Abstract
Stimulating microbial reduction of soluble U(VI) to insoluble U(IV) shows promise as a strategy for immobilizing uranium in uranium-contaminated subsurface environments. In order to learn more about which microorganisms might be involved in U(VI) reduction in situ, the changes in the microbial community when U(VI) reduction was stimulated with the addition of acetate were monitored in sediments from three different uranium-contaminated sites in the floodplain of the San Juan River in Shiprock, N.Mex. In all three sediments U(VI) reduction was accompanied by concurrent Fe(III) reduction and a dramatic enrichment of microorganisms in the family Geobacteraceae, which are known U(VI)- and Fe(III)-reducing microorganisms. At the point when U(VI) reduction and Fe(III) reduction were nearing completion, Geobacteraceae accounted for ca. 40% of the 16S ribosomal DNA (rDNA) sequences recovered from the sediments with bacterial PCR primers, whereas Geobacteraceae accounted for fewer than 5% of the 16S rDNA sequences in control sediments that were not amended with acetate and in which U(VI) and Fe(III) reduction were not stimulated. Between 55 and 65% of these Geobacteraceae sequences were most similar to sequences from Desulfuromonas species, with the remainder being most closely related to Geobacter species. Quantitative analysis of Geobacteraceae sequences with most-probable-number PCR and TaqMan analyses indicated that the number of Geobacteraceae sequences increased from 2 to 4 orders of magnitude over the course of U(VI) and Fe(III) reduction in the acetate-amended sediments from the three sites. No increase in Geobacteraceae sequences was observed in control sediments. In contrast to the predominance of Geobacteraceae sequences, no sequences related to other known Fe(III)-reducing microorganisms were detected in sediments. These results compare favorably with an increasing number of studies which have demonstrated that Geobacteraceae are important components of the microbial community in a diversity of subsurface environments in which Fe(III) reduction is an important process. The combination of these results with the finding that U(VI) reduction takes place during Fe(III) reduction and prior to sulfate reduction suggests that Geobacteraceae will be responsible for much of the Fe(III) and U(VI) reduction during uranium bioremediation in these sediments.
Microbial reduction of U(VI) to U(IV) may be an important reaction influencing uranium biogeochemistry in a variety of sedimentary environments and is a promising strategy for the bioremediation of uranium-contaminated groundwaters (15, 18). This microbial process is particularly significant because U(VI) is highly soluble, and therefore mobile, in most groundwaters (14), whereas the reduced form of uranium, U(IV), is generally insoluble and precipitates from most groundwaters. Therefore, stimulating microbial reduction of U(VI) could potentially stop the spread of uranium contamination in the subsurface and, if the bioremediation process is engineered properly, concentrate uranium into a discrete zone for subsequent recovery.
Rational engineering of uranium bioremediation requires detailed information on the microorganisms responsible for U(VI) reduction in the subsurface environment. Two major groups of microorganisms are known to be capable of U(VI) reduction, dissimilatory Fe(III)-reducing microorganisms (18) and sulfate-reducing microorganisms (17, 19). Recent studies that have investigated microbial bioremediation of uranium have focused on the potential role of sulfate-reducing microorganisms in this process (6). However, an investigation of the biogeochemistry of uranium-contaminated subsurface sediments in which microbial U(VI) reduction was stimulated with the addition of organic compounds indicated that U(VI) reduction took place concurrently with Fe(III) reduction and prior to sulfate reduction (10; K. T. Finneran, M. Housewright, and D. R. Lovley, submitted for publication). These results suggest that Fe(III)-reducing microorganisms rather than sulfate-reducing microorganisms are primarily responsible for the U(VI) reduction.
Although Fe(III)-reducing microorganisms can conserve energy to support growth from U(VI) reduction (18), microbially reducible Fe(III) oxides provided >3 orders of magnitude more electron-accepting capacity in uranium-contaminated subsurface sediments (10; Finneran et al., submitted). This finding suggests that when U(VI) reduction was stimulated, most of the growth of the Fe(III)-reducing microorganisms resulted from Fe(III) reduction rather than U(VI) reduction (10).
Phylogenetically distinct Fe(III)-reducing microorganisms may have fundamentally different mechanisms for conserving energy to support growth from the reduction of insoluble Fe(III) oxides. For example, Shewanella species (23, 25) and Geothrix fermentans (24) can reduce Fe(III) oxides that they cannot contact by releasing compounds that can shuttle electrons from the cell surface to Fe(III) oxide. These organisms can also solubilize Fe(III) from Fe(III) oxides, further alleviating the need for contact between the organisms and the Fe(III) oxides (23, 24). In contrast, Geobacter metallireducens does not produce electron shuttling compounds or Fe(III) chelators (22), and it specifically produces pili during growth on insoluble Fe(III) or Mn(IV) oxides but not during growth on soluble electron acceptors (7). This coupled with its chemotaxis to Fe(II) and Mn(II) suggests that Geobacter metallireducens searches for insoluble Fe(III) and Mn(IV) oxides and then establishes direct contact with the Fe(III) and Mn(IV) oxides in order to reduce them.
It is important to take the significant physiological differences, not only between Fe(III) reducers and sulfate reducers but also among different types of Fe(III)-reducing microorganisms, into account when effective bioremediation strategies in uranium-contaminated sites are designed. Therefore, it is necessary to know which microorganisms are involved in U(VI) reduction in order to fully understand microbial U(VI) reduction in subsurface environments. Here we report on studies with three geochemically different sediments from a uranium mill tailings site with uranium-contaminated groundwater. Previous studies have demonstrated that the addition of acetate stimulated microbial U(VI) reduction and effectively removed uranium from groundwater (10). The data presented here demonstrate that the period of U(VI) reduction was associated with a remarkable increase in 16S ribosomal DNA (rDNA) sequences closely related to the 16S rDNA sequences of Fe(III)-reducing microorganisms in the family Geobacteraceae.
MATERIALS AND METHODS
Sample collection and sediment incubations.
The molecular analyses reported in this study were conducted on the same sediment incubations in which the potential for microbial metal reduction to remove uranium from groundwater was previously investigated (10; Finneran et al., submitted). Sediments were collected from below the water table within the floodplain of the San Juan River in Shiprock, N.Mex., as previously described (10). The floodplain is contaminated with uranium from an adjacent uranium mill tailings pile which is a designated Department of Energy Uranium Mill Tailings Remedial Act site (12). Sediments were collected from three separate zones near wells 1103, 853, and 857.
Groundwater and sediment chemistry was analyzed as previously reported (10; Finneran et al., submitted). Briefly, sediment Fe(II) that could be extracted with 0.5 N HCl was measured with ferrozine, and total HCl-extractable iron was determined after reduction of Fe(III) to Fe(II) with hydroxylamine. Nitrate, nitrite, and sulfate were quantified with a DX-100 ion chromatograph and an AS4-SC IonPac column (Dionex Corp., Sunnyvale, Calif.). U(VI) dissolved in the groundwater was quantified with a kinetic phosphorescence analyzer (Chemchek Corp., La Brea, Calif.).
Uranium, nitrate, and sulfate concentrations at site 1103 were ca. 1 μM, 19 mM, and 49 mM, respectively. There was no detectable nitrate at site 853 or 857, but soluble U(VI) was also approximately 1 μM at both of these sites. Site 853 had ca. 19 mM sulfate; site 857 had ca. 28 mM sulfate. The concentrations of iron extractable in 0.5 N HCl were 12 μmol/g at site 1103, 6.4 μmol/g at site 857, and 7.1 μmol/g at site 853. At the time of collection all the HCl-extractable iron in site 1103 sediments was present as Fe(III). In site 857 sediments, less than 5% of the iron was present as Fe(II), and in site 853 sediments, the percent Fe(II) was 25%. The pH at all sites was between 6.8 and 7.1.
As previously described (10; Finneran et al., submitted), sediments from each site were incubated under strict anaerobic conditions, and the reduction of nitrate, U(VI), and Fe(III) was monitored over time. Some sediments were amended with acetate to stimulate the reduction of these electron acceptors. For the molecular studies reported here, sediment aliquots (ca. 3 g) were taken at appropriate time points with a sterile spatula and frozen at −70°C in sterile 2-ml tubes for molecular analysis.
Amplification of 16S rDNA sequences and construction of clone libraries.
Four separate clone libraries were constructed for each of the sampling sites. For sites 853 and 857, clone libraries were constructed from both nonamended and acetate-amended sediments collected at time zero and after 23 days of incubation. For site 1103, clone libraries were constructed from both nonamended and acetate-amended sediments collected after 2 and 37 days of incubation. DNA was extracted from the sediments with a fast DNA spin kit for soil (Bio101, Inc., Carlsbad, Calif.). 16S rDNA sequences were amplified with the two following primer combinations: 8 forward (9) with 519 reverse (13) and 338 forward (2) with 907 reverse (13). The total volume of each PCR mixture was 100 μl: ∼6 ng of DNA template, 10 μl of 10X buffer (15 mM MgCl2; Qiagen, Valencia, Calif.), 5 μl of buffer Q (Qiagen), 8 μl of a 0.25 μM deoxynucleoside triphosphate solution (Sigma Chemical Company, St. Louis, Mo.), 60 pmol of forward and reverse primers, 5 μl of dimethyl sulfoxide, and 3 U of Taq polymerase (Qiagen). To ensure sterility, the PCR mixtures were exposed to UV radiation for 10 min prior to the addition of template and Taq polymerase. PCR amplification was performed in a DNA Engine thermal cycler (MJ Research, Inc., Waltham, Mass.) with an initial denaturation step at 94°C for 4 min, followed by 35 cycles of 94°C (30 s), 50°C (30 s), and 72°C (45 s) with a final extension at 72°C for 7 min. PCR products amplified with the two primer sets were pooled prior to construction of clone libraries. Clone libraries were constructed from the pooled 16S rDNA inserts with a TOPO TA cloning kit, version K2 (Invitrogen, Carlsbad, Calif.), according to the manufacturer's instructions.
Restriction enzyme analysis and sequencing of 16S rDNA.
A total of 30 clones from each clone library were selected for restriction enzyme analysis. 16S rDNA was amplified from various clones using M13 forward and reverse primers (Invitrogen). The amplified inserts (∼500 ng) were digested for 16 h at 37°C with HhaI and MspI (New England Biolabs, Beverly, Mass.) according to the manufacturer's instructions. Restriction fragments were visualized on a 3% Metaphor agarose gel (BioWhittaker Molecular Applications, Rockland, Maine), and 16S rDNA inserts with similar banding patterns were assumed to be identical. Plasmid inserts were sequenced at the University of Massachusetts Amherst automated sequencing facility with the M13 forward primer.
Phylogenetic analysis of clones.
Sequences were compared to the GenBank and RDP databases with the BLAST (1), and SIMILARITY_RANK (21) algorithms. Representative sequences were manually aligned in the Wisconsin Package, version 10.2 (Genetics Computer Group, Madison, Wis.), and hypervariable regions were masked. The aligned sequences were imported into PAUP 4.0b 4a (Sinauer Associates, Sunderland, Mass.), where phylogenetic trees were inferred. Branching order was determined and compared by using a character-based (maximum parsimony) and a distance-based (HKY-85 four-parameter model) algorithm. Bootstrap analysis was performed with the distance-based HKY-85 four-parameter model, with 100 replicates.
The GenBank accession numbers of the sequences used as references are as follows: Desulfobacter hydrogenophilus, DSPRR1613; Desulfuromonas palmitatis, U28172; Desulfuromusa bakii, X79412; Pelobacter acidigallici, X77216; Geobacter metallireducens, L07834; Geobacter chapellei, U41561; Deferribacter thermophilus, DTU75602; Geovibrio ferrireducens, GF16SRR; Thermotoga maritima, AJ401017; Thermoterrabacterium ferrireducens, TFU76364; Bacillus infernus, U20384; Geothrix fermentans, GFU41563; Thiobacillus ferrooxidans, AB039820; Aeromonas hydrophila, AH16SRDN; Ferrimonas balearica, FB16S; Shewanella putrefaciens, AF170300; Ferribacterium limneticum, UBPR16S, and Sulfurospirillum barnesii, GBU41564.
Testing and design of Geobacteraceae-specific primers.
A manual alignment of 16S rDNA sequences from various Desulfuromonas, Pelobacter, and Desulfuromusa isolates and clones was performed with the Wisconsin Package, version 10.2. This alignment was then imported into Primer Express, version 1.0 (PE Applied Biosystems, Foster City, Calif.), and the forward primer Geobacteraceae-494F (5′-AGG AAG CAC CGG CTA ACT CC-3′) was designed from a consensus region identified by the program. A previously designed Geobacter-specific reverse primer, Geo825R (5), was used with Geobacteraceae-494F to yield an amplicon of approximately 330 bp. Optimum temperature and cycle parameters were determined in a gradient thermal cycler (MJ Research, Inc.). Optimum amplification parameters were found to be 94°C for 5 min, 35 cycles of 94°C (20 s), 51°C (20 s), and 72°C (30 s), followed by a final elongation step at 72°C for 7 min. Primers Geobacteraceae-494F and Geo825R were tested on DNA extracted from pure cultures of Desulfuromonas acetoxidans, Pelobacter carbinolicus, Geobacter sulfurreducens, Escherichia coli, and Desulfuromusa succinoxidans. 16S rRNA genes were amplified from all of the organisms with the exception of E. coli. In order to further evaluate the specificity of the primers, a clone library was constructed from 16S rDNA fragments amplified from the 37-day time point of the acetate-amended site 1103 sediment with Geobacteraceae-494F and Geo825R using the TOPO TA cloning kit, version K2 (Invitrogen). Thirty clones were selected, and all 30 clone inserts were most similar to 16S rDNA sequences from Desulfuromonas or Geobacter species.
Assessment of Desulfuromonas and Geobacter 16S rDNA sequences with MPN PCR and TaqMan quantitative analyses.
Prior to most-probable-number (MPN) and TaqMan analyses, the extracted DNA was further purified with the Wizard DNA clean-up system (Promega, Madison, Wis.). Five-tube MPN PCR analyses were performed, as previously described (5), with DNA extracted from sediment samples taken on days 0, 2, 5, 9, 23, 27, and 37 for site 1103 and days 0 and 23 for sites 853 and 857. Serial 10-fold dilutions were made, and 5-μl aliquots of the diluted DNA were amplified with Geobacteraceae-specific primers (Geobacteraceae-494F and Geo825R). PCR products were visualized on an ethidium bromide-stained agarose gel. The highest dilution that yielded product was noted, and a standard five-tube MPN chart was consulted to estimate the number of 16S rRNA genes in each sample.
Optimal TaqMan PCR conditions were determined using the manufacturer's guidelines. Each PCR consisted of a total volume of 50 μl and contained 60 pmol of Geobacteraceae-494F and Geo-825R, 5 μl of template, 5 μl of 10× SYBR green PCR buffer (PE Biosystems, Warrington, United Kingdom), 6 μl of 25 mM MgCl2 solution, 4 μl of 2.5 mM deoxynucleoside triphosphate mix, 3 μl of buffer Q solution (Qiagen), and 2.5 μl of dimethyl sulfoxide. Three different genomic DNA concentrations were used for each sample subjected to TaqMan PCR: ∼120 ng/μl (undiluted), ∼24 ng/μl (1:5 dilution), and ∼12 ng/μl (1:10 dilution). PCR amplification and detection were performed with the GeneAmp 5700 sequence detection system (PE Biosystems, Foster City, Calif.) with 1 cycle of 50°C for 2 min, 1 cycle of 95°C for 10 min, and 45 cycles of 95°C for 15 s and 56°C for 60 s. The standard curve was generated with known concentrations of purified PCR product from amplification of Desulfuromonas acetoxidans 16S rDNA with primers 494F and 825R. The template concentrations for the standard curve ranged from 8 × 104 to 8 × 10−5 ng/μl.
RESULTS
Reduction of U(VI), Fe(III), and nitrate observed previously.
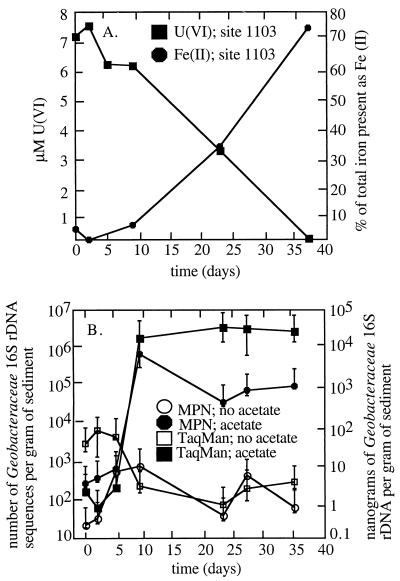
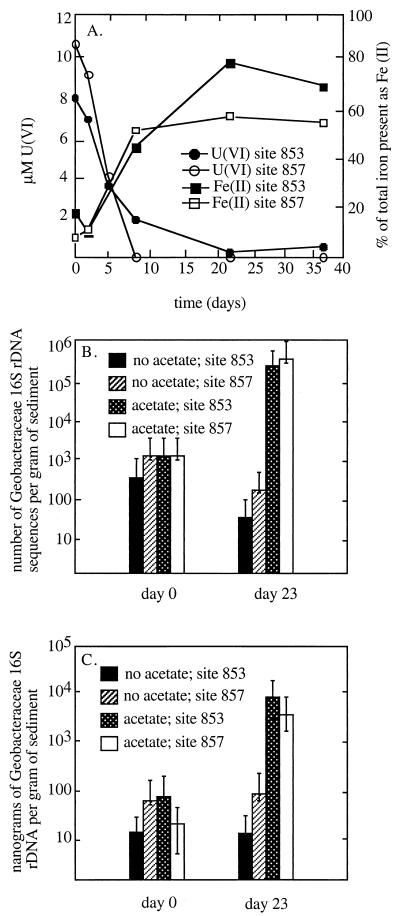
A previous study (Finneran et al., submitted) demonstrated that the addition of acetate stimulated the reduction of the ca. 5 mM nitrate initially present in site 1103 sediments. Following rapid reduction of nitrate in the first 5 days of incubation (Finneran et al., submitted), U(VI) and Fe(III) were reduced concurrently (Fig. 1A). Acetate also stimulated simultaneous reduction of U(VI) and Fe(III) in the sediments from sites 853 and 857 (10), which did not contain detectable nitrate (Fig. 2A).
FIG. 1.
U(VI) and Fe(II) concentrations in acetate-amended site 1103 sediments over time (data are from Finneran et al. [submitted]) (A) and Geobacteraceae 16S rDNA sequences quantified by five-tube MPN PCR and TaqMan (B). Each point is the average of five replicates. The error bars represent 95% confidence intervals.
FIG. 2.
U(VI) and Fe(II) concentrations at sites 853 and 857 over time (data are from reference 10) (A) as well as Geobacteraceae 16S rDNA sequences as quantified by five-tube MPN PCR (B) and TaqMan (C). Each point is the average of five replicates. The error bars represent 95% confidence intervals.
Analysis of clone libraries.
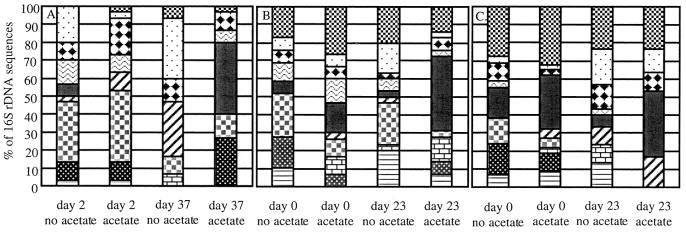
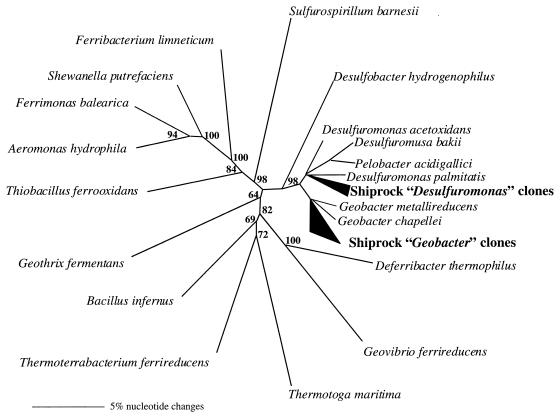
In order to evaluate potential changes in the microbial community associated with U(VI) and Fe(III) reduction in the sediments from site 1103, 16S rDNA sequences on day 2 and day 37 from acetate-amended and control sediments were analyzed. On day 2, which was prior to the start of U(VI) and Fe(III) reduction, the 16S rDNA sequences were diverse for both the acetate-amended and control sediments (Fig. 3A). By day 37, a time point at which Fe(III) reduction and U(VI) reduction were near completion in the acetate-amended sediments, the microbial composition of the acetate-amended sediment appeared to change significantly. Most notably, in the sediments in which metal reduction had been stimulated, there was a dramatic increase in the relative proportion of 16S rDNA sequences most closely related to 16S rDNA sequences of known Geobacteraceae. Geobacteraceae sequences increased from 3% of the cloned 16S rDNA sequences on day 2 to 40% on day 37 (Fig. 3A). The Geobacteraceae sequences detected in the clone libraries fell into two clusters, one most closely related to known species of the genus Desulfuromonas and the other most similar to known Geobacter species (Fig. 4). Of the Geobacteraceae sequences recovered on day 37, 65% were most closely related to known Desulfuromonas species, while the remaining 35% were most similar to Geobacter species. In contrast, in the control sediments not amended with acetate, no Geobacteraceae sequences were recovered on day 37. The other notable change in the acetate-amended sediments was an increase in 16S rDNA sequences closely related to the genus Azoarcus. The percentage of Azoarcus sequences increased from 4 to 27% in the acetate-amended sediments, whereas no Azoarcus sequences were detected in the control sediments on day 37.
FIG. 3.
Relative proportions of organisms in clone libraries from site 1103 (A), site 857 (B), and site 853 (C). , Firmicutes; , Clostridium/Bacillus; , Cytophagales; , Geobacteraceae; , other delta proteobacteria; , gamma proteobacteria; , Azoarcus; , other beta proteobacteria; , alpha proteobacteria; , other.
FIG. 4.
Phylogenetic tree constructed by maximum parsimony analysis showing the relationship of the Geobacteraceae clones to 16S rDNA sequences of previously described bacteria. Thermotoga maritima was used as the outgroup, and bootstrap analysis was done with 100 replicates.
Very few Azoarcus sequences were detected in the sediments from sites 853 and 857, which did not contain nitrate. However, Geobacteraceae sequences closely related to known Desulfuromonas and Geobacter species were again detected. As observed in the sediments from site 1103, the addition of acetate to the site 853 and 857 sediments resulted in a significant enrichment of Geobacteraceae that was associated with the stimulation of U(VI) and Fe(III) reduction (Fig. 3B and C). The percentage of Geobacteraceae on day 23, when U(VI) reduction and Fe(III) reduction were near completion, was similar to that observed in site 1103 sediments for both site 853 (37%) and site 857 (40%). In the acetate-amended 853 sediments, 55% of the Geobacteraceae sequences were most closely related to Desulfuromonas species, and the proportion most similar to Desulfuromonas species was 58% for the site 857 sediments.
Although the final percentage of Geobacteraceae sequences in the acetate-amended sediments was similar for all three sites, the overall increase in Geobacteraceae sequences was more dramatic at site 1103. Higher initial proportions of Geobacteraceae sequences were detected in sediments from sites 853 (28%) and 857 (13%) than site 1103 sediment (0%). This may be related to the fact that while there was no detectable Fe(II) at the start of the incubation of site 1103 sediments, sediments from sites 853 and 857 initially contained Fe(II) (10). This suggests that there were low levels of Fe(III) reduction in situ or during the initial storage of the sediments under anaerobic conditions prior to the start of the experiment.
Quantitative analysis of increase in Geobacteraceae associated with U(VI) and Fe(III) reduction.
In order to further evaluate the increase in Geobacteraceae associated with the stimulation of Fe(III) and U(VI) reduction, the number of Geobacteraceae sequences were quantified with both MPN PCR and TaqMan analyses. Both types of analyses indicated that the number of Geobacteraceae sequences in site 1103 sediments increased by several orders of magnitude, coincident with an increase in the rates of U(VI) and Fe(III) reduction in acetate-amended sediments (Fig. 1B). In contrast, in control sediments, in which metal reduction was not stimulated, the numbers of Geobacteraceae remained low. MPN PCR and the TaqMan assays indicated that, by the completion of Fe(III) and U(VI) reduction, the number of Geobacteraceae sequences in the sediments from sites 857 and 853 had increased by 2 orders of magnitude (Fig. 2B and C). However, there was no increase in Geobacteraceae sequences when acetate was not added to the sediments.
DISCUSSION
The results demonstrate that there was a significant, specific enrichment of microorganisms in the family Geobacteraceae associated with the stimulation of Fe(III) and U(VI) reduction and the removal of U(VI) from the groundwater in the three different sediments. As detailed below, this response is consistent with the physiological characteristics of Geobacteraceae that are available in pure culture and suggests that Fe(III)-reducing microorganisms, not sulfate-reducing microorganisms, were responsible for the U(VI) reduction in these sediments. These results, as well as the previously described predominance of Geobacteraceae in other sedimentary environments in which Fe(III) reduction is an important process (27, 28, 30, 32), suggest that Geobacteraceae are well adapted for growth in the subsurface. Therefore, studies on the physiology of Geobacteraceae are likely to yield insights into the mechanisms controlling the rate and extent of metal reduction in sedimentary environments and suggest strategies for stimulating this process.
Evaluation of methods.
Three methods for analyzing the changes in the microbial community were employed in an attempt to provide a reliable evaluation of community dynamics: clone libraries, MPN PCR, and TaqMan PCR. Each of these approaches has different potential biases. For example, when 16S rDNA clone libraries are constructed with universal PCR primers, nonspecific bacterial primers may favor the amplification of DNA from microorganisms with low concentrations of cytidine and guanosine nucleotides (26). In addition, PCR products may tend towards 1:1 ratios, despite the initial template concentration (26). Although it seems unlikely that such potential biases could have accounted for the large enrichment of Geobacteraceae sequences in the clone libraries associated with the stimulation of Fe(III) and U(VI) reduction, the response of the Geobacteraceae was further assessed with two quantitative techniques, MPN PCR and TaqMan PCR, which relied upon the use of Geobacteraceae-specific primers rather than nonspecific bacterial primers. Because these quantitative approaches can be adversely effected by such factors as cations present in the extracted DNA or quenching and autofluorescing by molecules other than DNA, such as humic acids (31), attempts were made to minimize these potential sources of error. Although background levels of autofluorescence were not detected in the extracted DNA, DNA was further purified with the Wizard clean-up system, and template was diluted sufficiently to reduce any residual contaminants prior to quantitative analysis.
Although it may be difficult to infer the absolute number of Desulfuromonas and Geobacter species present from the MPN PCR and TaqMan approaches, these techniques do provide a good indication of the relative numbers of target sequences in comparable sediment samples. The fact that the analysis of clone libraries, generated with nonspecific bacterial primers, as well as the MPN PCR and TaqMan results, obtained with Geobacteraceae-specific primers, each indicated a significant enrichment of Geobacteraceae sequences in all three sediment types clearly demonstrates that increased rates of metal reduction were associated with an increase in Geobacteraceae sequences.
Correspondence between metal reduction and growth of Geobacteraceae.
The finding that enhanced rates of Fe(III) and U(VI) reduction corresponded with a large increase in Geobacteraceae 16S rDNA sequences suggests that these organisms were responsible for much of the metal reduction in the sediments. It is likely that Fe(III) reduction accounts for most of the growth of the Geobacteraceae, as the extent of Fe(II) production and U(VI) loss indicated that >99% of electrons transferred to the metals went to Fe(III) reduction (10; Finneran et al., submitted). In fact, the extent of electron transfer to U(VI) was so low that it is conceivable that some minor population other than the Geobacteraceae may have been involved in some of the U(VI) reduction observed in these sediments. However, microorganisms in the family Geobacteraceae have been shown to readily reduce U(VI) in pure culture (16). Therefore, given the predominance of Geobacteraceae in the treated sediments examined in this study, it would be expected that organisms from this family would be responsible for the majority of U(VI) reduction. Geochemical and molecular community analysis of these sites suggested that sulfate-reducing microorganisms were not responsible for the U(VI) reduction observed in acetate-amended sediment. 16S rDNA sequences most similar to sulfate reducers from the genera Desulfotomaculum and Desulfovibrio accounted for a small portion (3 to 8%) of the microbial community at the start of the incubations and there was no apparent enrichment of sulfate reducers as the rates of metal reduction increased. The concurrent reduction of U(VI) and Fe(III), prior to sulfate reduction, in the Shiprock sediments is consistent with the order of microbial redox processes in marine sediments (3, 4)
A previous study suggested that sulfate-reducing microorganisms might be important for U(VI) reduction at the Shiprock site (6). However, the molecular analysis of the sediments focused only on the distribution of dissimilatory sulfite reductase genes, which are specific for sulfate-reducing microorganisms. Furthermore, the community analyses were performed on freshly collected aerobic sediments in which U(VI) reduction was not taking place. Chang et al. (6) suggested that a positive correlation between uranium concentrations and the prevalence of Desulfotomaculum sequences provided evidence for the potential significance of these organisms in the natural attenuation of uranium. However, it would be expected that the organisms responsible for U(VI) reduction would be in higher numbers where U(VI) was being reduced, not where the concentrations of U(VI) remain high. Therefore, analysis of the microbial community prior to the stimulation of U(VI) reduction is likely to provide little insight into which organisms are ultimately responsible for U(VI) reduction.
In contrast to sulfate, which was not reduced until after Fe(III) reduction and U(VI) reduction were complete, nitrate needs to be removed from the environment before Fe(III) and U(VI) are reduced (Finneran et al., submitted). The significant amount of nitrate that was available for microbial reduction in the site 1103 sediments may explain the increase in the proportion of 16S rDNA sequences most similar to Azoarcus species that was associated with nitrate reduction in the acetate-amended sediments. Azoarcus species are not known to function as dissimilatory metal-reducing microorganisms, but they are capable of anaerobic growth with nitrate serving as the terminal electron acceptor (29). Further evidence suggesting that the organisms closely related to Azoarcus were involved in nitrate reduction, but not Fe(III) reduction, was the finding that the increase in Azoarcus sequences was observed only in the nitrate-containing site 1103 sediments and not in the sediments from site 853 or 857.
Comparison to predominance of Geobacteraceae in other subsurface environments.
In light of the data presented here on the predominance of Geobacteraceae during the Fe(III) and U(VI) reduction phase in Shiprock sediments, these soils join the growing number of subsurface environments in which, molecular studies suggest, Geobacteraceae are important in Fe(III) reduction. For example, 16S rDNA sequences most similar to known Geobacter species were specifically enriched in the Fe(III)-reducing zone of a petroleum-contaminated aquifer (28). Analysis of 16S rDNA sequences also suggested that the growth of Geobacter species was stimulated in two field experiments in which Fe(III) reduction was enhanced with the addition of benzoate or aromatic hydrocarbons (32). Similar Geobacter sequences were enriched in laboratory incubations of subsurface sediments in which Fe(III) reduction was stimulated with the addition of a variety of organic compounds and/or electron shuttles (32). A molecular examination of microbial communities associated with freshwater aquatic sediments in which metal cycling was important indicated that microorganisms in the Geobacteraceae were the most abundant microbial group (30). Geobacter species also appeared to be the dominant microorganisms in the Fe(III) reduction zone of a landfill leachate-contaminated aquifer (27).
A potential explanation for the observed predominance of Geobacteraceae over other Fe(III)-reducing microorganisms in subsurface environments is differences in the mechanisms utilized to access insoluble Fe(III) oxides, which are the primary electron acceptor for the growth of dissimilatory metal-reducing microorganisms in these environments. Studies with Geobacter species have indicated that they specifically produce appendages to aid in locating and attaching to Fe(III) oxides (7) and that it is necessary for Geobacter species to directly contact Fe(III) oxide in order to reduce it (22). In contrast, other well-studied Fe(III)-reducing microorganisms, such as Shewanella species and Geothrix fermentans, may produce electron-shuttling compounds and/or Fe(III) chelators in order to alleviate the need to establish contact with Fe(III) oxide (23-25). Although the production of electron-shuttling compounds and Fe(III) chelators may be advantageous for the reduction of Fe(III) oxide in culture, it may not be an energetically favorable approach for Fe(III) oxide reduction in nutrient-poor subsurface environments (7, 23)
In previous molecular studies of subsurface environments, the predominant Geobacteraceae sequences fell into the Geobacter phylogenetic cluster (27, 28, 32). However, clone library analysis of the Shiprock sediments suggested that many of the Geobacteraceae sequences were most similar to Desulfuromonas species. This result may reflect the fact that previously studied subsurface environments had freshwater salinities whereas the groundwaters at the Shiprock sites were similar to those found in marine environments. In general, microorganisms in the Desulfuromonas cluster have been cultured from marine environments and prefer higher salinities than Geobacter cultures that have been recovered from freshwater environments (16).
Implications for research on remediation of subsurface environments.
These results suggest that when in situ bioremediation strategies for uranium-contaminated groundwaters, such as those found at the Shiprock site, are engineered, it is important to consider the physiological properties of the Geobacteraceae that are likely to be responsible for Fe(III) and U(VI) reduction. As yet, little is known about the physiology of Geobacter and Desulfuromonas species. However, the genome of one of these organisms, Geobacter sulfurreducens, is complete, and the sequencing of two other species, Geobacter metallireducens and Desulfuromonas acetoxidans, is under way. This information coupled with a recently developed genetic system for these organisms (8) and biochemical studies (11, 20; J. R. Lloyd, A. L. H. Myerson, L. Yin, and D. R. Lovley, submitted for publication) is expected to provide information that will be helpful in understanding metal reduction in subsurface environments.
Acknowledgments
We thank Todd Anderson and Phil Long for help in sample collection and advice.
This research was funded by the Natural and Accelerated Bioremediation Research (NABIR) program, Biological and Environmental Research (BER), U.S. Department of Energy (grant DE-FG02-97ER62475).
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson, R. F., M. Q. Fleisher, and A. P. LeHuray. 1989. Concentration, oxidation state, and particulate flux of uranium in the Black Sea. Geochim. Cosmochim. Acta 53:2215-2224. [Google Scholar]
- 4.Anderson, R. F., A. P. LeHuray, M. Q. Fleisher, and J. W. Murray. 1989. Uranium deposition in Saanich Inlet sediments, Vancouver Island. Geochim. Cosmochim. Acta 53:2205-2213. [Google Scholar]
- 5.Anderson, R. T., J. Rooney-Varga, C. V. Gaw, and D. R. Lovley. 1998. Anaerobic benzene oxidation in the Fe(III)-reduction zone of petroleum-contaminated aquifers. Environ. Sci. Technol. 32:1222-1229. [Google Scholar]
- 6.Chang, Y. J., A. D. Peacock, P. E. Long, J. R. Stephaen, J. P. McKinley, S. J. Macnaughton, A. K. M. A. Hussain, A. M. Saxton, and D. C. White. 2001. Diversity and characterization of sulfate-reducing bacteria in groundwater at a uranium mill tailings site. Appl. Environ. Microbiol. 67:3149-3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Childers, S. E., S. Ciufo, and D. R. Lovley. Geobacter metallireducens accesses Fe(III) oxide by chemotaxis. Nature, in press. [DOI] [PubMed]
- 8.Coppi, M. V., C. Leang, S. J. Sandler, and D. R. Lovley. 2001. Development of a genetic system for Geobacter sulfurreducens. Appl. Environ. Microbiol. 67:3180-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eden, P. E., T. M. Schmidt, R. P. Blakemore, and N. R. Pace. 1991. Phylogenetic analysis of Aquaspirillum magnetotacticum using polymerase chain reaction-amplified 16S rRNA-specific DNA. Int. J. Syst. Bacteriol. 41:324-325. [DOI] [PubMed] [Google Scholar]
- 10.Finneran, K. T., R. T. Anderson, K. P. Nevin, and D. R. Lovley. Potential for uranium bioremediation by microbial U(VI) reduction. J. Soil Sediment Contam., in press.
- 11.Galushko, A. S., and B. Schink. 2000. Oxidation of acetate through reactions of the citric acid cycle by Geobacter sulfurreducens in pure culture and in syntrophic coculture. Arch. Microbiol. 174:314-321. [DOI] [PubMed] [Google Scholar]
- 12.Ivanova, I. A., J. R. Stephen, Y. J. Chang, J. Bruggemann, P. E. Long, J. P. McKinley, G. A. Kowalchuk, D. C. White, and S. J. Macnaughton. 2000. A survey of 16S rRNA and amoA genes related to autotrophic ammonia-oxidizing bacteria of the beta-subdivision of the class proteobacteria in contaminated groundwater. Can. J. Microbiol. 46:1012-1020. [DOI] [PubMed] [Google Scholar]
- 13.Lane, D. L., B. Pace, G. J. Olsen, D. Stahl, M. L. Sogin, and N. R. Pace. 1985. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analysis. Proc. Nat. Acad. Sci. USA 82:6955-6959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Langmuir, D. 1978. Uranium solution-mineral equilibria at low temperatures with applications to sedimentary ore deposits. Geochim. Cosmochim. Acta 42:547-569. [Google Scholar]
- 15.Lovley, D. R. 1995. Bioremediation of organic and metal contaminants with dissimilatory metal reduction. J. Ind. Microbiol. 14:85-93. [DOI] [PubMed] [Google Scholar]
- 16.Lovley, D. R. 2000. Fe(III)- and Mn(IV)-reducing prokaryotes. In M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes. Springer-Verlag, Inc., New York, N.Y.
- 17.Lovley, D. R., and E. J. P. Phillips. 1992. Reduction of uranium by Desulfovibrio desulfuricans. Appl. Environ. Microbiol. 58:850-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lovley, D. R., E. J. P. Phillips, Y. A. Gorby, and E. R. Landa. 1991. Microbial reduction of uranium. Nature 350:413-416. [Google Scholar]
- 19.Lovley, D. R., E. E. Roden, E. J. P. Phillips, and J. C. Woodward. 1993. Enzymatic iron and uranium reduction by sulfate-reducing bacteria. Mar. Geol. 113:41-53. [Google Scholar]
- 20.Magnuson, T. S., N. Isoyama, A. L. Hodges-Myerson, G. Davidson, M. J. Maroney, G. G. Geesey, and D. R. Lovley. 2001. Isolation, characterization and gene sequence analysis of a membrane-associated 89 kDa Fe(III) reducing cytochrome c from Geobacter sulfurreducens. Biochem. J. 359:147-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, P. R. Saxman, R. J. Farris, G. M. Garrity, G. J. Olsen, T. M. Schmidt, and J. M. Tiedje. 2001. The RDP-II (Ribosomal Database Project). Nucleic Acids Res. 29:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nevin, K. P., and D. R. Lovley. 2000. Lack of production of electron-shuttling compounds or solubilization of Fe(III) during reduction of insoluble Fe(III) oxide by Geobacter metallireducens. Appl. Environ. Microbiol. 66:2248-2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nevin, K. P., and D. R. Lovley. Mechanisms for Fe(III) oxide reduction in sedimentary environments. Geomicrobiol. J., in press.
- 24.Nevin, K. P., and D. R. Lovley. 2002. Mechanisms for accessing insoluble Fe(III) oxide during dissimilatory Fe(III) reduction by Geothrix fermentans. Appl. Environ. Microbiol. 68:2294-2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Newman, D. K., and R. Kolter. 2000. A role for excreted quinones in extracellular electron transfer. Nature 405:94-97. [DOI] [PubMed] [Google Scholar]
- 26.Polz, M. F., and C. M. Cavanaugh. 1998. Bias in template-to-product ratios in multitemplate PCR. Appl. Environ. Microbiol. 64:3724-3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roling, W. F., B. M. van Breukelen, M. Braster, B. Lin, and H. W. van Verseveld. 2001. Relationships between microbial community structure and hydrochemistry in a landfill leachate-polluted aquifer. Appl. Environ. Microbiol. 67:4619-4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rooney-Varga, J. N., R. T. Anderson, J. L. Fraga, D. Ringelberg, and D. R. Lovley. 1999. Microbial communities associated with anaerobic benzene degradation in a petroleum-contaminated aquifer. Appl. Environ. Microbiol. 65:3056-3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song, B., M. M. Haggblom, J. Zhou, J. M. Tiedje, and N. J. Palleroni. 1999. Taxonomic characterization of denitrifying bacteria that degrade aromatic compounds and description of Azoarcus toluvorans sp. nov. and Azoarcus toluclasticus sp. nov. Int. J. Syst. Bacteriol. 49:1129-1140. [DOI] [PubMed] [Google Scholar]
- 30.Stein, L. Y., M. T. La Duc, T. J. Grundl, and K. H. Nealson. 2001. Bacterial and archaeal populations associated with freshwater ferromanganous micronodules and sediments. Environ. Microbiol. 3:10-18. [DOI] [PubMed] [Google Scholar]
- 31.Stults, J. R., O. L. Snoeyenbos-West, B. Methe, D. R. Lovley, and D. P. Chandler. 2001. Application of the 5′ fluorogenic exonuclease assay (TaqMan) for quantitative ribosomal DNA and rRNA analysis in sediments. Appl. Environ. Microbiol. 67:2781-2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Snoeyenbos-West, O. L., K. P. Nevin, R. T. Anderson, and D. R. Lovley. 2000. Enrichment of Geobacter species in response to stimulation of Fe(III) reduction in sandy aquifer sediments. Microb. Ecol. 39:153-167. [DOI] [PubMed] [Google Scholar]