Abstract
Aminocyclitols structurally related to streptamine, a 1,3-diaminocyclitol, are common components of the RNA-binding aminoglycoside antibiotics. The respective aminocyclitol cores of hygromycin B and spectinomycin are N3-methyl-2-deoxy-d-streptamine and N1,N3-dimethyl-2-epi-streptamine. Adenosyl[methyl-14C]methionine:2-deoxystreptamine N-methyltransferase activities were detected in extracts of early-stationary-phase mycelia of the hygromycin B producer Streptomyces hygroscopicus subsp. hygroscopicus ATCC 27438 and the spectinomycin producer Streptomyces flavopersicus ATCC 19756. Extracts of both strains methylated the N1- and N3-amino groups of 2-deoxystreptamine, streptamine, and 2-epi-streptamine; the N1-amino group of N3-methyl-2-deoxy-d-streptamine, and the N3-amino group of N1-ethyl-2-deoxy-d-streptamine, the semisynthetic aminocyclitol of netilmicin. The mono[14C]methyl derivatives of 2-deoxystreptamine, streptamine, and 2-epi-streptamine were excellent substrates for l-glutamine:aminocyclitol aminotransferase and thereby provided a sensitive assay for derepression of this key enzyme, a generic biosynthetic marker that we have shown to be the only enzyme common to the biosyntheses of all major aminoglycoside antibiotics. Other prospective uses for these methyl-labeled 2-deoxystreptamine analogs are also described.
The RNA-binding aminoglycoside antibiotics are synthesized by many species of filamentous actinomycete soil bacteria, which evidently continue to find them useful in the soil microenvironment, as is the case for streptomycin producers (2), despite the coexistence of competing microbes possessing an array of aminoglycoside-inactivating enzymes. The chemical structures of aminoglycoside antibiotics most commonly consist of an aminocyclitol core structurally related to streptamine (I, Fig. 1), which is bonded with one or two carbohydrates (10). Some of these aminocyclitol derivatives can bind tightly to specific sites on RNAs that occur in ribosomes, group I introns, human immunodeficiency virus, or even short RNA aptamers (11). Spectinomycin, which consists of an N1,N3-dimethyl-2-epi-streptamine (IX) core and a single sugar substituent, binds at a specific site on 16S rRNA (8). Hygromycin B, which contains an N3-methyl-2-deoxy-d-streptamine (IV) core, binds to specific sites on both 16S and 18S rRNAs (8). Consequently producers of hygromycin B and the related destomycin group (3, 10) can compete for nutrients not only with bacteria but also with protozoa, fungi, and invertebrates in the continually changing soil microenvironments. Because of assay problems it is not yet known whether aminocyclitols themselves have other functions during the life cycles of certain streptomycetes.
FIG. 1.
Structures of streptamine analogs examined as possible methyl acceptors. I, streptamine; II, 2-deoxystreptamine; III, 2-epi-streptamine; IV, N3-methyl-2-deoxy-d-streptamine; V, N1-methyl-2-deoxy-d-streptamine; VI, N1-ethyl-2-deoxy-d-streptamine; VII, N1-methyl-2-epi-d-streptamine; VIII, N3-methyl-2-epi-d-streptamine; IX, N1,N3-dimethyl-2-epi-streptamine. Vertical lines represent hydroxyl groups.
My laboratory has long been interested in the enzymatic synthesis of various aminocyclitols. By 1971 all of the enzymatic reactions specifically involved in synthesis of the streptidine core of streptomycin had been discovered (Fig. 2) (19-25). Subsequently the reactions specifically required for synthesis of the bluensidine core of bluensomycin were identified (14, 17). More recently, my laboratory has initiated studies of the enzymatic synthesis of the aminocyclitol cores of spectinomycin (IX) (18) and of hygromycin B (IV).
FIG. 2.
Enzymatic reactions previously discovered by my laboratory to be involved in biosynthesis of the streptidine core of streptomycin (13, 16, 21) and the bluensidine core of bluensomycin (14, 17). Enzymes U and V are absent in streptomycin producers, and enzymes H, I, and J are absent in bluensomycin producers (17). Note that in streptomycin producers two analogous series of reactions, in the same order, convert an -OH group to a guanidino group, with intriguing evolutionary implications (14, 17). Vertical lines represent hydroxyl groups. Enzyme D, l-glutamine:aminocyclitol aminotransferase, is the only enzyme common to the biosyntheses of all major aminoglycoside antibiotics (1, 9, 16, 18, 20, 24). KGN is 2-ketoglutaramate. N1-Amidino-d-streptamine (IGN) is the aminocyclitol core of one of the destomycin antibiotics (3), also with interesting evolutionary implications. Enzyme F, amidinotransferase, is an easily assayed (19, 24) generic biosynthetic marker for streptomycin producers (2). Enzyme W also phosphorylates streptomycin (16).
The primary goals of the present study were to (i) detect the N-methyltransferase activities involved in biosynthesis of the aminocyclitol cores of hygromycin B (IV) and spectinomycin (IX), (ii) determine their substrate specificities with respect to both the aminocyclitol rings and the specific amino groups methylated, and (iii) test the abilities of their 14C-methylated aminocyclitol products to serve as surrogate substrates for monitoring the degree of expression of derepressed enzymes involved in aminoglycoside antibiotic biosyntheses, with particular emphasis on the one enzyme common to the biosyntheses of all major aminoglycoside antibiotics: l-glutamine:aminocyclitol aminotransferase (enzyme D of Fig. 2) (1, 7, 14, 18, 22).
MATERIALS AND METHODS
Materials.
Streptomyces hygroscopicus subsp. hygroscopicus ATCC 27438, Streptomyces flavopersicus ATCC 19756, Streptomyces bikiniensis ATCC 11062, and Streptomyces griseocarneus ATCC 12628 came from the American Type Culture Collection. Adenosyl[methyl-14C]methionine (Ado[14C]Met), 53 mCi/mmol and 24 μCi/ml, came from Amersham, was diluted to 126,000 cpm/5 μl (on filter paper), and stored at −20°C. Streptamine (I) and 2-deoxystreptamine (II) were prepared from streptomycin and kanamycin A, respectively, as described previously (24). N3-Methyl-2-deoxy-d-streptamine (IV) was a gift from Richard Baltz and Kay Koch of Eli Lilly. Actinamine (IX) came from Brian Bannister of Upjohn. 2-epi-Streptamine (III) and dl-N-methyl-2-epi-streptamine (VII and VIII) were a gift from Richard C. Thomas (12) of Upjohn. A solution containing N1-ethyl-2-deoxy-d-streptamine (VI) was prepared by prolonged hydrolysis of pharmaceutical-grade netilmicin (Netromycin) with 6 N HCl at 100°C, following removal of sulfate with BaCl2. L-[guanidino-14C]arginine came from New England Nuclear and was evaporated to dryness and made up to 25 μCi/ml. A mixture containing 50% N1-methyl-2-deoxy-d-streptamine (V), 20% N3-methyl-2-deoxy-d-streptamine (IV), and 30% N1,N3-dimethyl-2-deoxystreptamine, obtained by hydrolysis of destomycin A, was kindly supplied by Shinishi Kondo of the Microbial Chemistry Research Foundation, Tokyo, Japan. All other compounds, including scyllo-inosose, came from Sigma Chemical Co.
Growth of cultures and preparation of extracts.
Stock cultures were grown at room temperature on slants of yeast-malt extract agar (Difco catalog no. 0770-01). After 10 to 13 days aliquots were transferred to 2-liter Erlenmeyer flasks containing 500 ml of 0.05% glucose-1% myo-inositol-1% Soytone-0.2% yeast extract and grown at 26°C on a New Brunswick rotary shaker (240 rpm) to early stationary phase (19), approximately 42 to 51 h. Mycelia were harvested by suction filtration or centrifugation. Washed mycelial pads were stored at −20°C and then sonicated with 2 volumes of cold water and centrifuged at 4°C for 20 min at 20,000 × g, and the supernatants were stored at −20°C. Each extract was tested for biosynthetic competence by activity of glutamine:aminocyclitol aminotransferase (18).
Assay for N-methyltransferase activity (Tables 1 to 4).
TABLE 1.
Methylation of 2-deoxystreptamine derivatives by extracts of the hygromycin B producer S. hygroscopicus with Ado[14C]Met as methyl donord
| Expt no. and reactanta | Final concn (μg/ml) in reaction mixture | N-[14C]methyl product formed (cpm/10 μl)b |
|---|---|---|
| Expt 1 | ||
| Ado[14C]Met + 2-deoxystreptamine (II) | 143 | 29,071 (40) |
| Expt 2 | ||
| Ado[14C]Met + N3-methyl-2-deoxy-d-streptamine (IV) | 167 | 3,007 (4) |
| Ado[14C]Met + N3-methyl-2-deoxy-d-streptamine | 33 | 369 (0) |
| Ado[14C]Met + N1-ethyl-2-deoxy-d-streptamine (VI)c | 29,103 (40) |
TABLE 4.
Methylation of other streptamine analogs by extracts of the spectinomycin producer with Ado[14C]Met as methyl donora
| Expt no. and reactant | Final concn (μg/ml) in reaction mixture | N-[14C]methyl product formed (cpm/10 μl)b |
|---|---|---|
| Expt 1 | ||
| Ado[14C]Met + streptamine (I) | 143 | 28,148 (39) |
| Expt 2 | ||
| Ado[14C]Met + 2-deoxystreptamine (II) | 143 | 21,145 (29) |
| Ado[14C]Met + N3-methyl-2-deoxy-d-streptamine (IV) | 167 | 13,362 (19) |
| Ado[14C]Met + N3-methyl-2-deoxy-d-streptamine | 33 | 5,983 (8) |
| Ado[14C]Met + N1-ethyl-2-deoxy-d-streptamine (VI) | 24,367 (34) |
Labeled reactants and products were separated by high-voltage paper electrophoresis at pH 3.6 (15).
Numbers in parentheses are percentages of transfer of labeled methyl group from donor to acceptor.
The assay mixtures employed were as follows: Ado[14C]Met, 126,000 cpm/5 μl, 10 μl; 0.1 M K phosphate, pH 7.4, or 0.2 M NaHCO3, 5 μl; streptamine analog, 1 mg/ml, 5 μl; H2O, 5 μl; and sonicate supernatant, 10 μl. The mixture was incubated at 34°C for 2 h. Tris buffers, pH 7.4 and 8.5, were also employed in preliminary experiments. The pH 7.4 phosphate buffer was used for streptamine and 2-epi-streptamine, while the NaHCO3 buffer was used for 2-deoxystreptamine and methylated streptamine analogs. After incubation, the N-[14C]methylstreptamine derivatives, with a 2+ charge at pH 3.6, were separated from reactants and other labeled products by high-voltage paper electrophoresis at pH 3.6 (ammonium formate, 0.2 ionic strength) with a Savant horizontal plate apparatus (30 V/cm) and Whatman no. 1 filter paper matrix, with a 46-cm path marked in 1-cm intervals. Picric acid was the visible reference compound during electrophoresis (15). After drying, 1-cm segments were cut, placed in scintillation vials containing 5 ml of Liquifluor (Fisher), and counted with a Beckman LS 3801 liquid scintillation counter. Compounds with 2+ charges were located by comparison with N1-[14C]amidino-d-streptamine (IGN, Fig. 2). Up to six assays can be run in parallel.
Batch isolation of N-[14C]methylstreptamine analogs.
Larger quantities of [14C]methyl-labeled streptamine analogs were prepared by scaling up the reaction mixtures 100-fold and using 0.2 M NaCO3 as buffer (even though it was not optimum for streptamine and 2-epi-streptamine). After incubation for 2 to 3 h, 8% trichloroacetic acid was added dropwise with shaking to the reaction mixtures. After 10 to 15 min, the tubes were centrifuged at 4°C and 10,000 × g for 15 min, and the supernatants were stored at −20°C. The thawed supernatants were added to 10-ml plastic columns containing 8 ml of AG-50 (H+), 200-400 mesh, from Bio-Rad Laboratories. The contents were eluted stepwise with water, 1 N HCl, 2 N HCl, and 3 N HCl in 3.5-ml portions. Aliquots of 5 μl were spotted on Whatman no. 1 squares (1 cm per side), dried, and counted as before. The N-14C-methylated streptamine analogs were eluted with the 3 N HCl fractions. The radioactive peaks were pooled and evaporated to dryness in vacuo over petri plates containing NaOH pellets. The residues were taken up in water and counted. Each labeled product had a different value of counts per minute per 5 μl.
Assay of N-[14C]methylstreptamine analogs as amino donors for glutamine:aminocyclitol aminotransferase by a pseudo-half-reaction with scyllo-inosose as amino acceptor (Table 5).
TABLE 5.
Abilities of the N-[14C]methyl streptamine analogs formed by N-methyltransferases from the hygromycin B and spectinomycin producers to serve as amino donors to scyllo-inosose in pseudo-half-reactions catalyzed by glutamine:aminocyclitol aminotransferase from the hygromycin B producera
| Expt no. and reactant | Keto product (cpm/10 μl) | Fraction reacted (%)b |
|---|---|---|
| Expt 1 (streptamine analogs methylated by hygromycin B producer as amino donors) | ||
| N-[14C]methyl-2-deoxystreptamine + scyllo-inosose | 12,060 | 50 |
| N-[14C]methyl streptamine + scyllo-inosose | 7,751 | 53 |
| N-[14C]methyl-2-epi-streptamine + scyllo-inosose | 26,350 | 52 |
| Expt 2 (streptamine analogs methylated by spectinomycin producer as amino donors) | ||
| N-[14C]methyl-2-deoxystreptamine + scyllo-inosose | 4,524 | 24 |
| N-[14C]methylstreptamine + scyllo-inosose | 11,216 | 28 |
| N-[14C]methyl-2-epi-streptamine + scyllo-inosose | 18,828 | 40 |
Radioactive reactants (2+ charge) were separated from labeled products (1+ charge) by high-voltage paper electrophoresis at pH 3.6 (15).
Fraction of total labeled streptamine derivatives that donated an amino group to scyllo-inosose, forming scyllo-inosamine (IN, Fig. 2). This is a reversible reaction, and donors are often a mixture of N1- and N3-methylated streptamines.
Assay mixtures were as follows: N-[14C]methyl-2-deoxy-d-streptamine product of S. hygroscopicus N-methyltransferase (88,000 cpm/5 μl) or other N-[14C]methylstreptamine analog, 10 μl; 0.1 M K phosphate buffer, pH 7.4, plus neutralized pyridoxal-P, 2.5 mg/ml, 5 μl; scyllo-inosose, 10 mg/ml, 10 μl; and S. hygroscopicus sonicate supernatant, 10 μl. The mixture was incubated at 34°C for 2 h. Labeled reactants and products were separated by high-voltage paper electrophoresis at pH 3.6 as before. The labeled amino donor with a 2+ charge was converted to a labeled keto product with a 1+ charge after its transamination. The fraction of label transferred was 50%.
N-[14C]methylstreptamine analogs as possible substrates for d-streptamine 6-kinase in extracts of a streptomycin producer (Table 6).
TABLE 6.
Abilities of N-[14C]methylstreptamine analogs formed by N-methyltransferases from producers of hygromycin B and spectinomycin to serve as phosphate acceptors, catalyzed by a streptamine kinase activity in extracts of the streptomycin producer S. bikiniensisa
| Expt no. and reactant | Phosphorylated product (cpm/10 μl)b | Rf value |
|---|---|---|
| Expt 1 (streptamine analogs methylated by the spectinomycin producer) | ||
| N-[14C]methylstreptamine + MgATP | 17,127 (26) | 0.36 |
| N-[14C]methyl-2-deoxystreptamine + MgATP | 0 | |
| N-[14C]methyl-2-epi-streptamine + MgATP | 0 | |
| Expt 2 (streptamine analogs methylated by the hygromycin B producer) | ||
| N-[14C]methylstreptamine + MgATP | 2,357 (7) | 0.35 |
| N-[14C]methyl-2-deoxystreptamine + MgATP | 0 | |
| N-[14C]methyl-2-epi-streptamine + MgATP | 0 |
Labeled reactants (Rf value, 0.80) and products were separated by ammoniacal phenol paper chromatography, with the indicated Rf values (15).
Assay mixtures were as follows: N-[14C]methyl-d-streptamine analog product of S. flavopersicus N-methyltransferase (124,000 cpm/5 μl), or a product of S. hygroscopicus N-methyltransferase, 10 μl; 0.5 M Tris (pH 7.4), 5 μl; l-ornithine HCl (30 mg/ml), 5 μl; neutralized MgATP (30 mg/ml), 5 μl; and S. bikiniensis sonicate, 10 μl. The mixtures were incubated for 2 h at 33°C. Labeled reactants and products were separated by ascending paper chromatography with an 80% phenol-20% H2O, NH4OH atmosphere. The fraction of total labeled substrate converted to 6-O-phosphoryl product was 27%.
Assay of N-alkylated streptamine analogs as substrates for a coupled streptamine kinase-amidinotransferase reaction (Table 7).
TABLE 7.
Use of a coupled streptamine kinase-arginine:streptamine-P amidinotransferase system to distinguish enantiomers of N-monoalkylated streptamine analogsa
| Reactant | Labeled productb (cpm/10 μl) | Rf |
|---|---|---|
| [guanidino-14C]arginine + MgATP + 2-deoxystreptamine | 16,066 | 0.39 |
| [guanidino-14C]arginine + MgATP + dl-N-methyl-2-deoxystreptamine (IV + V) | 18,741 | 0.58 |
| [guanidino-14C]arginine + MgATP + N1-ethyl-2-deoxy-d-streptamine (VI) | 26,067 | 0.73 |
| [guanidino-14C]arginine + MgATP + N3-methyl-2-deoxy-d-streptamine (IV) | 0 | |
| [guanidino-14C]arginine + MgATP + 2-epi-streptamine (III) | 27,144 | 0.25 |
| [guanidino-14C]arginine + MgATP + dl-N-monomethyl-2-epi-streptamine (VII + VIII) | 20,080 | 0.48 |
Assay mixtures were as follows: [guanidino-14C]arginine (25,000 cpm/5 μl), 5 μl; 0.5 M Tris (pH 7.4), 5 μl; Li2-carbamoyl-P, 10 mg/ml, 5 μl; neutralized MgATP, 30 mg/ml, 5 μl; diluted N1-ethyl-2-deoxy-d-streptamine preparation or streptamine analog, 5 mg/ml, 5 μl; and S. griseocarneus sonicate supernatant, 10 μl. The reaction mixture was incubated at 34°C for 2 h. Labeled reactants and products were separated by paper chromatography with an 80% phenol-20% H2O, NH4OH atmosphere (15, 18).
RESULTS AND DISCUSSION
Search for enzymes that can methylate 2-deoxystreptamine (II) in extracts of a hygromycin B producer.
Cultures of S. hygroscopicus were grown on complex media in batch culture, harvested during early stationary phase (19), and sonicated, and the sonicate supernatants were assayed for their ability to catalyze transfer of the methyl group of adenosyl[methyl-14C]methionine (Ado[14C]Met) to 2-deoxystreptamine. The results are shown in experiment 1 of Table 1. An impressive 40% of the donor methyl label was transferred to 2-deoxystreptamine.
The question next arose as to whether the N1- or N3-amino group was preferentially methylated. The results are shown in experiment 2 of Table 1, employing monoalkylated 2-deoxystreptamine derivatives of known structures as potential methyl acceptors. Whereas N3-methyl-2-deoxy-d-streptamine (IV) is a weak acceptor, N1-ethyl-2-deoxy-d-streptamine (VI) is as active an acceptor as is 2-deoxystreptamine. It can therefore be concluded that, since reaction 1 occurs, reaction 2 also occurs. Reactions 1 and 2 are as follows:
 |
(1) |
 |
(2) |
Reaction 2 represents the final step in biosynthesis of the aminocyclitol core (IV) of hygromycin B.
Abilities of other aminocyclitols to undergo methylation by extracts of a hygromycin B producer.
The results of experiment 1 of Table 2 show that streptamine (I) can be readily methylated in vitro, despite the fact that N-methylated streptamine cores have not yet been detected among the isolated destomycins (10). scyllo-Inosamine (IN, Fig. 2), even at 50 times the concentration of streptamine, did not inhibit methylation of streptamine. Surprisingly, 2-epi-streptamine (III) is an excellent methyl acceptor, as good as 2-deoxystreptamine is. However, a mixture of N1- and N3-methyl-2-epi-d-streptamine enantiomers (VII and VIII) was not significantly methylated. It is concluded that, since reaction 1 occurs, both reaction 3 and reaction 4 can also be catalyzed in good yields by this hygromycin B producer. Reactions 3 and 4 are as follows:
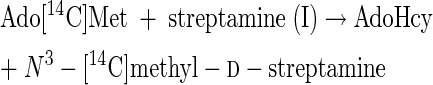 |
(3) |
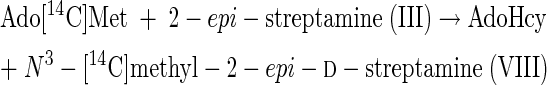 |
(4) |
TABLE 2.
Methylation of other streptamine analogs by extracts of the hygromycin B producer, with Ado[14C]Met as methyl donora
| Expt no. and reactant(s) | Final concn (μg/ml) in reaction mixture | N-[14C]methyl product formed (cpm/10 μl)b |
|---|---|---|
| Expt 1 | ||
| Ado[14C]Met + streptamine (I) | 143 | 17,312 (24) |
| Expt 2 | ||
| Ado[14C]Met + 2-epi-streptamine (III) | 143 | 37,549 (52) |
| Ado[14C]Met + 2-epi-streptamine + N3-methyl-2-deoxystreptamine (IV) | 143, 713c | 3,821 (5) |
| Ado[14C]Met + dl-N-methyl-2-epi-streptamine (VII + VIII) | 286 | 602 (1) |
| Ado[14C]Met + dl-N-methyl-2-epi-streptamine | 57 | 0 |
Labeled reactants and products were separated by high-voltage paper electrophoresis at pH 3.6 (15).
Numbers in parentheses are percentages of transfer of labeled methyl group from donor to acceptor.
Respectively.
Search for enzymes that can methylate 2-epi-streptamine (III) in extracts of a spectinomycin producer.
Biosynthetically competent extracts of S. flavopersicus, which could convert myo-[14C]inositol (mI, Fig. 2) plus NAD+, oxalacetate, and l-glutamine or N3-methyl-2-deoxy-d-streptamine (IV) to scyllo-[14C]inosamine (IN, Fig. 2) (18), were tested for their ability to methylate 2-epi-streptamine (III). The results are shown in experiment 1 of Table 3. A remarkable 51% of the donor methyl label was transferred to 2-epi-streptamine. When a mixture of N1- and N3-methyl-2-epi-d-streptamine enantiomers (VII and VIII) was the methyl acceptor, 27% of the methyl label was transferred. When the N1,N3-dimethyl-2-epi-streptamine (IX) core of spectinomycin was tested, no labeled methyl group was transferred, thereby ruling out any O-methylation.
TABLE 3.
Methylation of 2-epi-streptamine (III) by extracts of the spectinomycin producer S. flavopersicus with Ado[14C]Met as methyl donora
| Expt no. and reactant | Final concn (μg/ml) in reaction mixture | N-[14C]methyl product formed (cpm/10 μl)b |
|---|---|---|
| Expt 1 | ||
| Ado[14C]Met + 2-epi-streptamine (III) | 114 | 36,640 (51) |
| Expt 2 | ||
| Ado[14C]Met + dl-N-methyl-2-epi-streptamine (VII + VIII) | 286 | 19,552 (27) |
| Ado[14C]Met + dl-N-methyl-2-epi-streptamine | 57 | 8,443 (12) |
Labeled reactants and products were separated by high-voltage paper electrophoresis at pH 3.6 (15).
Numbers in parentheses are percentages of transfer of labeled methyl group from donor to acceptor.
Abilities of other aminocyclitol derivatives to be methylated by extracts of the spectinomycin producer.
Experiment 1 of Table 4 shows that streptamine is a very good methyl acceptor. Most importantly for our third goal and contrary to our expectations (see L. Slechta and J. H. Coats, Abstr. 14th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 294, 1974), 2 deoxystreptamine (II), N1-ethyl-2-deoxy-d-streptamine (VI), and N3-methyl-2-deoxy-d-streptamine (IV) were all excellent methyl acceptors as well.
From the results of Tables 3 and 4 it can be inferred that the following reactions can be catalyzed by extracts of this spectinomycin producer.
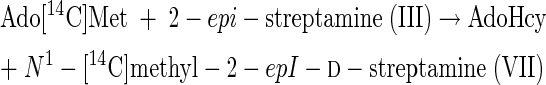 |
(5) |
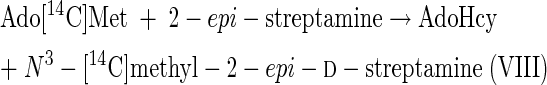 |
(6) |
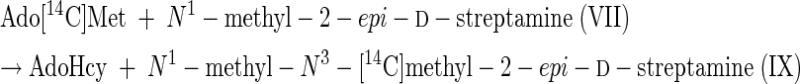 |
(7) |
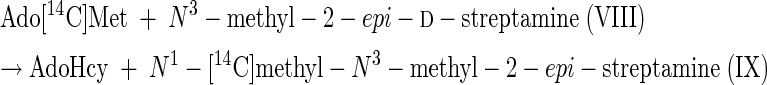 |
(8) |
 |
(3) |
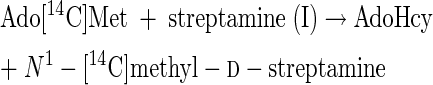 |
(9) |
 |
(10) |
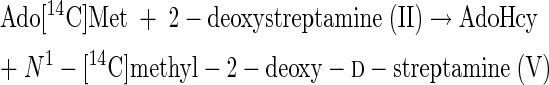 |
(11) |
 |
(2) |
 |
(12) |
 |
(1) |
Reactions 5 to 8 represent the final steps in the biosynthesis of the aminocyclitol core (IX) of spectinomycin. Although it is beyond the scope of this study to identify the gene or genes that code for these N-methyltransferase activities, it is suggested that a comparison of the results of purifications based on the assays represented by reactions 1 and 12 should resolve the question of whether one or two different enzymes, or isozymes, are involved. The scyllo configurations and symmetry of streptamine and 2-deoxystreptamine are such that it is possible that flip-flopping the rings could permit one enzyme to add both methyl groups.
Abilities of N-[14C]methylstreptamine analogs to serve as surrogate substrates for l-glutamine:aminocyclitol aminotransferase.
The N-[14C]methyl derivatives of 2-deoxystreptamine, streptamine, and 2-epi-streptamine produced in 100-fold scaled-up incubations with each of the two N-methyltransferase preparations were separated from unreacted methyl donor and other labeled products by cation-exchange column chromatography. The isolated labeled streptamine analogs were then tested for their ability to serve as amino donors in pseudo-half-reactions catalyzed by glutamine:aminocyclitol aminotransferases in the same mycelial extracts. The results are shown in Table 5, with extracts of the hygromycin B producer as the source of aminotransferase. Similar results were obtained when extracts of the spectinomycin producer were the source of aminotransferase, with yields of 19 to 24%.
All six labeled aminocyclitol isolates contained high levels of active amino donors in this assay, which also provided the first evidence for this aminocyclitol aminotransferase in the destomycin family of antibiotics. Since we have previously shown that nonlabeled N3-methyl-2-deoxy-d-streptamine (IV) is an excellent amino donor for aminocyclitol aminotransferases in producers of streptomycin (18), spectinomycin (18), and bluensomycin (18), as well as producers of the 2-deoxystreptamine antibiotics gentamicin and neomycin (6, 7, 16), the results of Table 5 can be represented by reactions 13 to 15.
 |
(13) |
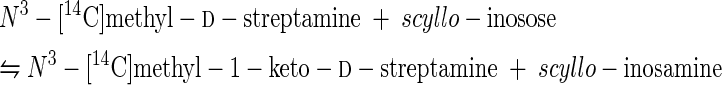 |
(14) |
 |
(15) |
Abilities of N-[14C]methylstreptamine analogs to be phosphorylated by a d-streptamine 6-kinase in extracts of a streptomycin producer.
Streptomycin and bluensomycin producers have been found to possess a total of four different aminocyclitol kinase activities (enzymes E, J, V, and W, Fig. 2). In addition there is a kinase that phosphorylates streptamine and 2-deoxystreptamine at position 6 (13, 23, 25) that is different from enzyme W (Fig. 2) (20) but might be the same as enzyme E (Fig. 2). Kinases E, J, and V are each different enzymes (17). It was of interest to determine whether any of the six N-[14C]methylstreptamine analogs used for the above transaminations might provide an easily obtained substrate for assays of d-streptamine 6-kinase activity. The results are shown in Table 6. At these low surrogate substrate concentrations the only reaction with a high-enough kcat/Km value was that shown as reaction 16.
 |
(16) |
This relative specificity raised the question of whether at higher substrate concentrations other N1-alkylated d-streptamine analogs with lower kcat/Km values could be phosphorylated by this kinase (21). These higher concentrations would require use of nonlabeled streptamine derivatives and a coupled d-streptamine 6-kinase-[guanidino-14C]arginine:d-streptamine-6-P amidinotransferase assay to detect the products (13, 21). The results of such experiments are shown in Table 7. Whereas N3-methyl-2-deoxy-d-streptamine (IV) was inactive in this assay, N1-ethyl-2-deoxy-d-streptamine (VI) and the N1-methyl-2-epi-d-streptamine (VII) enantiomer of the dl-N-methyl-2-epi-streptamine racemic mixture (VII + VIII) were indeed sequentially phosphorylated and transamidinated (enzyme F, Fig. 2), as depicted in reactions 17 to 22.
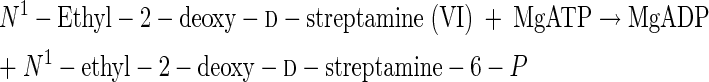 |
(17) |
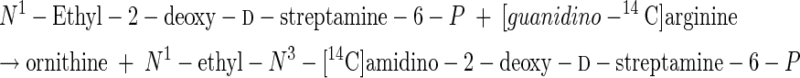 |
(18) |
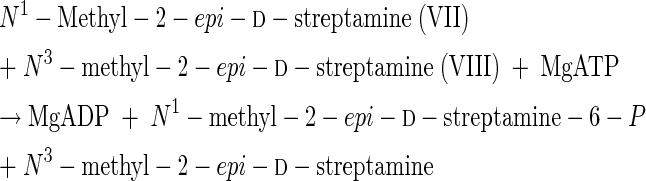 |
(19) |
 |
(20) |
 |
(21) |
 |
(22) |
These results confirm our earlier conclusions that this kinase phosphorylates the hydroxyl group para to a free amino group, which can then be amidinated by arginine:aminocyclitol-P amidinotransferase (enzyme F, Fig. 2) (21, 23-25). This ability to distinguish between N3- and N1-methyl-2-deoxy-d-streptamine enantiomers (IV and V) and separate them would have also been useful during the controversies concerning the respective structures of the aminocyclitol cores of hygromycin B and destomycin A (5). (Indeed, the 12th edition of the Merck Index [1996] still had the wrong structure for destomycin A.)
Some other potential uses for streptamine N-methyltransferases.
Streptamine and its analogs have properties compatible with possible roles as proprietary nutrient reserves (17), osmolytes, and/or extended-range pH buffers (pKas of 7.0 and 9.0; see SigmaUltra buffer no. 9410; Sigma Chemical Co.) during some stages in the feast-or-famine life cycles of streptomycetes in soils. Streptamine N-methyltransferases can provide the first specific assays for evaluation of such postulated roles.
In the biosynthesis of aminoglycoside antibiotics the next step following synthesis of the aminocyclitol cores usually involves an NDP-sugar:streptamine glycosyltransferase. The structure of kanamycin A (10) suggests that glucose itself is the first substituent on its 2-deoxystreptamine core, and mutasynthesis experiments have shown that N-methyl-2-deoxystreptamines can be incorporated into a number of antibiotics (4, 9). Reaction of a biosynthetically competent (aminotransferase-positive) extract of a kanamycin producer with the N-[14C]methyl-2-deoxy-d-streptamine products of spectinomycin N-methyltransferase(s) plus an NDP-glucose should produce the predicted labeled conjugate. The labeled conjugate in turn could readily serve as a substrate for the detection of later biosynthetic reactions. It remains to be seen whether insertion of a 2-deoxystreptamine N1-methyltransferase gene into producers of other 2-deoxystreptamine antibiotics would result in antibiotics useful against certain resistant organisms.
REFERENCES
- 1.Chen, Y.-M., and J. B. Walker. 1977. Transaminations involving keto- and amino-inositols and glutamine in actinomycetes which produce gentamicin and neomycin. Biochem. Biophys. Res. Commun. 77:688-692. [DOI] [PubMed] [Google Scholar]
- 2.Huddleston, A. S., N. Cresswell, M. Cristina, P. Neves, J. E. Beringer, S. Baumberg, D. E. Thomas, and E. M. H. Wellington. 1997. Molecular detection of streptomycin-producing streptomycetes in Brazilian soils. Appl. Environ. Microbiol. 63:1288-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ikeda, Y., S. Kondo, F. Kanai, T. Sawa, M. Hamada, T. Takeuchi, and H. Umezawa. 1985. A new destomycin-family antibiotic produced by Saccharopolyspora hirsuta. J. Antibiot. 38:436-438. [DOI] [PubMed] [Google Scholar]
- 4.Kojima, M., and A. Satoh. 1973. Microbial semi-synthesis of aminoglycosidic antibiotics by mutants of S. ribosidificus and S. kanamyceticus. J. Antibiot. 26:184-185. [DOI] [PubMed] [Google Scholar]
- 5.Kondo, S., K. Iinuma, H. Naganawa, and Y. Sekizawa. 1975. Structural studies on destomycins A and B. J. Antibiot. 28:79-82. [DOI] [PubMed] [Google Scholar]
- 6.Lucher, L. A. 1983. Ph.D. thesis. Rice University, Houston, Tex. University Microfilms International, Ann Arbor, Mich.
- 7.Lucher, L. A., Y.-M. Chen, and J. B. Walker. 1989. Reactions catalyzed by purified l-glutamine:keto-scyllo-inositol aminotransferase, an enzyme required for biosynthesis of aminocyclitol antibiotics. Antimicrob. Agents Chemother. 33:452-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moazed, D., and H. F. Noller. 1987. Interaction of antibiotics with functional sites in 16S ribosomal RNA. Nature 327:389-394. [DOI] [PubMed] [Google Scholar]
- 9.Rinehart, K. L., Jr. 1980. Biosynthesis and mutasynthesis of aminocyclitol antibiotics. Am. Chem. Soc. Symp. Ser. 125:335-370. [PubMed] [Google Scholar]
- 10.Rinehart, K. L., Jr., and L. S. Shield. 1980. Aminocyclitol antibiotics: an introduction. Am. Chem. Soc. Symp. Ser. 125:1-11. [Google Scholar]
- 11.Schroeder, R., C. Waldsich, and H. Wank. 2000. Modulation of RNA function by aminoglycoside antibiotics. EMBO J. 19:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thomas, R. C. 1982. Spectinomycin modification. I. Catalytic N-demethylation of spectinomycin. J. Antibiot. 35:1202-1207. [DOI] [PubMed] [Google Scholar]
- 13.Walker, J. B. 1971. Enzymatic reactions involved in streptomycin biosynthesis and metabolism. Lloydia (J. Nat. Prod.) 34:363-371. [PubMed] [Google Scholar]
- 14.Walker, J. B. 1974. Biosynthesis of the monoguanidinated inositol moiety of bluensomycin, a possible evolutionary precursor of streptomycin. J. Biol. Chem. 249:2397-2404. [PubMed] [Google Scholar]
- 15.Walker, J. B. 1975. Pathways of biosynthesis of the guanidinated inositol moieties of streptomycin and bluensomycin (eight articles). Methods Enzymol. 43:429-470. [DOI] [PubMed] [Google Scholar]
- 16.Walker, J. B. 1980. Biosynthesis of aminoglycoside antibiotics. Dev. Ind. Microbiol. 21:103-113. [Google Scholar]
- 17.Walker, J. B. 1990. Possible evolutionary relationships between streptomycin and bluensomycin biosynthetic pathways: detection of novel inositol kinase and O-carbamoyltransferase activities. J. Bacteriol. 172:5844-5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walker, J. B. 1995. Enzymatic synthesis of aminocyclitol moieties of aminoglycoside antibiotics from inositol by Streptomyces spp.: detection of glutamine:aminocyclitol aminotransferase and diaminocyclitol aminotransferase activities in a spectinomycin producer. J. Bacteriol. 177:818-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walker, J. B., and V. S. Hnilica. 1964. Developmental changes in arginine:X amidinotransferase activity in streptomycin-producing strains of Streptomyces. Biochim. Biophys. Acta 89:473-482. [DOI] [PubMed] [Google Scholar]
- 20.Walker, J. B., and M. S. Walker. 1967. Streptomycin biosynthesis. Enzymatic synthesis of O-phosphorylstreptidine from streptidine and adenosinetriphosphate. Biochim. Biophys. Acta 148:335-341. [DOI] [PubMed] [Google Scholar]
- 21.Walker, J. B., and M. S. Walker. 1967. Enzymatic synthesis of streptidine from scyllo-inosamine. Biochemistry 6:3821-3829. [DOI] [PubMed] [Google Scholar]
- 22.Walker, J. B., and M. S. Walker. 1969. Streptomycin biosynthesis. Transamination reactions involving inosamines and inosadiamines. Biochemistry 8:763-770. [DOI] [PubMed] [Google Scholar]
- 23.Walker, J. B., and M. S. Walker. 1973. Phosphate transfer from dihydrostreptomycin 6-phosphate to inosamines, streptamine, and 2-deoxystreptamine. J. Biol. Chem. 248:2441-2446. [PubMed] [Google Scholar]
- 24.Walker, M. S., and J. B. Walker. 1966. Enzymic studies on the biosynthesis of streptomycin. Transamidination of inosamine and streptamine derivatives. J. Biol. Chem. 241:1262-1270. [PubMed] [Google Scholar]
- 25.Walker, M. S., and J. B. Walker. 1971. Streptomycin biosynthesis: separation and substrate specificities of phosphatases acting on guanidinodeoxy-scyllo-inositol phosphate and streptomycin(streptidino)phosphate. J. Biol. Chem. 246:7034-7040. [PubMed] [Google Scholar]




