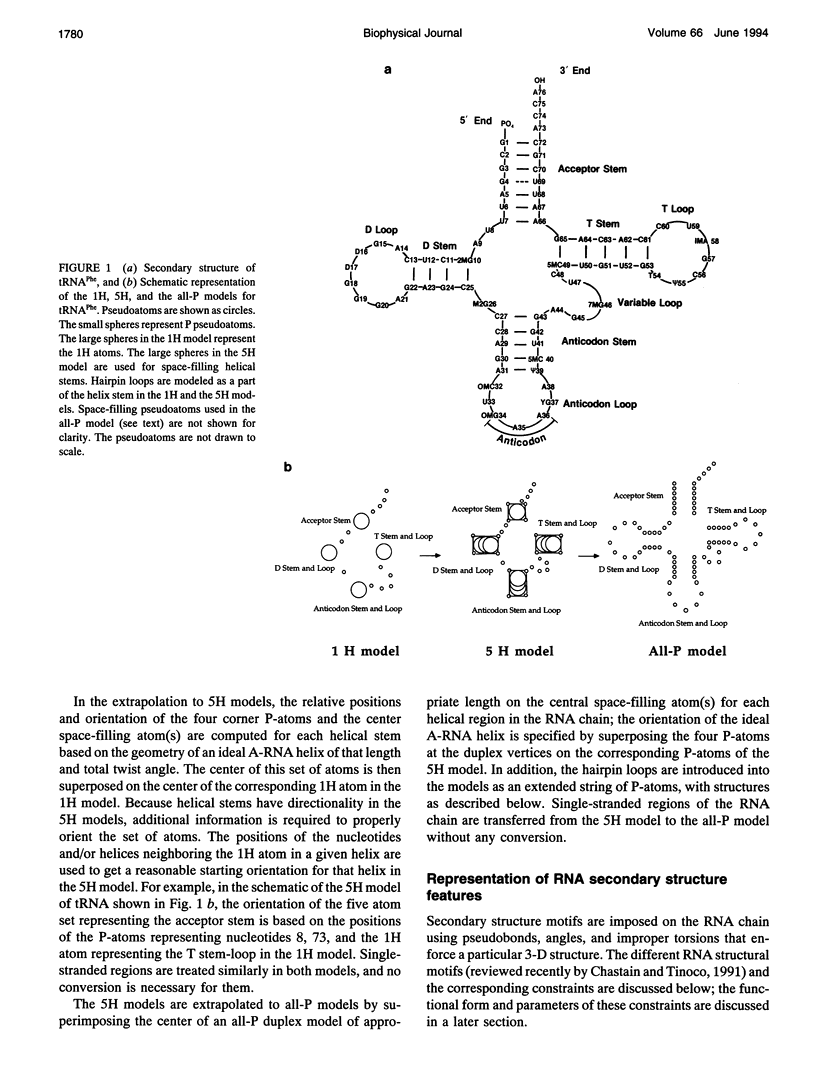
Abstract
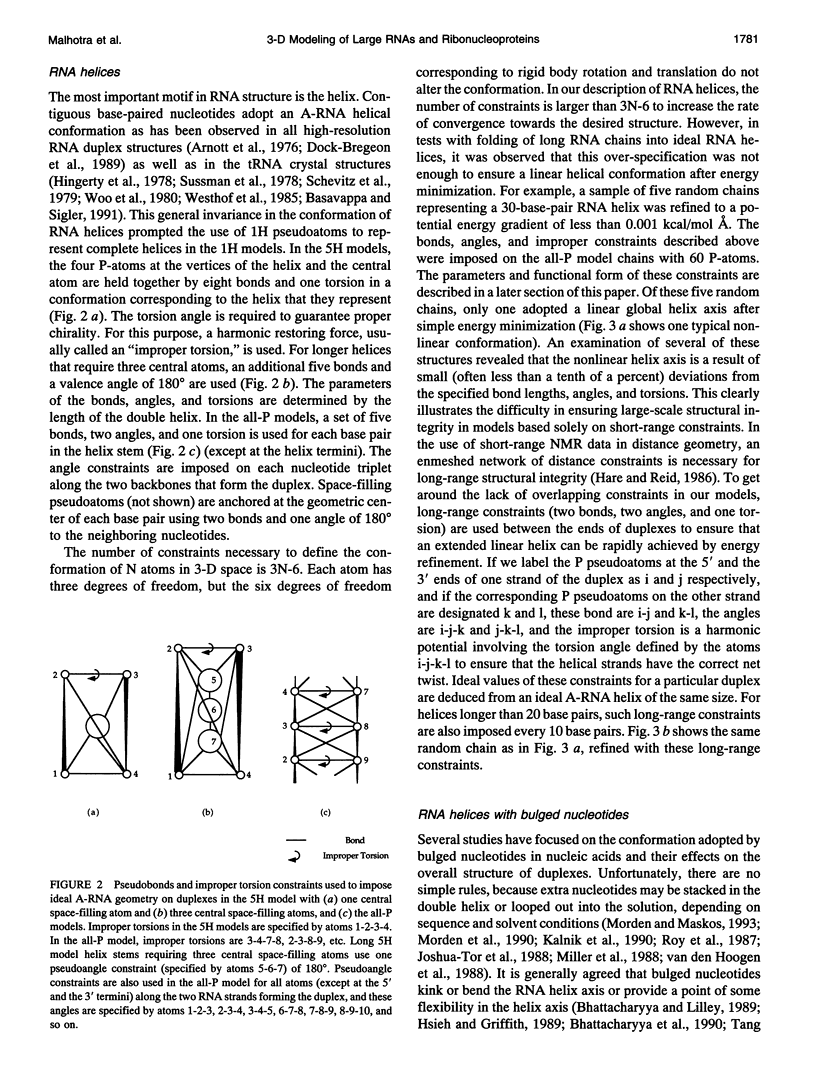
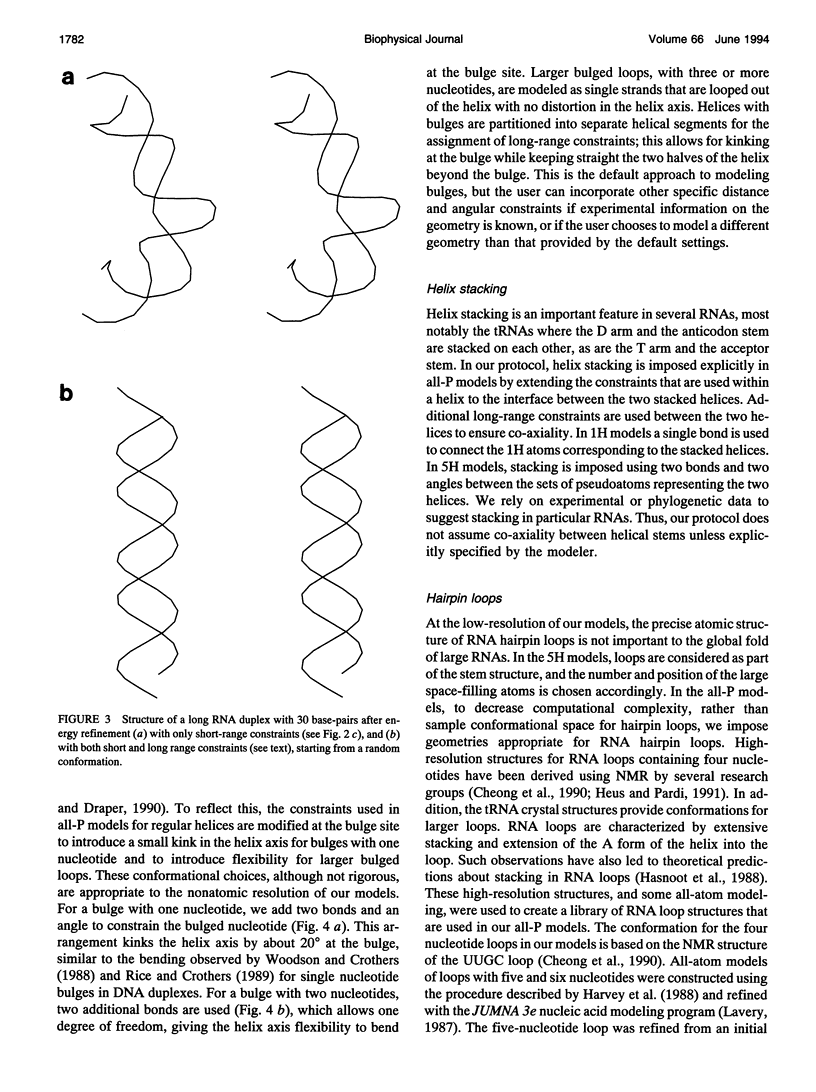
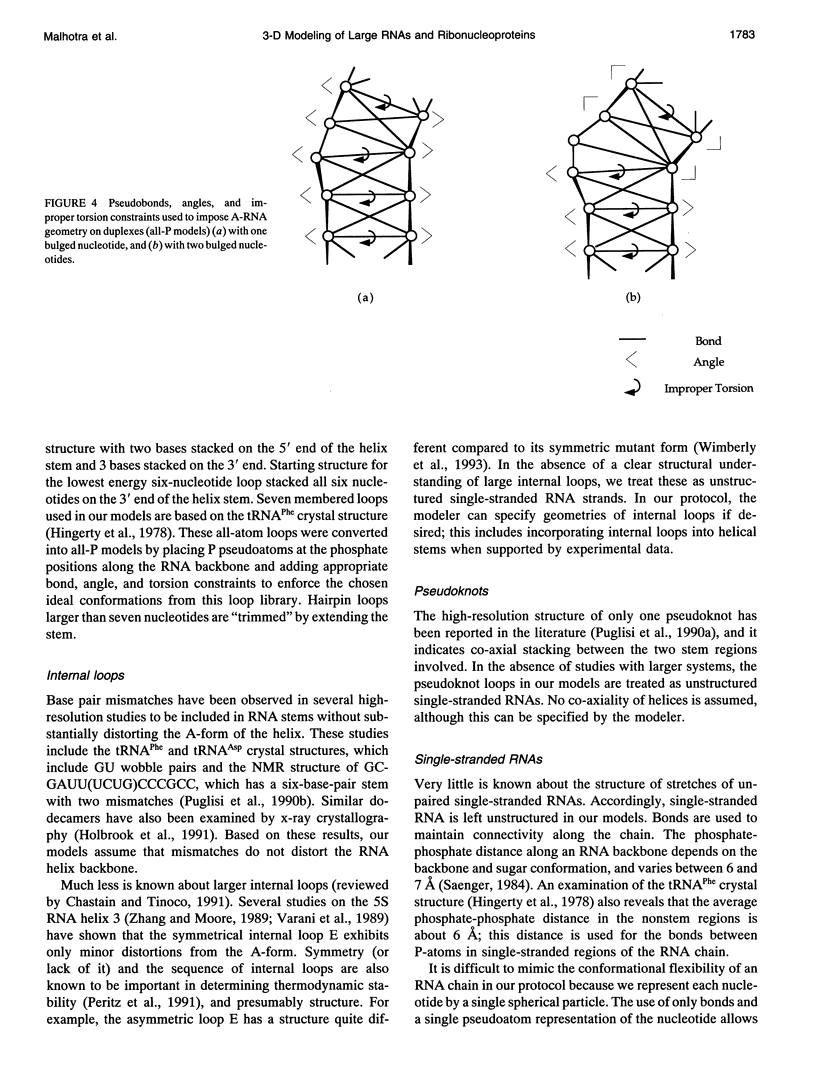
There is a growing body of low-resolution structural data that can be utilized to devise structural models for large RNAs and ribonucleoproteins. These models are routinely built manually. We introduce an automated refinement protocol to utilize such data for building low-resolution three-dimensional models using the tools of molecular mechanics. In addition to specifying the positions of each nucleotide, the protocol provides quantitative estimates of the uncertainties in those positions, i.e., the resolution of the model. In typical applications, the resolution of the models is about 10-20 A. Our method uses reduced representations and allows us to refine three-dimensional structures of systems as big as the 16S and 23S ribosomal RNAs, which are about one to two orders of magnitude larger than nucleic acids that can be examined by traditional all-atom modeling methods. Nonatomic resolution structural data--secondary structure, chemical cross-links, chemical and enzymatic footprinting patterns, protein positions, solvent accessibility, and so on--are combined with known motifs in RNA structure to predict low-resolution models of large RNAs. These structural constraints are imposed on the RNA chain using molecular mechanics-type potential functions with parameters based on the quality of experimental data. Surface potential functions are used to incorporate shape and positional data from electron microscopy image reconstruction experiments into our models. The structures are optimized using techniques of energy refinement to get RNA folding patterns. In addition to providing a consensus model, the method finds the range of models consistent with the data, which allows quantitative evaluation of the resolution of the model. The method also identifies conflicts in the experimental data. Although our protocol is aimed at much larger RNAs, we illustrate these techniques using the tRNA structure as an example and test-bed.
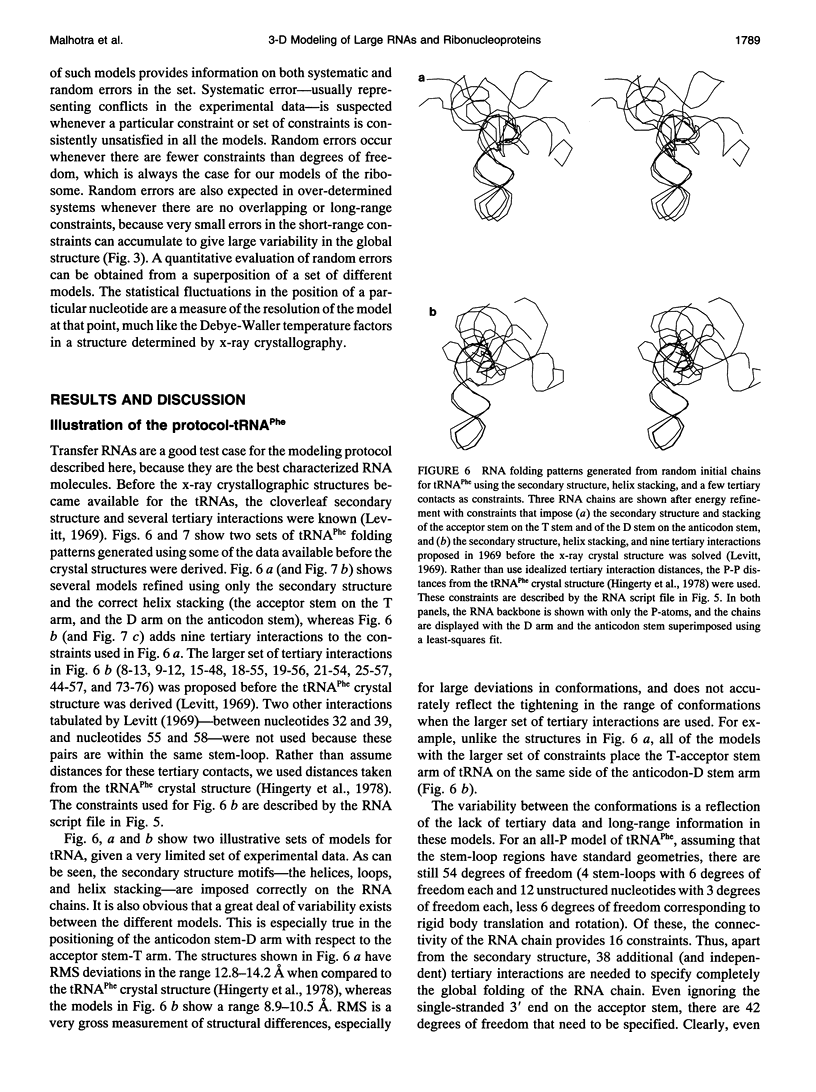
Full text
PDF













Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Basavappa R., Sigler P. B. The 3 A crystal structure of yeast initiator tRNA: functional implications in initiator/elongator discrimination. EMBO J. 1991 Oct;10(10):3105–3111. doi: 10.1002/j.1460-2075.1991.tb07864.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman H. M., Olson W. K., Beveridge D. L., Westbrook J., Gelbin A., Demeny T., Hsieh S. H., Srinivasan A. R., Schneider B. The nucleic acid database. A comprehensive relational database of three-dimensional structures of nucleic acids. Biophys J. 1992 Sep;63(3):751–759. doi: 10.1016/S0006-3495(92)81649-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya A., Lilley D. M. The contrasting structures of mismatched DNA sequences containing looped-out bases (bulges) and multiple mismatches (bubbles). Nucleic Acids Res. 1989 Sep 12;17(17):6821–6840. doi: 10.1093/nar/17.17.6821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya A., Murchie A. I., Lilley D. M. RNA bulges and the helical periodicity of double-stranded RNA. Nature. 1990 Feb 1;343(6257):484–487. doi: 10.1038/343484a0. [DOI] [PubMed] [Google Scholar]
- Brimacombe R., Atmadja J., Stiege W., Schüler D. A detailed model of the three-dimensional structure of Escherichia coli 16 S ribosomal RNA in situ in the 30 S subunit. J Mol Biol. 1988 Jan 5;199(1):115–136. doi: 10.1016/0022-2836(88)90383-x. [DOI] [PubMed] [Google Scholar]
- Brimacombe R. The emerging three-dimensional structure and function of 16S ribosomal RNA. Biochemistry. 1988 Jun 14;27(12):4207–4214. doi: 10.1021/bi00412a001. [DOI] [PubMed] [Google Scholar]
- Brunel C., Romby P., Westhof E., Ehresmann C., Ehresmann B. Three-dimensional model of Escherichia coli ribosomal 5 S RNA as deduced from structure probing in solution and computer modeling. J Mol Biol. 1991 Sep 5;221(1):293–308. doi: 10.1016/0022-2836(91)80220-o. [DOI] [PubMed] [Google Scholar]
- Brünger A. T., Kuriyan J., Karplus M. Crystallographic R factor refinement by molecular dynamics. Science. 1987 Jan 23;235(4787):458–460. doi: 10.1126/science.235.4787.458. [DOI] [PubMed] [Google Scholar]
- Capel M. S., Kjeldgaard M., Engelman D. M., Moore P. B. Positions of S2, S13, S16, S17, S19 and S21 in the 30 S ribosomal subunit of Escherichia coli. J Mol Biol. 1988 Mar 5;200(1):65–87. doi: 10.1016/0022-2836(88)90334-8. [DOI] [PubMed] [Google Scholar]
- Celander D. W., Cech T. R. Visualizing the higher order folding of a catalytic RNA molecule. Science. 1991 Jan 25;251(4992):401–407. doi: 10.1126/science.1989074. [DOI] [PubMed] [Google Scholar]
- Chastain M., Tinoco I., Jr Structural elements in RNA. Prog Nucleic Acid Res Mol Biol. 1991;41:131–177. doi: 10.1016/S0079-6603(08)60008-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong C., Varani G., Tinoco I., Jr Solution structure of an unusually stable RNA hairpin, 5'GGAC(UUCG)GUCC. Nature. 1990 Aug 16;346(6285):680–682. doi: 10.1038/346680a0. [DOI] [PubMed] [Google Scholar]
- Connolly M. L. Solvent-accessible surfaces of proteins and nucleic acids. Science. 1983 Aug 19;221(4612):709–713. doi: 10.1126/science.6879170. [DOI] [PubMed] [Google Scholar]
- Covell D. G., Jernigan R. L. Conformations of folded proteins in restricted spaces. Biochemistry. 1990 Apr 3;29(13):3287–3294. doi: 10.1021/bi00465a020. [DOI] [PubMed] [Google Scholar]
- Crippen G. M. Prediction of protein folding from amino acid sequence over discrete conformation spaces. Biochemistry. 1991 Apr 30;30(17):4232–4237. doi: 10.1021/bi00231a018. [DOI] [PubMed] [Google Scholar]
- Darsillo P., Huber P. W. The use of chemical nucleases to analyze RNA-protein interactions. The TFIIIA-5 S rRNA complex. J Biol Chem. 1991 Nov 5;266(31):21075–21082. [PubMed] [Google Scholar]
- Dock-Bregeon A. C., Chevrier B., Podjarny A., Johnson J., de Bear J. S., Gough G. R., Gilham P. T., Moras D. Crystallographic structure of an RNA helix: [U(UA)6A]2. J Mol Biol. 1989 Oct 5;209(3):459–474. doi: 10.1016/0022-2836(89)90010-7. [DOI] [PubMed] [Google Scholar]
- Ehresmann C., Baudin F., Mougel M., Romby P., Ebel J. P., Ehresmann B. Probing the structure of RNAs in solution. Nucleic Acids Res. 1987 Nov 25;15(22):9109–9128. doi: 10.1093/nar/15.22.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Expert-Bezançon A., Wollenzien P. L. Three-dimensional arrangement of the Escherichia coli 16 S ribosomal RNA. J Mol Biol. 1985 Jul 5;184(1):53–66. doi: 10.1016/0022-2836(85)90043-9. [DOI] [PubMed] [Google Scholar]
- Frank J., Penczek P., Grassucci R., Srivastava S. Three-dimensional reconstruction of the 70S Escherichia coli ribosome in ice: the distribution of ribosomal RNA. J Cell Biol. 1991 Nov;115(3):597–605. doi: 10.1083/jcb.115.3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J., van Heel M. Correspondence analysis of aligned images of biological particles. J Mol Biol. 1982 Oct 15;161(1):134–137. doi: 10.1016/0022-2836(82)90282-0. [DOI] [PubMed] [Google Scholar]
- Gautheret D., Major F., Cedergren R. Modeling the three-dimensional structure of RNA using discrete nucleotide conformational sets. J Mol Biol. 1993 Feb 20;229(4):1049–1064. doi: 10.1006/jmbi.1993.1104. [DOI] [PubMed] [Google Scholar]
- Haasnoot C. A., Hilbers C. W., van der Marel G. A., van Boom J. H., Singh U. C., Pattabiraman N., Kollman P. A. On loop folding in nucleic acid hairpin-type structures. J Biomol Struct Dyn. 1986 Apr;3(5):843–857. doi: 10.1080/07391102.1986.10508468. [DOI] [PubMed] [Google Scholar]
- Hadwiger M. A., Fox G. E. Explicit distance geometry: identification of all the degrees of freedom in a large RNA molecule. J Biomol Struct Dyn. 1991 Feb;8(4):759–779. doi: 10.1080/07391102.1991.10507843. [DOI] [PubMed] [Google Scholar]
- Happ C. S., Happ E., Nilges M., Gronenborn A. M., Clore G. M. Refinement of the solution structure of the ribonucleotide 5'r(GCAUGC)2: combined use of nuclear magnetic resonance and restrained molecular dynamics. Biochemistry. 1988 Mar 8;27(5):1735–1743. doi: 10.1021/bi00405a053. [DOI] [PubMed] [Google Scholar]
- Hare D. R., Reid B. R. Three-dimensional structure of a DNA hairpin in solution: two-dimensional NMR studies and distance geometry calculations on d(CGCGTTTTCGCG). Biochemistry. 1986 Sep 9;25(18):5341–5350. doi: 10.1021/bi00366a053. [DOI] [PubMed] [Google Scholar]
- Harvey S. C., Luo J., Lavery R. DNA stem-loop structures in oligopurine-oligopyrimidine triplexes. Nucleic Acids Res. 1988 Dec 23;16(24):11795–11809. doi: 10.1093/nar/16.24.11795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey S. C. Treatment of electrostatic effects in macromolecular modeling. Proteins. 1989;5(1):78–92. doi: 10.1002/prot.340050109. [DOI] [PubMed] [Google Scholar]
- Heus H. A., Pardi A. Structural features that give rise to the unusual stability of RNA hairpins containing GNRA loops. Science. 1991 Jul 12;253(5016):191–194. doi: 10.1126/science.1712983. [DOI] [PubMed] [Google Scholar]
- Hinds D. A., Levitt M. A lattice model for protein structure prediction at low resolution. Proc Natl Acad Sci U S A. 1992 Apr 1;89(7):2536–2540. doi: 10.1073/pnas.89.7.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hingerty B., Brown R. S., Jack A. Further refinement of the structure of yeast tRNAPhe. J Mol Biol. 1978 Sep 25;124(3):523–534. doi: 10.1016/0022-2836(78)90185-7. [DOI] [PubMed] [Google Scholar]
- Holbrook S. R., Cheong C., Tinoco I., Jr, Kim S. H. Crystal structure of an RNA double helix incorporating a track of non-Watson-Crick base pairs. Nature. 1991 Oct 10;353(6344):579–581. doi: 10.1038/353579a0. [DOI] [PubMed] [Google Scholar]
- Hsieh C. H., Griffith J. D. Deletions of bases in one strand of duplex DNA, in contrast to single-base mismatches, produce highly kinked molecules: possible relevance to the folding of single-stranded nucleic acids. Proc Natl Acad Sci U S A. 1989 Jul;86(13):4833–4837. doi: 10.1073/pnas.86.13.4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard J. M., Hearst J. E. Computer modeling 16 S ribosomal RNA. J Mol Biol. 1991 Oct 5;221(3):889–907. doi: 10.1016/0022-2836(91)80182-t. [DOI] [PubMed] [Google Scholar]
- Hubbard J. M., Hearst J. E. Predicting the three-dimensional folding of transfer RNA with a computer modeling protocol. Biochemistry. 1991 Jun 4;30(22):5458–5465. doi: 10.1021/bi00236a019. [DOI] [PubMed] [Google Scholar]
- Joshua-Tor L., Rabinovich D., Hope H., Frolow F., Appella E., Sussman J. L. The three-dimensional structure of a DNA duplex containing looped-out bases. Nature. 1988 Jul 7;334(6177):82–84. doi: 10.1038/334082a0. [DOI] [PubMed] [Google Scholar]
- Kalnik M. W., Norman D. G., Li B. F., Swann P. F., Patel D. J. Conformational transitions in thymidine bulge-containing deoxytridecanucleotide duplexes. Role of flanking sequence and temperature in modulating the equilibrium between looped out and stacked thymidine bulge states. J Biol Chem. 1990 Jan 15;265(2):636–647. [PubMed] [Google Scholar]
- Kim S. H., Cech T. R. Three-dimensional model of the active site of the self-splicing rRNA precursor of Tetrahymena. Proc Natl Acad Sci U S A. 1987 Dec;84(24):8788–8792. doi: 10.1073/pnas.84.24.8788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick S., Gelatt C. D., Jr, Vecchi M. P. Optimization by simulated annealing. Science. 1983 May 13;220(4598):671–680. doi: 10.1126/science.220.4598.671. [DOI] [PubMed] [Google Scholar]
- Levitt M. A simplified representation of protein conformations for rapid simulation of protein folding. J Mol Biol. 1976 Jun 14;104(1):59–107. doi: 10.1016/0022-2836(76)90004-8. [DOI] [PubMed] [Google Scholar]
- Levitt M. Detailed molecular model for transfer ribonucleic acid. Nature. 1969 Nov 22;224(5221):759–763. doi: 10.1038/224759a0. [DOI] [PubMed] [Google Scholar]
- Levitt M., Warshel A. Computer simulation of protein folding. Nature. 1975 Feb 27;253(5494):694–698. doi: 10.1038/253694a0. [DOI] [PubMed] [Google Scholar]
- Major F., Turcotte M., Gautheret D., Lapalme G., Fillion E., Cedergren R. The combination of symbolic and numerical computation for three-dimensional modeling of RNA. Science. 1991 Sep 13;253(5025):1255–1260. doi: 10.1126/science.1716375. [DOI] [PubMed] [Google Scholar]
- Malhotra A., Tan R. K., Harvey S. C. Prediction of the three-dimensional structure of Escherichia coli 30S ribosomal subunit: a molecular mechanics approach. Proc Natl Acad Sci U S A. 1990 Mar;87(5):1950–1954. doi: 10.1073/pnas.87.5.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzler W. J., Hare D. R., Pardi A. Limited sampling of conformational space by the distance geometry algorithm: implications for structures generated from NMR data. Biochemistry. 1989 Aug 22;28(17):7045–7052. doi: 10.1021/bi00443a040. [DOI] [PubMed] [Google Scholar]
- Miller M., Harrison R. W., Wlodawer A., Appella E., Sussman J. L. Crystal structure of 15-mer DNA duplex containing unpaired bases. Nature. 1988 Jul 7;334(6177):85–86. doi: 10.1038/334085a0. [DOI] [PubMed] [Google Scholar]
- Moore P. B. The ribosome returns. Nature. 1988 Jan 21;331(6153):223–227. doi: 10.1038/331223a0. [DOI] [PubMed] [Google Scholar]
- Morden K. M., Gunn B. M., Maskos K. NMR studies of a deoxyribodecanucleotide containing an extrahelical thymidine surrounded by an oligo(dA).oligo(dT) tract. Biochemistry. 1990 Sep 18;29(37):8835–8845. doi: 10.1021/bi00489a047. [DOI] [PubMed] [Google Scholar]
- Morden K. M., Maskos K. NMR studies of an extrahelical cytosine in an A.T rich region of a deoxyribodecanucleotide. Biopolymers. 1993 Jan;33(1):27–36. doi: 10.1002/bip.360330104. [DOI] [PubMed] [Google Scholar]
- Nagano K., Harel M., Takezawa M. Prediction of three-dimensional structure of Escherichia coli ribosomal RNA. J Theor Biol. 1988 Sep 17;134(2):199–256. doi: 10.1016/s0022-5193(88)80202-9. [DOI] [PubMed] [Google Scholar]
- Oakes M. I., Kahan L., Lake J. A. DNA-hybridization electron microscopy tertiary structure of 16 S rRNA. J Mol Biol. 1990 Feb 20;211(4):907–918. doi: 10.1016/0022-2836(90)90083-X. [DOI] [PubMed] [Google Scholar]
- Oakes M. I., Lake J. A. DNA-hybridization electron microscopy. Localization of five regions of 16 S rRNA on the surface of 30 S ribosomal subunits. J Mol Biol. 1990 Feb 20;211(4):897–906. doi: 10.1016/0022-2836(90)90082-W. [DOI] [PubMed] [Google Scholar]
- Olson W. K., Flory P. J. Spatial configurations of polynucleotide chains. I. Steric interactions in polyribonucleotides: a virtual bond model. Biopolymers. 1972 Jan;11(1):1–23. doi: 10.1002/bip.1972.360110102. [DOI] [PubMed] [Google Scholar]
- Peritz A. E., Kierzek R., Sugimoto N., Turner D. H. Thermodynamic study of internal loops in oligoribonucleotides: symmetric loops are more stable than asymmetric loops. Biochemistry. 1991 Jul 2;30(26):6428–6436. doi: 10.1021/bi00240a013. [DOI] [PubMed] [Google Scholar]
- Puglisi J. D., Wyatt J. R., Tinoco I., Jr Conformation of an RNA pseudoknot. J Mol Biol. 1990 Jul 20;214(2):437–453. doi: 10.1016/0022-2836(90)90192-O. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puglisi J. D., Wyatt J. R., Tinoco I., Jr Solution conformation of an RNA hairpin loop. Biochemistry. 1990 May 1;29(17):4215–4226. doi: 10.1021/bi00469a026. [DOI] [PubMed] [Google Scholar]
- Rice J. A., Crothers D. M. DNA bending by the bulge defect. Biochemistry. 1989 May 16;28(10):4512–4516. doi: 10.1021/bi00436a058. [DOI] [PubMed] [Google Scholar]
- Richards F. M. Areas, volumes, packing and protein structure. Annu Rev Biophys Bioeng. 1977;6:151–176. doi: 10.1146/annurev.bb.06.060177.001055. [DOI] [PubMed] [Google Scholar]
- Romby P., Moras D., Bergdoll M., Dumas P., Vlassov V. V., Westhof E., Ebel J. P., Giegé R. Yeast tRNAAsp tertiary structure in solution and areas of interaction of the tRNA with aspartyl-tRNA synthetase. A comparative study of the yeast phenylalanine system by phosphate alkylation experiments with ethylnitrosourea. J Mol Biol. 1985 Aug 5;184(3):455–471. doi: 10.1016/0022-2836(85)90294-3. [DOI] [PubMed] [Google Scholar]
- Roy S., Sklenar V., Appella E., Cohen J. S. Conformational perturbation due to an extra adenosine in a self-complementary oligodeoxynucleotide duplex. Biopolymers. 1987 Dec;26(12):2041–2052. doi: 10.1002/bip.360261206. [DOI] [PubMed] [Google Scholar]
- Schellman J. A. Flexibility of DNA. Biopolymers. 1974 Jan;13(1):217–226. doi: 10.1002/bip.1974.360130115. [DOI] [PubMed] [Google Scholar]
- Schevitz R. W., Podjarny A. D., Krishnamachari N., Hughes J. J., Sigler P. B., Sussman J. L. Crystal structure of a eukaryotic initiator tRNA. Nature. 1979 Mar 8;278(5700):188–190. doi: 10.1038/278188a0. [DOI] [PubMed] [Google Scholar]
- Sharp K. A., Honig B. Electrostatic interactions in macromolecules: theory and applications. Annu Rev Biophys Biophys Chem. 1990;19:301–332. doi: 10.1146/annurev.bb.19.060190.001505. [DOI] [PubMed] [Google Scholar]
- Skolnick J., Kolinski A. Simulations of the folding of a globular protein. Science. 1990 Nov 23;250(4984):1121–1125. doi: 10.1126/science.250.4984.1121. [DOI] [PubMed] [Google Scholar]
- Stern S., Powers T., Changchien L. M., Noller H. F. RNA-protein interactions in 30S ribosomal subunits: folding and function of 16S rRNA. Science. 1989 May 19;244(4906):783–790. doi: 10.1126/science.2658053. [DOI] [PubMed] [Google Scholar]
- Stern S., Weiser B., Noller H. F. Model for the three-dimensional folding of 16 S ribosomal RNA. J Mol Biol. 1988 Nov 20;204(2):447–481. doi: 10.1016/0022-2836(88)90588-8. [DOI] [PubMed] [Google Scholar]
- Sussman J. L., Holbrook S. R., Warrant R. W., Church G. M., Kim S. H. Crystal structure of yeast phenylalanine transfer RNA. I. Crystallographic refinement. J Mol Biol. 1978 Aug 25;123(4):607–630. doi: 10.1016/0022-2836(78)90209-7. [DOI] [PubMed] [Google Scholar]
- Tan R. K., Harvey S. C. Molecular mechanics model of supercoiled DNA. J Mol Biol. 1989 Feb 5;205(3):573–591. doi: 10.1016/0022-2836(89)90227-1. [DOI] [PubMed] [Google Scholar]
- Tang R. S., Draper D. E. Bulge loops used to measure the helical twist of RNA in solution. Biochemistry. 1990 Jun 5;29(22):5232–5237. doi: 10.1021/bi00474a003. [DOI] [PubMed] [Google Scholar]
- Varani G., Wimberly B., Tinoco I., Jr Conformation and dynamics of an RNA internal loop. Biochemistry. 1989 Sep 19;28(19):7760–7772. doi: 10.1021/bi00445a036. [DOI] [PubMed] [Google Scholar]
- Vorobjev YuN Block-units method for conformational calculations of large nucleic acid chains. I. Block-units approximation of atomic structure and conformational energy of polynucleotides. Biopolymers. 1990 Oct-Nov;29(12-13):1503–1518. doi: 10.1002/bip.360291202. [DOI] [PubMed] [Google Scholar]
- Vorobjev Y. N. Block-units method for conformational calculations of large nucleic acid chains. II. The two-hierarchical approach and its application to conformational arrangement of the unusual T psi C loop of rabbit tRNA(Val). Biopolymers. 1990 Oct-Nov;29(12-13):1519–1529. doi: 10.1002/bip.360291203. [DOI] [PubMed] [Google Scholar]
- WATSON J. D., CRICK F. H. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953 Apr 25;171(4356):737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- Westhof E., Dumas P., Moras D. Crystallographic refinement of yeast aspartic acid transfer RNA. J Mol Biol. 1985 Jul 5;184(1):119–145. doi: 10.1016/0022-2836(85)90048-8. [DOI] [PubMed] [Google Scholar]
- Westhof E., Romby P., Romaniuk P. J., Ebel J. P., Ehresmann C., Ehresmann B. Computer modeling from solution data of spinach chloroplast and of Xenopus laevis somatic and oocyte 5 S rRNAs. J Mol Biol. 1989 May 20;207(2):417–431. doi: 10.1016/0022-2836(89)90264-7. [DOI] [PubMed] [Google Scholar]
- Wimberly B., Varani G., Tinoco I., Jr The conformation of loop E of eukaryotic 5S ribosomal RNA. Biochemistry. 1993 Feb 2;32(4):1078–1087. doi: 10.1021/bi00055a013. [DOI] [PubMed] [Google Scholar]
- Woo N. H., Roe B. A., Rich A. Three-dimensional structure of Escherichia coli initiator tRNAfMet. Nature. 1980 Jul 24;286(5771):346–351. doi: 10.1038/286346a0. [DOI] [PubMed] [Google Scholar]
- Woodson S. A., Crothers D. M. Structural model for an oligonucleotide containing a bulged guanosine by NMR and energy minimization. Biochemistry. 1988 May 3;27(9):3130–3141. doi: 10.1021/bi00409a004. [DOI] [PubMed] [Google Scholar]
- Zhang P., Moore P. B. An NMR study of the helix V-loop E region of the 5S RNA from Escherichia coli. Biochemistry. 1989 May 30;28(11):4607–4615. doi: 10.1021/bi00437a015. [DOI] [PubMed] [Google Scholar]
- van den Hoogen Y. T., van Beuzekom A. A., de Vroom E., van der Marel G. A., van Boom J. H., Altona C. Bulge-out structures in the single-stranded trimer AUA and in the duplex (CUGGUGCGG).(CCGCCCAG). A model-building and NMR study. Nucleic Acids Res. 1988 Jun 10;16(11):5013–5030. doi: 10.1093/nar/16.11.5013. [DOI] [PMC free article] [PubMed] [Google Scholar]




