATGATTCGGAGCCAGATGTTAGTA | AF336365 (6734-6707) |
| Tri7n/delp1 | CCGGTGGGCCTAGTTTAAAGTTCAATCT | AF336365 (4429-4402) |
| Tri7n/delp2 | CACTAAACTGAATCCTTGGCGAAAAAC | AF336365 (6400-6426) |
| Nwtri13/p1 | GGCTGATAGGGCGGTCTTGAAAATGAAC | AY064209 (18-46) |
| Nwtri13/p2 | CCTGGGAATTCAATGGTGTCAAGA | AY064209 (3660-3637) |
| Deltri13/p1 | GAAGATCTACTTTGAGCTGTTGCCTTGTCCTA | AY064209 (853-830)c |
| Deltri13/p2 | GAAGATCTGCCACAGCCACCAGACCGATAGAG | AY064209 (2505-2528)c |
| Plasmids | ||
| pNTri7H | Fungal transformation vector (HygBR, AmpR, KanR) carrying an intact 88-1 Tri7 ORF (9.4 kb) | This study |
| pdelN7H | Fungal transformation vector (HygBR, AmpR, KanR) carrying partial sequences of Tri3 and Tri8 used for deletion of Tri7 via double crossover (9.4 kb) | This study |
| pNTri13G | Fungal transformation vector (GenR, AmpR, KanR) carrying an intact 88-1 Tri13 ORF (9.8 kb) | This study |
| pdelN13G | Fungal transformation vector (GenR, AmpR, KanR) carrying partial 5′ and 3′ flanking sequences of Tri13 used for deletion via double crossover (7.3 kb) | This study |
| pBCATPH | Plasmid carrying ChrR and HygBR (5.5 kb) | 43 |
| pBCGT | Plasmid carrying ChrR and GenR (6.2 kb) | This study |
| pII99 | Plasmid carrying GenR and AmpR (5.3 kb) | 31 |
| pII99-1 | pII99 derivative lacking the EcoRI site (5.3 kb) | This study |
AmpR, resistant to ampicillin; KanR, resistant to kanamycin; ChrR, resistant to chloramphenicol.
The number in parentheses indicates the primer position in the deposited sequence.
A BglII recognition site (AGATCT) was added to each 5′ end.
Amplification, cloning, and sequencing of Tri13.
We amplified Tri13 genes from the 88-1 and H-11 strains by using several sets of primers that were based on the known sequences of the Tri12 gene from G. zeae and the Tri13 and Tri14 genes from F. sporotrichioides (39). Most of the primer pairs led to successful amplification of fragments of the expected sizes. These PCR products were sufficient for construction of contigs for both strains. PCR products of the expected sizes were cloned into pCR2.1TOPO by using a TOPO TA cloning kit (Invitrogen, San Diego, Calif.). Sequencing of the inserts in pCR2.1TOPO was initiated with M13 reverse and forward primers and then extended with specific primers corresponding to the newly sequenced regions. DNA sequencing was performed at the National Instrumentation Center for Environmental Management (Seoul National University, Suwon, Korea) with an ABI377 automated DNA sequencer (Applied Biosystems Inc., Foster City, Calif.). Primers for sequencing were designed by using the PrimerSelect program (DNASTAR, Inc., Madison, Wis.). Sequences were assembled using the SeqMan program (DNASTAR, Inc.) and analyzed with the MegAlign and MapDraw programs (DNASTAR, Inc.). BLAST (4) searches were performed against the National Center for Biotechnology Information and GenBank databases.
Plasmid construction.
For insertion of Tri7 from 88-1 into the genome of H-11, a plasmid containing an intact copy of Tri7 was prepared. Tri7 was amplified from 88-1 genomic DNA by using primers Ntri7/p1 and Ntri7/p2 and cloned into pCR2.1TOPO as described above. The Tri7 insertion vector pNTri7H (9.4 kb) was then created by subcloning a 2.1-kb hygromycin B resistance gene (HygBR) taken from pBCATPH into the XbaI site of the pCR2.1TOPO-Tri7 vector described above. For deletion of Tri7 from the genome of 88-1 via double crossover, a plasmid harboring a 3.4-kb fragment carrying the 5′ and 3′ flanking sequences of the 88-1 Tri7 ORF was constructed. This fragment, which carried 0.9 and 2.5 kb of the 5′ and 3′ flanking sequences, respectively, was obtained by inverse PCR with primers Tri7n/delp1 and Tri7n/delp2, using NheI-digested and self-ligated 88-1 genomic DNA as a template. The inverse PCR product was cloned into pCR2.1TOPO along with the HygBR cassette, creating a 9.4-kb plasmid designated pdelN7H.
Plasmids used for insertion or deletion of the 88-1 Tri13 gene were prepared by the same strategies described above for Tri7. For an insertion plasmid, primers NWTri13/p1 and NWTri13/p2 were used to amplify the entire Tri13 ORF from 88-1 genomic DNA. The 3.6-kb amplified product was cloned into pCR2.1TOPO and then removed from the cloning vector by digestion with EcoRI. This EcoRI fragment was subcloned into an EcoRI site of pBCGT, which contains a gene conferring resistance to Geneticin (GenR), to create the 9.8-kb plasmid pNTri13G. To create a deletion plasmid, 88-1 genomic DNA was digested with EcoRI, self-ligated, and used as a template for inverse PCR with primers delTri13/p1 and delTri13/p2, each including a BglII recognition sequence. The PCR amplified a 2.0-kb fragment carrying the 5′ and 3′ flanking sequences of the 88-1 Tri13 gene. The amplified product was cleaned by phenol extraction, digested with BglII, and ligated into BglII-digested pII99-1 containing GenR and no EcoRI site. The resulting 7.3-kb plasmid was designated pdelN13G (Table 1).
Fungal transformation.
For sporulation, mycelial plugs of each strain were inoculated into CMC liquid medium (15 g of carboxylmethyl cellulose, 1 g of yeast extract, 0.5 g of MgSO4, 1 g of NH4NO3, and 1 g of KH2PO4 per liter) at 25°C with shaking (100 rpm) for 3 days. Fungal conidia produced in CMC culture were inoculated into 100 ml of YPG liquid medium (3 g of yeast extract, 10 g of peptone, and 20 g of glucose per liter) at 106 per ml and grown for 12 h with shaking at 25°C. Mycelia were harvested by filtration through sterile Whatman no. 2 filter paper and incubated in 80 ml of 1 M NH4Cl containing Driselase (10 mg/ml) (InterSpex Products, Inc., San Mateo, Calif.) to generate protoplasts. Further steps in transformation were as previously described (45). Each transformant was transferred to fresh potato dextrose agar medium amended with the desired antibiotics and purified by single-conidium isolation. For insertion of the 88-1 Tri7 or Tri13 gene, circular pNTri7H or pNTri13G was transformed into protoplasts of H-11. For gene deletions, plasmids pdelN7H and pdelN13G were linearized by digestion with NheI and EcoRI, respectively, prior to transformation.
Trichothecene analysis.
Transgenic and wild-type strains of G. zeae were screened for trichothecene production on rice medium. Rice cultures were harvested after 3 weeks of incubation at 25°C and extracted as previously described (36). A portion of each extract was reacted with trimethylsilating reagent and analyzed with a JEOL JMS-AX 505 gas chromatograph-mass spectrometer in full-scan mode using a DB-5 fused silica column (0.25 mm [inside diameter] by 30 m; 0.25-μm film) (J & W Scientific, Folsom, Calif.). The column temperature was maintained at 120°C for 5 min and then increased to 270°C at 5°C per min. The injector, ion source, and interface temperatures were 280, 200, and 250°C, respectively. The ionizing voltage was 70 eV.
Nucleotide sequence accession numbers.
The sequences of the Tri13 genes obtained from G. zeae 88-1 and H-11 have been deposited in GenBank under accession numbers AY064209 and AY064210, respectively.
RESULTS
Comparative sequence analysis of Tri13.
The putative ORF of a Tri13 homolog from G. zeae strain 88-1 was identified by sequence comparison with the cDNA sequence of Tri13 from F. sporotrichioides (GenBank accession number AF330109). The Tri13 ORF is located in the trichothecene gene cluster of G. zeae 88-1 immediately upstream of Tri12, as it is in F. sporotrichioides. This 1,853-bp ORF is interrupted once by a putative intron of 62 bp. The Tri13 genes from 88-1 and F. sporotrichioides are 78 and 80% identical at the nucleotide and amino acid levels, respectively; both exhibit similarities to a putative cytochrome P450 monooxygenase. Seventeen amino acids at the N terminus of the F. sporotrichioides TRI13 protein are missing in the corresponding region of the putative TRI13 protein of 88-1 (data not shown).
G. zeae strain H-11 carries a Tri13 homolog that is strikingly different from the Tri13 homologs of 88-1 and F. sporotrichioides. The H-11 Tri13 gene is only 65 and 61% identical to the Tri13 genes from G. zeae 88-1 and F. sporotrichioides, respectively. In addition, alignment of these nucleotide sequences reveals many alterations present only in the H-11 gene. This gene appears to have incurred several substitutions, insertions, and deletions, causing a deficient translation start and frameshifts in the putative TRI13 amino acid sequence. Nucleotide alignment of Tri13 fragments amplified from genomic DNAs of other G. zeae isolates revealed that these features found in H-11 Tri13 are highly conserved among DON-producing isolates of the G. zeae strains tested from Korea and the United States (data not shown).
Molecular manipulations of Tri7 and Tri13. (i) Deletion of either the Tri7 or Tri13 ORF from the G. zeae genome.
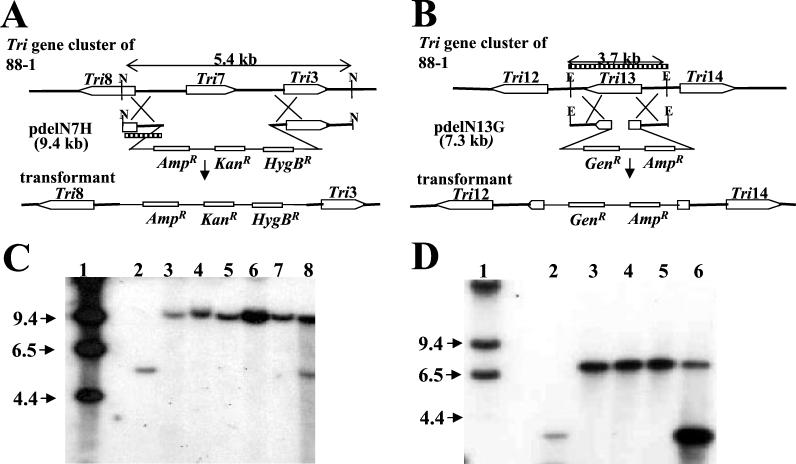
The region containing the entire Tri7 ORF or an internal portion of the Tri13 ORF in the genome of strain 88-1 was deleted by targeted gene replacement using linearized pdelN7H and pdelN13G, respectively, via double crossover between homologous regions (Fig. 1A and B). The desired transformants sustaining a deletion of either Tri7 or Tri13 were identified by gel blot analysis (Fig. 1C and D). In transformants TxNΔ7-2, TxNΔ7-3, TxNΔ7-4, and TxNΔ7-5, a single 9.4-kb band (in lieu of a 5.4-kb native band) was observed to hybridize with a Tri7 probe, indicating that a 2.0-kb region including the entire Tri7 ORF was deleted and replaced with the vector pdelN7H (Fig. 1A and C). The transformants TxNΔ13-1, TxNΔ13-2, and TxNΔ13-3 contained the intact Tri7 ORF but carried a 520-bp deletion of the Tri13 ORF, which was replaced with the vector pdelN13G (Fig. 1B and D).
FIG. 1.
Schemes for deletion of either Tri7 or Tri13 from the genome of G. zeae 88-1 (A and B) and gel blots of genomic DNAs from Tri7 (C) and Tri13 (D) deletion transformants digested with NheI and EcoRI, respectively. (A and B) Probes used for hybridization are marked as cross-hatched. N, NheI; E, EcoRI; AmpR, ampicillin resistance gene; KanR, kanamycin resistance gene; HygBR, hygromycin B resistance gene; GenR, geneticin resistance gene. (C) Lane 1, lambda DNA cut with HindIII; lane 2, wild-type 88-1; lanes 3 to 7, Tri7 deletion strains of 88-1, TxNΔ7-1, TxNΔ7-2, TxNΔ7-3, TxNΔ7-4, and TxNΔ7-5, respectively; lane 8, transformant TxNΔ7-6 carrying the transforming vector at an ectopic site. (D) Lane 1, lambda DNA cut with HindIII; lane 2, wild-type 88-1; lanes 3 to 5, Tri13 deletion strains of 88-1 , TxNΔ13-1, TxNΔ13-2, and TxNΔ13-3, respectively; lane 6, transformant TxNΔ13-4 carrying the transforming vector at an ectopic site. Band sizes in kilobases are marked with arrows on the left.
(ii) Heterologous expression of intact Tri7 and/or Tri13 ORFs.
The DON-producing H-11 strain of G. zeae was transformed with the circular vectors pNTri7H and pNTri13G, either singly or in sequential combination (Table 1). Vector pNTri7H carries the Tri7 ORF from 88-1 and the HygBR gene; vector pNTri13G carries the Tri13 ORF from 88-1 and the GenR gene. The resulting HygBR GenS, HygBS GenR, or HygBR GenR transformants were purified by single-conidium isolation, and integration events were examined by gel blot analysis.
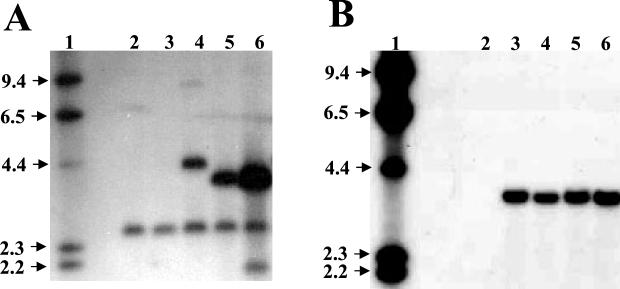
The probe, a 3.6-kb EcoRI fragment from pNTri13G carrying the 88-1 Tri13 ORF, hybridized with genomic DNAs from all HygBS GenR transformants tested. In EcoRI-digested genomic DNAs of transformants TxN13-2 and TxN13-3, the 3.6-kb fragment of the heterologous Tri13 ORF hybridized along with a 2.5-kb fragment of the native Tri13, indicating that the intact 88-1 Tri13 ORF integrated at an ectopic site of the recipient genome (Fig. 2A). In EcoRI-digested genomic DNAs from all HygBR GenS transformants tested, a 3.4-kb probe carrying the 88-1 Tri7 ORF hybridized with a single 3.6-kb fragment of the heterologous Tri7 but not with a native Tri7, probably because the probe shared only 80% nucleotide identity with the native Tri7 (data not shown). The same probe identified HygBR GenR transformants carrying both intact 88-1 Tri7 and Tri13 ORFs at ectopic sites that were created by retransformation of transformant TxN13-2 with pNTri7H (Fig. 2B).
FIG. 2.
DNA gel blots of EcoRI-digested genomic DNAs from G. zeae H-11 transformants carrying heterologous Tri13 alone (A) or both Tri13 and Tri7 (B). (A) A 3.6-kb EcoRI fragment containing the entire Tri13 ORF of 88-1 was used as a probe. Lane 1, lambda-HindIII markers; lanes 2 and 3, wild-type H-11 showing a 2.5-kb native Tri13 band; lane 4, transformant TxN13 containing a truncated copy of the 88-1 Tri13 gene; lanes 5 and 6, transformants TxN13-2 and TxN13-3, each carrying an intact copy of the 88-1 Tri13 gene. (B) Transformants carrying both the 88-1 Tri13 and Tri7 genes were obtained by retransformation of TxN13-2 with pNTri7H. The probe was a 3.4-kb PCR product containing the entire Tri7 gene of 88-1. Lane 1, lambda-HindIII markers; lane 2, wild-type H-11; lanes 3 to 6, TxN713-1, TxN713-2, TxN713-3, and TxN713-4, respectively (transformants carrying an intact copy of the 88-1 Tri7 gene). Band sizes in kilobases are marked with arrows on the left.
Trichothecene production by transgenic strains. (i) Transgenic G. zeae 88-1 strains with either Tri7 or Tri13 deleted.
G. zeae 88-1 produced NIV and 4-ANIV in rice cultures (Fig. 3A), whereas all of the transgenic 88-1 strains tested that had sustained a deletion of the Tri7 ORF produced NIV but no 4-ANIV (Fig. 3C). In contrast, other 88-1 strains resulting from ectopic integration of the transforming vector (pdelN7H) produced NIV and 4-ANIV similarly to the wild-type strain (88-1) (Fig. 3D).
FIG. 3.
Total ion chromatograms of extracts of G. zeae cultures. (A) Wild-type 88-1; (B) wild-type H-11; (C) transgenic 88-1 with Tri7 deleted (TxNΔ7-1); (D) transgenic 88-1 carrying an ectopic integration of pdelN7H (TxNΔ7-6); (E) transgenic 88-1 with Tri13 deleted (TxNΔ13-1); (F) transgenic 88-1 carrying an ectopic integration of pdelN13G (TxNΔ13-4); (G) transgenic H-11 carrying Tri13 from 88-1 (TxN13-2); (H) transgenic H-11 carrying both Tri13 and Tri7 from 88-1 (TxN713-1).
Transgenic strains of 88-1 with targeted deletions of the Tri13 ORF from the 88-1 genome exhibited more dramatic changes in trichothecene production. All of the transgenic strains tested that had sustained deletions of the Tri13 ORF produced neither NIV nor 4-ANIV. Instead, they produced DON (Fig. 3E). Other strains resulting from ectopic integrations of pdelN13G produced NIV and 4-ANIV similarly to wild-type 88-1 (Fig. 3F).
(ii) Transgenic G. zeae H-11 strains carrying heterologous Tri7 and/or Tri13 ORFs.
G. zeae H-11 produced DON and 15-ADON in rice cultures (Fig. 3B). Transgenic G. zeae strains of H-11 carrying intact copies of the 88-1 Tri7 ORF produced DON and 15-ADON similarly to wild-type H-11 (data not shown). However, heterologous expression of the 88-1 Tri13 ORF in the genetic background of H-11 caused changes in trichothecene production. GC-MS analysis showed that transgenic H-11 strains carrying the 88-1 Tri13 ORF produced only NIV; neither 4-ANIV nor DON was produced (Fig. 3G). Furthermore, transgenic H-11 strains carrying intact copies of both the 88-1 Tri7 and Tri13 ORFs produced both NIV and 4-ANIV (Fig. 3H).
DISCUSSION
We located the Tri13 gene in the trichothecene gene cluster and found that the gene differs drastically between two chemotypes of G. zeae. In G. zeae strain H-11, several mutations are present in the nucleotide sequence of Tri13. These mutations were found in all DON-producing G. zeae field isolates tested, suggesting that the lack of a viable Tri13 gene is specific to the DON chemotype. Taken together with previous results concerning Tri7 (22), these results suggest that both the Tri7 and Tri13 genes are nonfunctional in all DON chemotypes. In a previous experiment, disruption of Tri7 in F. sporotrichioides caused accumulation of HT-2 toxin rather than T-2 toxin in fungal liquid culture, suggesting that Tri7 is required for acetylation of the hydroxyl group at C-4 of the F. sporotrichioides T-2 toxin (5).
The functions of Tri7 and Tri13 in trichothecene production by G. zeae have not yet been conclusively determined. To confirm their functions, we employed molecular manipulations, including gene deletion and insertions, as described in this study. When the Tri13 gene in the NIV-producing G. zeae strain 88-1 was deleted, DON, instead of NIV, was detected in rice cultures. This result indicates that the TRI13 protein is responsible for the oxygenation at C-4 during synthesis of NIV. In addition, heterologous expression of the functional copy of Tri13 in the genetic background of the DON chemotype H-11 showed that Tri13 is sufficient for conversion from DON production to NIV production in transgenic H-11 strains.
The same molecular strategies were used to confirm that the G. zeae TRI7 protein is involved in acetylation of the oxygen at C-4 of NIV, as in F. sporotrichioides (5). Heterologous expression of both the Tri13 and Tri7 genes in H-11 caused production of both NIV and 4-ANIV. Therefore, these functional analyses have confirmed that the Tri13 gene is the determinant for the DON-NIV switching in G. zeae and that the Tri7 gene is responsible for further modification of NIV. Further confirmation of the enzymatic activities of these proteins and their roles in trichothecene biosynthesis awaits detailed biochemical studies.
Transgenic strains produced toxins in quantities similar to those produced by recipient strains, although the kinds of trichothecenes were switched with respect to each other. H-11 produced approximately 5 times more trichothecenes than did 88-1. Transgenic H-11 carrying the 88-1 Tri13 gene produced more NIV than did wild-type 88-1. The amount of DON produced by transgenic 88-1 with Tri13 deleted was much less than that produced by the wild-type H-11. Studies of field isolates also revealed higher levels of DON production by DON chemotypes than of NIV production by NIV chemotypes (36). This difference may be attributed to differences in regulatory factors such as the Tri6 and Tri10 genes (34, 39) and/or in another quantitative genetic element(s).
In addition to functional studies, the transgenic strains created in this study will be useful in evaluating the relative contributions of the two types of trichothecenes in G. zeae pathogenesis toward cereals. Previous studies using a DON-deficient G. zeae mutant showed that DON was responsible for reduced virulence by G. zeae toward wheat (29, 33). However, the role of NIV in the virulence of G. zeae has not been quantitatively analyzed, although NIV is known to be less phytotoxic than DON (11). Combinations of isogenic strains differing only in trichothecene production, such as 88-1 and a transgenic 88-1 Tri13 deletion strain or H-11 and a transgenic H-11 strain carrying the 88-1 Tri13 gene, would be appropriate for these studies.
All DON chemotype isolates of G. zeae examined thus far carry defective sequences for both Tri7 and Tri13, which raises two questions regarding the population of G. zeae in Korea. First, is the presence of both defective genes in the trichothecene gene clusters common to all DON-producing isolates? Based on our functional studies, which suggest that Tri7 is not directly involved in DON-NIV switching, we can expect to find NIV- and DON-producing isolates of G. zeae that carry functional Tri13 but not Tri7 genes and functional Tri7 but not Tri13 genes, respectively. In the former case, the isolate should not be able to produce 4-ANIV because it has a defective Tri7 gene. The latter case may be rare, because a functional Tri7 gene may be dispensable, and thus mutations could accumulate. A PCR assay using primers derived from Tri7 (22) and Tri13 designed to reveal polymorphisms between the two chemotypes would be useful in testing these possibilities. This PCR assay would also provide a more reliable method to determine the chemotypes of G. zeae field isolates.
The second question is whether polymorphisms in Tri7 and Tri13 between DON and NIV chemotypes reflect the genetic diversity of Korean G. zeae populations. No polymorphisms were found in the gene clusters including Tri7 and Tri13 among the Korean NIV-producing isolates tested (unpublished data). In contrast, in the DON-producing isolates, the gene clusters differed at the two Tri genes. Significant conservation of this structural difference between the Tri gene clusters in the two chemotypes would suggest the presence of chemotype-specific lineages in Korea. According to the description by O'Donnell et al. (32), a preliminary study showed that Korean G. zeae populations from barley were dominated by a single lineage (lineage 6) and that those from maize were dominated by lineage 7, but lineage 3 was a relatively common component (K. A. Zeller, J. I. Vargas, Y.-W. Lee, R. L. Bowden, and J. F. Leslie, Abstr. National Fusarium Head Blight Forum, abstr. 163, 2001). It is likely that at least lineages 6 and 7 in Korean populations are specific to the NIV and DON chemotypes, respectively. Confirmation of this hypothesis will require a phylogenetic study of Korean G. zeae populations.
Acknowledgments
This study was supported by a grant (M1-01-KG-01-0001-01-K07-01-028-1-0) from the Crop Functional Genomics Center of the 21st Century Frontier Research Program funded by the Korean Ministry of Science and Technology and by a grant (2000-2-22100-004-3) from the Korean Science and Engineering Foundation. T.L. and Y.K.H. were supported by postdoctoral and graduate fellowships, respectively, from the Korean Ministry of Education through the Brain Korea 21 project.
We thank T. Tsuge, Nagoya University, Nagoya, Japan, for providing plasmid pII99.
REFERENCES
- 1.Abbas, H. K., C. J. Mirocha, T. Kommedahl, R. F. Vesonder, and P. Golinski. 1989. Production of trichothecene and non-trichothecene mycotoxins by Fusarium species isolated from maize in Minnesota. Mycopathologia 108:55-58. [DOI] [PubMed] [Google Scholar]
- 2.Alexander, N. J., T. M. Hohn, and S. P. McCormick. 1988. The TRI11 gene of Fusarium sporotrichioides encodes a cytochrome P-450 monooxygenase required for C-15 hydroxylation in trichothecene biosynthesis. Appl. Environ. Microbiol. 64:221-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alexander, N. J., S. P. McCormick, and T. M. Hohn. 1999. TRI12, a trichothecene efflux pump from Fusarium sporotrichioides: gene isolation and expression in yeast. Mol. Gen. Genet. 261:977-984. [DOI] [PubMed] [Google Scholar]
- 4.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 5.Brown, D., S. P. McCormick, N. J. Alexander, R. H. Proctor, and A. E. Desjardins. 2001. A genetic and biochemical approach to study trichothecene diversity in Fusarium sporotrichioides and Fusarium graminearum. Fung. Genet. Biol. 32:121-133. [DOI] [PubMed] [Google Scholar]
- 6.Cook, R. J. 1968. Fusarium root and foot rot of cereals in the Pacific Northwest. Phytopathology 78:1673-1677. [Google Scholar]
- 7.Cook, R. J. 1981. Fusarium diseases of wheat and other small grains in North America, p. 39-52. In P. E. Nelson and T. A. Toussoun (ed.), Fusarium diseases, biology, and taxonomy. The Pennsylvania State University Press, University Park.
- 8.Correll, J. C., C. J. R. Klittich, and J. F. Leslie. 1987. Nitrate nonutilizing mutants and their use in vegetative compatibility tests. Phytopathology 77:1640-1646. [Google Scholar]
- 9.Desjardins, A. E., T. M. Hohn, and S. P. McCormick. 1993. Trichothecene biosynthesis in Fusarium species: chemistry, genetics, and significance. Microbiol. Rev. 57:595-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Desjardins, A. E., H. K. Manadhar, R. D. Plattner, C. M. Maragos, K. Shrestha, and S. P. McCormick. 2000. Occurrence of Fusarium species and mycotoxins in Nepalese maize and wheat and the effect of traditional processing methods on mycotoxin levels. J. Agric. Food Chem. 48:1377-1383. [DOI] [PubMed] [Google Scholar]
- 11.Eudes, F., A. Comeau, S. Rioux, and J. Collin. 2000. Phytotoxicité de huit mycotoxines associéés à la fusariose de l'épi chez le blé. Can. J. Plant Pathol. 22:286-292. [Google Scholar]
- 12.Hohn, T. M., and P. D. Beremand. 1989. Isolation and nucleotide sequence of a sesquiterpene cyclase gene from trichothecene-producing fungus Fusarium sporotrichioides. Gene 79:131-138. [DOI] [PubMed] [Google Scholar]
- 13.Hohn, T. M., A. E. Desjardins, and S. P McCormick. 1995. The Tri4 gene of Fusarium sporotrichioides encodes a cytochrome P450 monooxygenase involved in trichothecene biosynthesis. Mol. Gen. Genet. 248:95-102. [DOI] [PubMed] [Google Scholar]
- 14.Hohn, T. M., S. P. McCormick, N. J. Alexander, A. E. Desjardins, and R. H. Proctor. 1998. Function and biosynthesis of trichothecenes produced by Fusarium species, p. 17-24. In K. Kohmoto and O. C. Yoder (ed.), Molecular genetics of host-specific toxins in plant diseases. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 15.Ichinoe, M., R. Amano, N. Morooka, T. Yoshizawa, T. Suzuki, and M. Kurisu. 1980. Geographic difference of toxigenic fungi of Fusarium species. Proc. Jpn. Assoc. Mycotoxicol. 11:20-22. [Google Scholar]
- 16.Ichinoe, M., H. Kurata, Y. Sugiura, and Y. Ueno. 1983. Chemotaxonomy of Gibberella zeae with special reference to production of trichothecenes and zearalenone. Appl. Environ. Microbiol. 46:1364-1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kerenyi, Z., K. Zeller, L. Hornok, and J. F. Leslie. 1999. Molecular standardization of mating type terminology in the Gibberella fujikuroi species complex. Appl. Environ. Microbiol. 65:4071-4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim, J.-C., H.-J. Kang, D.-H. Lee, Y.-W. Lee, and T. Yoshizawa. 1993. Natural occurrence of Fusarium mycotoxins (trichothecenes and zearalenone) in barley and corn in Korea. Appl. Environ. Microbiol. 59:3798-3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kimura, M., I. Kaneko, M. Komiyama, A. Takatsuki, H. Koshino, K. Yoneyama, and I. Yamaguchi. 1998. Trichothecene 3-O-acetyltransferase protects both the producing organism and transformed yeast from related mycotoxins. Cloning and characterization of Tri101. J. Biol. Chem. 273:1654-1661. [DOI] [PubMed] [Google Scholar]
- 20.Kimura, M., G. Matsumoto, Y. Shingu, K. Yoneyama, and I. Yamaguchi. 1998. The mystery of the trichothecene 3-O-acetyltransferase gene. Analysis of the region around Tri101 and characterization of its homologue from Fusarium sporotrichioides. FEBS Lett. 435:163-168. [DOI] [PubMed] [Google Scholar]
- 21.Kommedahl, T., and C. E. Windels. 1981. Root-, stalk-, and ear-infecting Fusarium species on corn in the USA, p. 94-103. In P. E. Nelson, T. A. Toussoun, and R. J. Cook (ed.), Fusarium diseases, biology, and taxonomy. The Pennsylvania State University Press, University Park.
- 22.Lee, T., D.-W. Oh, H.-S. Kim, J. Lee, Y.-H. Kim, S.-H. Yun, and Y.-W. Lee. 2001. Identification of deoxynivalenol- and nivalenol-producing chemotypes of Gibberella zeae using PCR. Appl. Environ. Microbiol. 67:2966-2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu, Z., and N. C. Mishra. 1995. A single-tube method for plasmid mini-prep from large numbers of clones for direct screening by size or restriction digestion. BioTechniques 18:214-217. [PubMed] [Google Scholar]
- 24.Logrieco, A., A. Bottalico, and C. Altomare. 1988. Chemotaxonomic observation on zearalenone and trichothecene production by Gibberella zeae from cereals in southern Italy. Mycologia 80:892-895. [DOI] [PubMed] [Google Scholar]
- 25.Manka, M., A. Visconti, J. Chelkoski, and A. Bottalico. 1985. Pathogenicity of Fusarium isolates from wheat, rye, and triticale towards seedlings and their ability to produce trichothecenes and zearalenone. Phytopathol. Z. 113:24-29. [Google Scholar]
- 26.Marasas, W. F. O., P. E. Nelson, and T. A. Toussoun. 1984. Toxigenic Fusarium species: identity and mycotoxicology. The Pennsylvania State University Press, University Park.
- 27.Matsumoto, G., J. Wuchiyama, Y. Shingu, M. Kimura, K. Yoneyama, and I. Yamaguchi. 1999. The trichothecene biosynthesis regulating gene from the type B producer Fusarium strains: sequence of Tri6 and its expression in Escherichia coli. Biosci. Biotechnol. Biochem. 63:2001-2004. [DOI] [PubMed] [Google Scholar]
- 28.McCormick, S. P., T. M. Hohn, and A. E. Desjardins. 1996. Isolation and characterization of Tri3, a gene encoding 15-O-acetyltransferase from Fusarium sporotrichioides. Appl. Environ. Microbiol. 62:353-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCormick, S. P., T. M. Hohn, A. E. Desjardins, R. H. Proctor, and N. J. Alexander. 1998. Role of toxins in plant microbial interactions, p. 17-30. In J. T. Romeo, K. R. Downum, and R. Verpoorte (ed.), Recent advances in phytochemistry, vol. 32. Phytochemical signals and plant-microbe interactions. Plenum Press, New York, N.Y.
- 30.Mirocha, C. J., H. K. Abbas, C. E. Windels, and W. Xie. 1989. Variation in deoxynivalenol, 15-acetyldeoxynivalenol, 3-acetyldeoxynivalenol, and zearalenone production by Fusarium graminearum isolates. Appl. Environ. Microbiol. 55:1315-1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Namiki, F., M. Matsunaga, M. Okuda, I. Inoue, K. Nishi, Y. Fujita, and T. Tsuge. 2001. Mutation of an arginine biosynthesis gene causes reduced pathogenicity in Fusarium oxysporum f. sp. melonis. Mol. Plant-Microbe Interact. 14:580-584. [DOI] [PubMed] [Google Scholar]
- 32.O'Donnell, K., H. C. Kistler, B. K. Tacke, and H. H. Casper. 2000. Gene genealogies reveal global phylogeographic structure and reproductive isolation among lineages of Fusarium graminearum, the fungus causing wheat scab. Proc. Natl. Acad. Sci. USA 97:7905-7910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Proctor, R. H., T. M. Hohn, and S. P. McCormick. 1995. Reduced virulence of Gibberella zeae caused by disruption of a trichothecene toxin biosynthetic gene. Mol. Plant-Microbe Interact. 8:593-601. [DOI] [PubMed] [Google Scholar]
- 34.Proctor, R. H., T. M. Hohn, S. P. McCormick, and A. E. Desjardins. 1995. Tri6 encodes an unusual zinc finger protein involved in regulation of trichothecene biosynthesis in Fusarium sporotrichioides. Appl. Environ. Microbiol. 61:1923-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 36.Seo, J.-A., J.-C. Kim, D.-H. Lee, and Y.-W. Lee. 1996. Variation in 8-ketotrichothecenes and zearalenone production by Fusarium graminearum isolates from corn and barley in Korea. Mycopathologia 134:31-37. [DOI] [PubMed] [Google Scholar]
- 37.Sugiura, Y., Y. Watanabe, T. Tanaka, S. Yamamoto, and Y. Ueno. 1990. Occurrence of Gibberella zeae strains that produce both nivalenol and deoxynivalenol. Appl. Environ. Microbiol. 56:3047-3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sydenham, E. W., W. F. O. Marasas, P. G. Thiel, G. S. Shephard, and J. J. Nieuwenhuis. 1991. Production of mycotoxins by selected Fusarium graminearum and F. crookwellense isolates. Food Addit. Contam. 8:31-41. [DOI] [PubMed] [Google Scholar]
- 39.Tag, A. G., G. F. Garifullia, A. W. Peplow, C. Ake, Jr., T. D. Phillips, T. M. Horn, and M. N. Beremand. 2001. A novel regulatory gene, Tri10, controls trichothecene toxin production and gene expression. Appl. Environ. Microbiol. 67:5294-5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tanaka, T., A. Hasegawa, S. Yamamoto, U. S. Lee, Y. Sugiura, and Y. Ueno. 1988. Worldwide contamination of cereals by the Fusarium mycotoxins nivalenol, deoxynivalenol and zearalenone. I. Survey of 19 countries. J. Agric. Food Chem. 36:979-983. [Google Scholar]
- 41.Wuchiyama, J., M. Kimura, and I. Yamaguchi. 2000. A trichothecene efflux pump encoded by Tri102 in the biosynthesis gene cluster of Fusarium graminearum. J. Antibiot. 53:196-200. [DOI] [PubMed] [Google Scholar]
- 42.Yoshizawa, T., and Y. Z. Jin. 1995. Natural occurrence of acetylated derivatives of deoxynivalenol and nivalenol in wheat and barley in Japan. Food Addit. Contam. 12:689-694. [DOI] [PubMed] [Google Scholar]
- 43.Yun, S.-H. 1998. Molecular genetics and manipulation of pathogenicity and mating determinants in Mycosphaerella zeae-maydis and Cochliobolus heterostrophus. Ph.D. thesis. Cornell University, Ithaca, N.Y.
- 44.Yun, S.-H., T. Arie, I. Kaneko, O. C. Yoder, and B. G. Turgeon. 2000. Molecular organization of mating type loci in heterothallic, homothallic, and asexual Gibberella/Fusarium species. Fung. Genet. Biol. 31:7-20. [DOI] [PubMed] [Google Scholar]
- 45.Yun, S.-H., M. L. Berbee, O. C. Yoder, and B. G. Turgeon. 1999. Evolution of the fungal self-fertile reproductive life style from self-sterile ancestors. Proc. Natl. Acad. Sci. USA 96:5592-5597. [DOI] [PMC free article] [PubMed] [Google Scholar]