Abstract
Toll-like receptors (TLRs) 2 and 4 have recently been identified as possible signal transducers for various bacterial ligands. To investigate the roles of TLRs in the recognition of periodontopathic bacteria by the innate immune system, a Chinese hamster ovary (CHO) nuclear factor-κB (NF-κB)-dependent reporter cell line, 7.7, which is defective in both TLR2- and TLR4-dependent signaling pathways was transfected with human CD14 and TLRs. When the transfectants were exposed to freeze-dried periodontopathic bacteria, Actinobacillus actinomycetemcomitans, Porphyromonas gingivalis, Capnocytophaga ochracea, and Fusobacterium nucleatum, and a non-oral bacterium, Escherichia coli, all species of the bacteria induced NF-κB-dependent CD25 expression in 7.7/huTLR2 cells. Although freeze-dried A. actinomycetemcomitans, F. nucleatum, and E. coli also induced CD25 expression in 7.7/huTLR4 cells, freeze-dried P. gingivalis did not. Similarly, lipopolysaccharides (LPS) extracted from A. actinomycetemcomitans, F. nucleatum, and E. coli induced CD25 expression in 7.7/huTLR4 cells, but LPS from P. gingivalis and C. ochracea did not. Furthermore, LPS from P. gingivalis and C. ochracea attenuated CD25 expression in 7.7/huTLR4 cells induced by repurified LPS from E. coli. LPS from P. gingivalis and C. ochracea also inhibited the secretion of interleukin-6 (IL-6) from U373 cells, the secretion of IL-1β from human peripheral blood mononuclear cells, and ICAM-1 expression in human gingival fibroblasts induced by repurified LPS from E. coli. These findings indicated that LPS from P. gingivalis and C. ochracea worked as antagonists for human TLR4. The antagonistic activity of LPS from these periodontopathic bacteria may be associated with the etiology of periodontal diseases.
It is generally accepted that most periodontal diseases are caused by bacteria in dental plaque (10). More than 300 species of bacteria colonize in the subgingival area, and their cell wall components can trigger immune activation (17). Those compounds induce a series of proinflammatory cytokines from host tissue (40), which induce alveolar bone resorption and production of matrix metalloproteinases that destroy the gingival connective tissue (9, 27).
Recently, two members of the Toll-like receptor (TLR) family, TLR2 and TLR4, have been identified as possible signaling receptors for bacterial cell wall components. The expression of TLR2 in Chinese hamster ovary (CHO) fibroblasts or human embryonic kidney cells (HEK293), which are TLR2 deficient (6, 14), conferred responsiveness to various bacterial components, such as peptidoglycan, lipoprotein, and lipoarabinomannan (19, 21, 31, 41). tlr4 cloned from lipopolysaccharide (LPS)-resistant C3H/HeJ mice harbored a point mutation that rendered it nonfunctional (28). Subsequently, in vivo roles of TLR2 and TLR4 were investigated (33). While TLR4-deficient mice were unresponsive to LPS, macrophages from TLR2-deficient mice lacked the response to gram-positive bacterial cell wall, strengthening the hypothesis that TLR4 is a principal signal transducer for LPS and TLR2 is a signal transducer for other bacterial components, such as peptidoglycan and lipoprotein.
In the present study, we investigated the roles of human TLR2 and TLR4 in the recognition of periodontopathic bacteria using a mutant CHO/CD14 reporter cell line, 7.7, which has a defect in its LPS-signaling pathway (2). As CHO cells do not express a functional transcript for TLR2 (6), 7.7 has a defect in both TLR2- and TLR4-dependent signaling pathways. Although the exact defect in signal transduction in 7.7 remains unknown, 7.7 transfected with human TLR4 (7.7/huTLR4) responds to high concentrations of LPS, and 7.7 transfected with human TLR2 (7.7/huTLR2) was as sensitive as CHO/CD14/huTLR2 to bacterial lipoprotein (19). These transfectants that expressed human TLRs were exposed to the following freeze-dried periodontopathic bacteria: Porphyromonas gingivalis, Actinobacillus actinomycetemcomitans, Capnocytophaga ochracea, and Fusobacterium nucleatum (11, 12). A non-oral bacterium, Escherichia coli, was used as a control. The results indicated that human TLR2 expression in 7.7 enhanced the response to each freeze-dried bacterium but that human TLR4 expression failed to enhance the response to freeze-dried P. gingivalis and C. ochracea, as determined by nuclear factor-κB (NF-κB)-dependent CD25 expression. As LPS is a potent TLR4 ligand, we extracted LPS from each of the bacteria and exposed 7.7/huTLR4 cells to each of the LPS. The results revealed a unique ability of LPS from P. gingivalis and C. ochracea to act as antagonists for human TLR4. The antagonistic activities of those LPS were also analyzed in the U373 human astrocytoma cell line, freshly isolated human peripheral blood mononuclear cells (PBMC), and human gingival fibroblasts.
MATERIALS AND METHODS
Reagents.
Phosphate-buffered saline (PBS), Ham’s F-12, RPMI 1640, Dulbecco’s modified Eagle’s medium (DMEM), α-MEM, penicillin-streptomycin, G418, and trypsin-EDTA were obtained from Gibco BRL (Rockville, Md.). Fetal bovine serum (FBS) was obtained from Biological Industry (Kibbutz Beit Haemek, Israel). Hygromycin B was obtained from Calbiochem (San Diego, Calif.). Anti-CD25 monoclonal antibody (MAb) conjugated with fluorescein isothiocyanate (FITC) was obtained from Becton Dickinson (Bedford, Mass.). Anti-ICAM-1 (CD54) MAb conjugated with FITC was obtained from Beckman Coulter (Fullerton, Calif.). Freeze-dried E. coli K12 and LPS from E. coli O111:B4 were obtained from Sigma (St. Louis, Mo.). Ficoll-Paque was obtained from Pharmacia (Uppsala, Sweden). An enzyme-linked immunosorbent assay (ELISA) kit, Cytoscreen, for interleukin-1β (IL-1β) and IL-6 was obtained from Biosource (Camarillo, Calif.). An enhanced colloidal gold kit was obtained from Bio-Rad (Hercules, Calif.).
Bacterial strains and growth conditions.
P. gingivalis 381, C. ochracea 25, and F. nucleatum ATCC 10953 cells were grown in GAM broth supplemented with vitamin K3 (5 μg/ml) and hemin (5 μg/ml) at 37°C for 3 days under anaerobic conditions (10% H2, 10% CO2, 80% N2), and A. actinomycetemcomitans Y4 cells were grown in Todd-Hewitt broth with 1% yeast extract under the same conditions (4, 35). The organisms were harvested by centrifugation, washed three times with distilled water, and freeze dried. Some of the freeze-dried periodontopathic bacteria and freeze-dried E. coli cells were used in experiments, and the remaining bacteria were used to prepare LPS. Staphylococcus aureus IID671 (a gift from N. Ohara, Department of Oral Bacteriology, Nagasaki University) was grown in LPS-free α-MEM. The cells were washed twice with PBS, and the cell density was determined by limiting dilution. Bacteria were resuspended in PBS, killed by incubation at 95°C for 20 min, and stored at −20°C until use.
Preparations of LPS.
LPS was purified according to the procedure described by Koga et al. (15) unless otherwise mentioned. Briefly, LPS was extracted from five species of microorganisms using the hot-phenol water procedure and then ultracentrifuged, treated with pronase and nuclease P1, and heated at 100°C for 5 min. To ensure the cells were activated by LPS but not contaminated protein, repurified LPS from E. coli was used only in the antagonistic experiments. The repurified LPS was prepared by the procedure described by Manthey et al. (20). Five milligrams of LPS from E. coli was resuspended in 1 ml of endotoxin-free water at room temperature containing 0.2% triethylamine (TEA). The sample was split into two 500-μl aliquots, and one aliquot was stored at −20°C without further manipulation (unpurified LPS). Deoxycholate was added to the remaining aliquot to a final concentration of 0.5%, and then 500 μl of water-saturated phenol was added. The sample was vortexed intermittently for 5 min, and phases were allowed to separate at room temperature for 5 min. The sample was placed on ice for 10 min and then centrifuged at 4°C for 2 min at 14,000 × g. The top aqueous layer was transferred to a new tube, and the phenol phase was subjected to reextraction with 500 μl of 0.2% TEA-0.5% deoxycholate. The aqueous phases were pooled and reextracted with 1 ml of water-saturated phenol. The pooled aqueous phases were adjusted to 75% ethanol and 30 mM sodium acetate and were allowed to precipitate at −20°C for 1 h. The precipitate was centrifuged at 4°C for 10 min at 14,000 × g, washed in 1 ml of cold 100% ethanol, and air dried. The precipitate was resuspended in the original volume (500 μl) of 0.2% TEA. Twenty-five micrograms of each of the LPS was subjected to sodium dodecyl sulfate-12% polyacrylamide gel electrophoresis and transferred to a polyvinylidene difluoride membrane. When the membrane was stained with colloidal gold, no protein band was detected in either the repurified LPS lane or the lanes of LPS from five species of bacteria, while there was a faint band in the lane of a commercial preparation of E. coli O111:B4 LPS, indicating that our preparations of LPS were highly purified. LPS was prepared at 1 to 10 mg/ml in PBS and stored at −20°C. Before use, the suspensions were thawed and sonicated in a Silentsonic (Sharp, Osaka, Japan) for 1 min.
Cell lines.
CHO reporter cells were grown as adherent monolayers at 37°C in a 5% saturated CO2 atmosphere, and they were passaged at least twice weekly to maintain logarithmic growth. The engineering of the CD14-expressing CHO reporter cell line CHO/CD14.elam.tac, also known as clone 3E10, has been previously described in detail (2). This clonal line expresses surface CD25 antigen under the control of a region from the human E-selectin promoter containing the nuclear factor-κB (NF-κB) binding site. 7.7 is a mutant derived from CHO/CD14 with a defect in the LPS-signaling pathway. The exact defect in signal transduction remains unknown, but 7.7 responds normally to tumor necrosis factor alpha (TNF-α) and IL-1β, and it expresses normal levels of mRNA for hamster TLR4 (30). CHO/CD14 and 7.7 cells were stably transfected with cDNA for human TLR2 or TLR4 as described elsewhere (19) and were grown in the presence of G418 (1 mg/ml) and hygromycin B (400 U/ml).
U373 cells were grown in DMEM supplemented with 10% FBS, penicillin (100 U/ml), and streptomycin (100 μg/ml) at 37°C in a 5% saturated CO2 atmosphere.
Human peripheral blood mononuclear cells (PBMC) were isolated by gradient centrifugation of heparinized blood on endotoxin-free Ficoll-Paque according to the manufacturer’s protocol. The cells were resuspended in RPMI 1640 medium supplemented with 10% human serum and used for experiments.
Human gingival fibroblasts were prepared from explants of clinically normal gingival tissue obtained at the time of extraction of a noninfected third molar from a patient according to the procedure described by Hayashi et al. (5). Informed consent was obtained prior to inclusion in this study. The cells were grown in DMEM supplemented with 10% FBS, penicillin (100 U/ml), and streptomycin (100 μg/ml) at 37°C in a 5% saturated CO2 atmosphere.
Flow cytometric analysis.
CHO transfectants and human gingival fibroblasts were plated in 24-well tissue culture dishes at a density of 105 cells per well (19). After overnight incubation, confluent monolayers of CHO cells and human gingival fibroblasts were stimulated with various ligands: LPS (10 ng/ml to 100 μg/ml), heat-killed S. aureus (1 × 108 to 2 × 108 CFU/ml), freeze-dried bacteria (0.1 to 300 μg/ml), and combinations of these ligands. Following incubation for 18 h, the cells were treated with trypsin/EDTA for 1 min and the detached cells were assessed by flow cytometry for the presence of surface CD25 or ICAM-1 as described previously (41).
Cytokine assays.
U373 cells were plated at a density of 3 × 104 cells per well in a 96-well dish. After overnight incubation, confluent monolayers of U373 cells were stimulated with LPS from indicated species of bacteria (10 to 100 ng/ml) with or without the addition of LPS from P. gingivalis or C. ochracea (0, 100, and 1,000 ng/ml) to the culture medium. Following incubation for 18 h, cell-free supernatants were harvested and analyzed for IL-6 release by ELISA.
Freshly isolated human PBMC were plated at a density of 4 × 105 cells per well in a 96-well dish (19). The cells were stimulated with LPS from indicated species of bacteria (1 to 10 ng/ml) immediately following the addition of LPS from P. gingivalis or C. ochracea (0, 10, and 100 ng/ml) to the culture medium. Following incubation for 18 h, cell-free supernatants were harvested and analyzed for IL-1β release by ELISA.
RESULTS
Activation of 7.7/huTLR2 and 7.7/huTLR4 cells by freeze-dried periodontopathic bacteria.
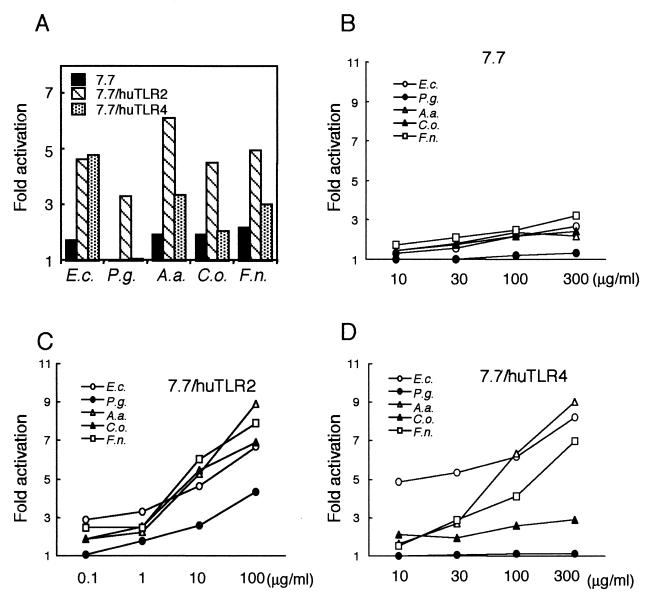
In order to examine how the TLR signaling complex recognizes periodontopathic bacteria, LPS-nonresponsive mutant CHO/CD14 reporter cells, 7.7 cells, were transfected with human TLR2 or TLR4 and exposed to freeze-dried periodontopathic bacteria for 18 h. While four of the five species of bacteria each (100 μg/ml) weakly activated the NF-κB reporter construct in 7.7 cells, the expression of human TLR2 in 7.7 cells induced two to three times higher levels of expression of CD25 (Fig. 1A). Although freeze-dried P. gingivalis cells induced only marginal levels of CD25 expression in 7.7 cells, the expression of human TLR2 significantly enhanced the response to the organisms. The expression of human TLR4 in 7.7 cells also induced two to three times higher levels of expression of CD25 in response to freeze-dried E. coli, A. actinomycetemcomitans, and F. nucleatum but failed to confer responsiveness to freeze-dried P. gingivalis. Although we stimulated the cells with up to 300 μg of freeze-dried bacteria/ml, CD25 expression levels in 7.7/huTLR4 cells were no higher than those in 7.7 when stimulated with freeze-dried P. gingivalis (Fig. 1B and D). We could not stimulate the cells with freeze-dried P. gingivalis at concentrations of higher than 300 μg/ml, because the cells did not survive with the organisms for 18 h. It is also notable that CD25 expression levels in 7.7/huTLR4 cells were no higher than those in 7.7 cells when stimulated with freeze-dried C. ochracea.
FIG. 1.
TLR2 and/or TLR4 mediated cellular activation by freeze-dried periodontopathic bacteria. (A) LPS-nonresponsive mutant CHO/CD14, 7.7, 7.7/huTLR2, and 7.7/huTLR4 cells were exposed to 100 μg of freeze-dried bacteria/ml for 18 h. The cells were stained with FITC-labeled anti-CD25 MAb and subjected to flow cytometric analysis for NF-κB-driven CD25 expression. (B to D) 7.7 (B), 7.7/huTLR2 (C), and 7.7/huTLR4 (D) cells were exposed to indicated doses of freeze-dried bacteria for 18 h. The cells were stained with FITC-labeled anti-CD25 MAb and subjected to flow cytometric analysis for NF-κB-driven CD25 expression. Activation is expressed as the fold induction of mean channel fluorescence in comparison to unstimulated cells. Representative results of one of three experiments performed are shown. E.c., freeze-dried E. coli; P.g., freeze-dried P. gingivalis; A.a., freeze-dried A. actinomycetemcomitans; C.o., freeze-dried C. ochracea; F.n., freeze-dried F. nucleatum.
Activation of 7.7/huTLR4 cells by LPS from E. coli, A. actinomycetemcomitans, and F. nucleatum but not by LPS from P. gingivalis or C. ochracea.
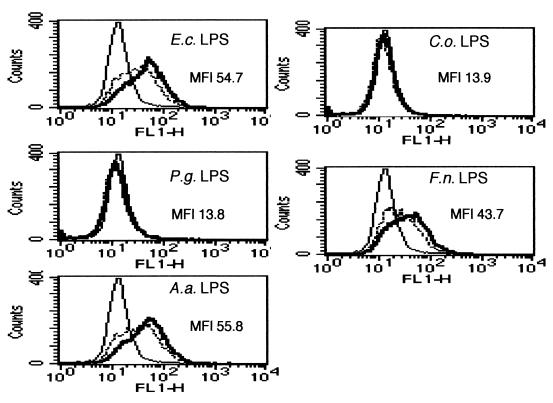
As LPS is the most potent inducer of TLR4-mediated activation, we exposed 7.7 and 7.7/huTLR4 cells to LPS extracted from the five species of bacteria. While none of the LPS from the five species of bacteria activated the NF-κB reporter construct in 7.7 cells (data not shown), the expression of human TLR4 in 7.7 cells conferred responsiveness to LPS from E. coli, A. actinomycetemcomitans, and F. nucleatum (Fig. 2). However, it failed to confer responsiveness to LPS from P. gingivalis and C. ochracea. No CD25 expression was observed in 7.7/huTLR4 cells when stimulated with up to 100 μg of LPS/ml from P. gingivalis and C. ochracea (data not shown). These results indicated that LPS was at least one of the components that determined the responsiveness of 7.7/huTLR4 cells to the microorganisms.
FIG. 2.
TLR4 mediated cellular activation by LPS from periodontopathic bacteria. 7.7/huTLR4 cells were exposed to either PBS (thin line) or LPS from indicated species of bacteria (dotted line, 1 μg/ml; thick line, 10 μg/ml). After 18 h of incubation, the cells were stained with FITC-labeled anti-CD25 MAb and subjected to flow cytometric analysis to determine transgene expression. Representative results of one of three experiments performed are shown. E.c., E. coli; P.g., P. gingivalis; A.a., A. actinomycetemcomitans; C.o., C. ochracea; F.n., F. nucleatum.
Inhibitory effects of LPS from P. gingivalis and C. ochracea on activation of 7.7/huTLR4 cells but not 7.7/huTLR2 cells.
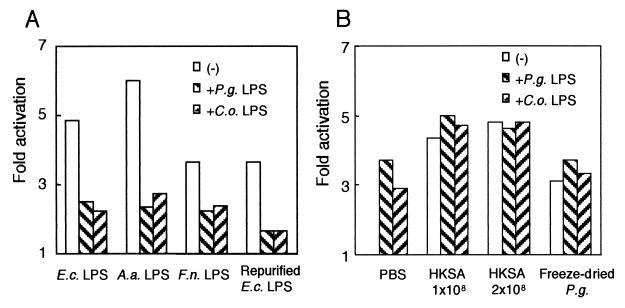
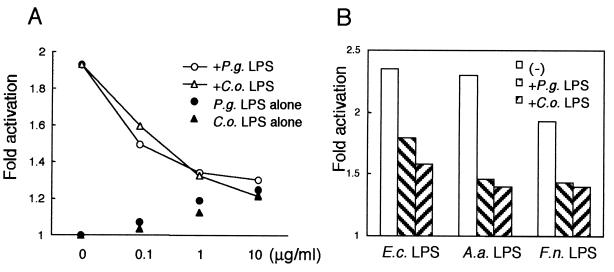
It is known that lipid IVa, a lipid A precursor, and Rhodobacter spheroides lipid A lack the ability to stimulate human cells and work as potent LPS antagonists (18, 29). We examined whether LPS from P. gingivalis and C. ochracea can act similarly as LPS antagonists. When 7.7/huTLR4 cells were stimulated with LPS from E. coli, A. actinomycetemcomitans, and F. nucleatum (1 μg/ml), the addition of LPS from P. gingivalis or C. ochracea (10 μg/ml) attenuated the expression of CD25 (Fig. 3A). To ensure that the cells were activated by LPS but not a small amount of contaminated protein, 7.7/huTLR4 cells were also exposed to repurified LPS from E. coli (1 μg/ml). The addition of LPS from P. gingivalis or C. ochracea (10 μg/ml) attenuated the expression of CD25 in 7.7/huTLR4 cells stimulated with repurified LPS from E. coli.
FIG. 3.
LPS from P. gingivalis and C. ochracea blocked NF-κB activation in 7.7/huTLR4 cells but not in 7.7/huTLR2 cells. (A) 7.7/huTLR4 cells were exposed to either PBS or various LPS: LPS from E. coli, A. actinomycetemcomitans, and F. nucleatum (1 μg/ml) and repurified LPS from E. coli (1 μg/ml), with or without LPS from P. gingivalis or C. ochracea (10 μg/ml). (B) 7.7/huTLR2 cells were exposed to PBS, heat-killed S. aureus (1 × 108 to 2 × 108 CFU/ml), or freeze-dried P. gingivalis (10 μg/ml) with or without LPS from P. gingivalis or C. ochracea (10 μg/ml). After 18 h of incubation, the cells were stained with FITC-labeled anti-CD25 MAb and subjected to flow cytometric analysis to determine transgene expression. Activation is expressed as the fold induction of mean channel fluorescence in comparison to unstimulated cells. Representative results of one of three experiments performed are shown. E.c., E. coli; P.g., P. gingivalis; A.a., A. actinomycetemcomitans; C.o., C. ochracea; F.n., F. nucleatum; HKSA, heat-killed S. aureus.
Next, we examined whether LPS from P. gingivalis and C. ochracea could inhibit other signaling pathways. When 7.7/huTLR2 cells were exposed to heat-killed S. aureus (1 × 108 to 2 × 108 CFU/ml, equivalent to 18.5 to 37 μg/ml), four- to fivefold higher levels of activation were induced as reported previously (19). Although LPS from P. gingivalis or C. ochracea themselves (10 μg/ml) induced the expression of CD25 in 7.7/huTLR2 cells, the addition of those LPS did not inhibit the activation in 7.7/huTLR2 cells induced by heat-killed S. aureus (Fig. 3B). Similarly, LPS from P. gingivalis or C. ochracea did not inhibit the expression of CD25 in 7.7/huTLR2 cells induced by freeze-dried P. gingivalis, indicating that LPS from P. gingivalis and C. ochracea did not inhibit TLR2-mediated signal transduction.
Inhibitory effects of LPS from P. gingivalis and C. ochracea on LPS-induced secretion of IL-6 from U373 cells.
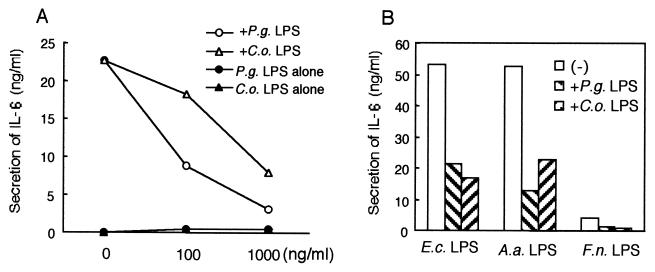
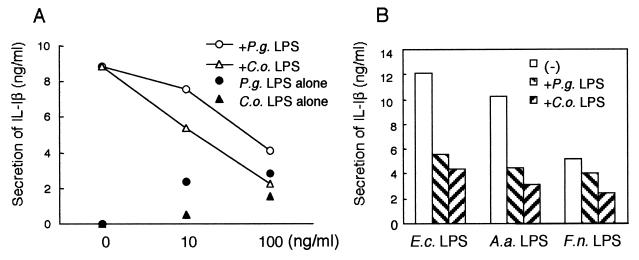
In order to determine whether our findings in CHO reporter cell lines reflect the signal transduction systems used by other physiological cell types, we stimulated a human astrocytoma cell line, U373, which has been reported to expresses TLR4 but not TLR2 (7, 36). As shown in Fig. 4A, repurified LPS from E. coli (50 ng/ml) induced a significant level of IL-6 release from U373 cells, while LPS from P. gingivalis and C. ochracea induced only minimal levels of it. The addition of LPS from P. gingivalis or C. ochracea (100 and 1,000 ng/ml) to the culture evidently attenuated the IL-6 secretion by U373 cells stimulated with repurified LPS from E. coli. As shown in Fig. 4B, IL-6 secretion by U373 cells stimulated with LPS from E. coli (10 ng/ml), A. actinomycetemcomitans (10 ng/ml), or F. nucleatum (100 ng/ml) was also attenuated by the addition of LPS from P. gingivalis or C. ochracea (1,000 ng/ml), indicating that LPS from P. gingivalis and C. ochracea worked as antagonists in U373 cells.
FIG. 4.
LPS from P. gingivalis and C. ochracea blocked the secretion of IL-6 from U373 cells. (A) U373 cells were plated at a density of 3 × 104 cells/well in 96-well dishes. The next day, cells were exposed to repurified LPS from E. coli (50 ng/ml) with or without indicated concentrations of LPS from P. gingivalis or C. ochracea. Some of the cells were exposed to LPS from P. gingivalis or C. ochracea alone. After incubation for 18 h, the supernatants were harvested and assayed for IL-6 by ELISA. (B) U373 cells were exposed to LPS from E. coli (10 ng/ml), A. actinomycetemcomitans (10 ng/ml), or F. nucleatum (100 ng/ml) with or without LPS from P. gingivalis and C. ochracea (1,000 ng/ml). After incubation for 18 h, the supernatants were harvested and assayed for IL-6 by ELISA. Representative results of one of three experiments performed are shown. E.c., E. coli; P.g., P. gingivalis; A.a., A. actinomycetemcomitans; C.o., C. ochracea, F.n., F. nucleatum.
Inhibitory effects of LPS from P. gingivalis and C. ochracea on LPS-induced secretion of IL-1β by PBMC and on LPS-induced ICAM-1 expression in human gingival fibroblasts.
Next, we stimulated freshly isolated human PBMC with LPS from periodontopathic bacteria. In contrast to U373 cells, PBMC express both TLR2 and TLR4, and they secrete IL-1β when stimulated with high concentrations of LPS from P. gingivalis or C. ochracea (Fig. 5A). Nevertheless, when the cells were stimulated with a low concentration of repurified LPS from E. coli (5 ng/ml), the addition of LPS from P. gingivalis or C. ochracea (10 and 100 ng/ml) inhibited the IL-1β production from PBMC dose dependently. Similarly, LPS from P. gingivalis or C. ochracea also inhibited IL-1β production from PBMC induced by LPS from E. coli, A. actinomycetemcomitans, or F. nucleatum (Fig. 5B).
FIG. 5.
LPS from P. gingivalis and C. ochracea blocked the secretion of IL-1β from PBMC. (A) Human PBMC from healthy volunteers were isolated by gradient centrifugation, resuspended in RPMI 1640 medium supplemented with 10% human serum, and plated at a density of 4 × 105 cells/well in 96-well dishes. The cells were exposed to repurified LPS from E. coli (5 ng/ml) with or without indicated concentrations of LPS from P. gingivalis or C. ochracea. Some of the cells were exposed to LPS from P. gingivalis or C. ochracea alone. (B) Human PBMC were exposed to LPS from E. coli (1 ng/ml), A. actinomycetemcomitans (1 ng/ml), or F. nucleatum (10 ng/ml) with or without LPS from P. gingivalis or C. ochracea (100 ng/ml). After incubation for 18 h, the supernatants were harvested and assayed for IL-1β by ELISA. Representative results of one of three experiments performed are shown. E.c., E. coli; P.g., P. gingivalis; A.a., A. actinomycetemcomitans; C.o., C. ochracea; F.n., F. nucleatum.
We also examined the effects of LPS from P. gingivalis or C. ochracea using a human gingival fibroblast cell line. As observed for PBMC, those cells expressed ICAM-1 when stimulated with high concentrations of LPS from P. gingivalis or C. ochracea (Fig. 6A). However, those LPS dose dependently inhibited the induction of ICAM-1 expression in gingival fibroblasts stimulated with repurified LPS from E. coli (100 ng/ml). ICAM-1 expression in gingival fibroblasts induced by LPS from E. coli, A. actinomycetemcomitans, or F. nucleatum (10 ng/ml) was also inhibited by the addition of LPS from P. gingivalis or C. ochracea (10 μg/ml) (Fig. 6B).
FIG. 6.
LPS from P. gingivalis and C. ochracea blocked the induction of ICAM-1 in the gingival fibroblasts. (A) Human gingival fibroblasts were plated at a density of 105 cells/well in 24-well dishes. The next day, cells were exposed to repurified LPS from E. coli (100 ng/ml) with or without indicated concentrations of LPS from P. gingivalis or C. ochracea. Some of the cells were exposed to LPS from P. gingivalis or C. ochracea alone. (B) Human gingival fibroblasts were exposed to LPS from E. coli, A. actinomycetemcomitans, or F. nucleatum (10 ng/ml) with or without LPS from P. gingivalis or C. ochracea (10 μg/ml). After incubation for 18 h, the cells were stained with FITC-labeled anti-ICAM-1 MAb and subjected to flow cytometric analysis. Activation is expressed as the fold induction of mean channel fluorescence in comparison to unstimulated cells. Representative results of one of three experiments performed are shown. E.c., E. coli; P.g., P. gingivalis; A.a., A. actinomycetemcomitans; C.o., C. ochracea, F.n., F. nucleatum.
Species-specific recognition of LPS from P. gingivalis and C. ochracea.
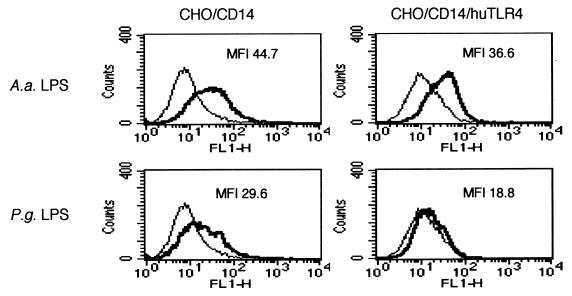
Although our present results strongly indicated that LPS from P. gingivalis and C. ochracea are antagonists for human TLR4, it has been reported that a synthetic lipid A of P. gingivalis activated C3H/HeN macrophages but not C3H/HeJ macrophages, indicating that mouse TLR4 played an essential role in the response to this compound (24). To address the question of whether the antagonistic activities of LPS from P. gingivalis and C. ochracea are species specific, CHO/CD14 cells which express a functional hamster TLR4 transcript were transfected with human TLR4. Although LPS from P. gingivalis (10 μg/ml) induced CD25 expression in CHO/CD14 cells, the overexpression of human TLR4 resulted in the loss of sensitivity (Fig. 7). Similarly, LPS from C. ochracea (10 μg/ml) induced CD25 expression in CHO/CD14 cells, but the expression of human TLR4 resulted in the loss of sensitivity (data not shown). In contrast, LPS from A. actinomycetemcomitans, E. coli, and F. nucleatum induced CD25 expression in both CHO/CD14 and CHO/CD14/huTLR4 cells (Fig. 7 and data not shown).
FIG. 7.
Expression of human TLR4 abrogated the responsiveness to LPS from P. gingivalis but not LPS from A. actinomycetemcomitans in CHO/CD14 cells. CHO/CD14 and CHO/CD14/huTLR4 cells were exposed to LPS from P. gingivalis or LPS from A. actinomycetemcomitans (10 μg/ml). After 18 h of incubation, the cells were stained with FITC-labeled anti-CD25 MAb and subjected to flow cytometric analysis to determine transgene expression. Representative results of one of three experiments performed are shown. P.g., P. gingivalis; A.a., A. actinomycetemcomitans.
DISCUSSION
When 7.7/huTLR2 cells were exposed to freeze-dried bacteria, NF-κB was activated dose dependently in response to all species of freeze-dried microorganisms used in this study (Fig. 1C), indicating that human TLR2 played an important role in the recognition of these bacteria. Although the expression of human TLR4 in 7.7 cells also conferred responsiveness to freeze-dried E. coli, A. actinomycetemcomitans, and F. nucleatum, it did not affect the response to freeze-dried P. gingivalis and C. ochracea, indicating two possibilities: (i) the whole bacterial components of P. gingivalis and C. ochracea were not recognized by human TLR4, or (ii) some microbial components present in these bacteria inhibited TLR4-mediated activation. LPS is known to be one of the most potent bacterial components in inducing TLR4-mediated signaling. In view of our findings that stable expression of human TLR4 in mutant 7.7 led to a bypass reversion of the LPS-nonresponder phenotype, we hypothesized that the unresponsiveness of 7.7/huTLR4 cells to freeze-dried P. gingivalis and C. ochracea was due to the unique characteristics of their LPS. In fact, LPS from P. gingivalis and C. ochracea did not induce human TLR4-mediated signaling. Conversely, those LPS inhibited the activation of 7.7/huTLR4 cells induced by LPS from the other bacteria, indicating that those LPS worked as antagonists for human TLR4. Although it remains unknown whether P. gingivalis and C. ochracea possess any component to induce TLR4-mediated signaling, it has become clear that their LPS contributed to the unresponsiveness of 7.7/huTLR4 cells to these organisms.
LPS from P. gingivalis has been studied by many investigators, while little is known about LPS from C. ochracea. Ogawa et al. and Tanamoto et al. analyzed the chemical structure of P. gingivalis lipid A and reported that the absence of ester-linked phosphate at the 4′ position of glucosamine disaccharide and the presence of branched and relatively longer fatty acids are unique features (16, 23). Tanamoto et al. also demonstrated that P. gingivalis lipid A induced TNF-α release from C3H/HeJ macrophages to the same extent as that from C3H/HeN macrophages (34), indicating that the induction of TNF-α release was irrelevant to the tlr4 mutation in the C3H/HeJ mouse. Using repurified LPS from P. gingivalis, Kirikae et al. confirmed that it could induce TNF-α release from C3H/HeJ macrophages (13), and Hirschfeld et al. showed that repurified P. gingivalis LPS induced IL-6 production from U87 cells transfected with human TLR2 but not TLR4, indicating that it does not signal through human TLR4 (8). Furthermore, Ogawa et al. demonstrated clear antagonistic effects of P. gingivalis lipid A and LPS against IL-1β production by PBMC stimulated with E. coli LPS or synthetic E. coli-type lipid A (compound 506) (26). In addition, Darveau et al. demonstrated that LPS from P. gingivalis blocked E. coli LPS-induced E-selectin expression in human umbilical cord vein endothelial cells (HUVEC) (1), which strongly express TLR4 but only very weakly express TLR2 (3). In accordance with the reported unique features of P. gingivalis LPS, our present results using a genetically engineered 7.7/huTLR4 cell line clearly demonstrated that LPS from P. gingivalis and C. ochracea inhibit TLR4-mediated signaling but not TLR2-mediated signaling. Furthermore, the inhibitory effect on the secretion of IL-6 by U373 cells strongly supports the hypothesis that LPS from P. gingivalis and C. ochracea work as antagonists for TLR4 in physiological human cells.
In contrast to 7.7/huTLR4 and U373 cells, human PBMC and gingival fibroblasts were weakly activated by LPS from P. gingivalis and C. ochracea, consistent with previous reports by other investigators (25, 32, 37). Since those cells express both TLR2 and TLR4 (22, 39), the TLR2-signaling complex might be activated in these cells, as observed in 7.7/huTLR2 cells (Fig. 3B). Although we could not detect any protein in our P. gingivalis and C. ochracea LPS preparation with colloidal gold staining, we cannot be certain what molecule is responsible for TLR2-mediated activation. Like LPS from Leptospira interrogans, which has recently been reported to act as a TLR2 agonist (38), LPS from P. gingivalis and C. ochracea may act in a similar way. However, very small amounts of contaminants may have remained in P. gingivalis and C. ochracea LPS preparations. These contaminants might activate human PBMC and gingival fibroblasts through TLR2.
Recently, P. gingivalis lipid A was synthesized and characterized (24). There was a slight difference between the natural P. gingivalis lipid A and the synthetic compound in cytokine-inducing activity, and synthetic P. gingivalis lipid A induced cytokine production in C3H/HeN macrophages but not in C3H/HeJ macrophages, indicating that functional mouse TLR4 was necessary for the response to this compound (24). However, we demonstrated in the present study that the overexpression of human TLR4 in CHO/CD14 cells, which express a functional hamster TLR4, abrogated the sensitivity to LPS from P. gingivalis and C. ochracea, indicating that these LPS are antagonists for human TLR4 but not for hamster TLR4. These pharmacological behaviors are quite similar to those of a lipid A analogue, lipid IVa, and R. spheroides lipid A (18, 29). Investigators have suggested that the species-dependent discrimination of lipid A substructures is fully attributable to the species origin of TLR4, consistent with our results. The species-specific activity of P. gingivalis LPS might be related to the sensitivity of the animals to periodontal diseases.
In the present study, LPS from two of four periodontopathic bacteria worked as antagonists for human TLR4. Although the precise effect of the antagonistic LPS on bacterial growth in the gingival sulcus and the prevalence of these unique bacteria in patients remain to be elucidated, the antagonistic activity would be a great advantage for the microorganisms to escape from the innate immune system. In spite of the potent proinflammatory activity of LPS, gram-negative bacteria predominate in moderate to severe periodontal lesions (11). The antagonistic LPS may play a role in this paradoxical situation and may be associated with the progression of periodontal diseases.
Acknowledgments
This work was supported in part by Grants-in-Aid for Encouragement of Young Scientists from the Ministry of Education, Science, Sports and Culture of Japan, 12771332 and 12771333 (A.Y. and T.K.). Y.H. is supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, 13672192. D.T.G. is supported by NIH grants RO1 GM54060, AI PO150305, and AI32725.
Editor: R. N. Moore
REFERENCES
- 1.Darveau, R. P., M. D. Cunningham, T. Bailey, C. Seachord, K. Ratcliffe, B. Bainbridge, M. Dietsch, R. C. Page, and A. Aruffo. 1995. Ability of bacteria associated with chronic inflammatory disease to stimulate E-selectin expression and promote neutrophil adhesion. Infect. Immun. 63: 1311–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Delude, R. L., A. Yoshimura, R. R. Ingalls, and D. T. Golenbock. 1998. Construction of a lipopolysaccharide reporter cell line and its use in identifying mutants defective in endotoxin, but not TNF-α, signal transduction. J. Immunol. 161: 3001–3009. [PubMed] [Google Scholar]
- 3.Faure, E., O. Equils, P. A. Sieling, L. Thomas, F. X. Zhang, C. J. Kirschning, N. Polentarutti, M. Muzio, and M. Arditi. 2000. Bacterial lipopolysaccharide activates NF-κB through Toll-like receptor 4 (TLR-4) in cultured human dermal endothelial cells. Differential expression of TLR-4 and TLR-2 in endothelial cells. J. Biol. Chem. 275: 11058–11063. [DOI] [PubMed] [Google Scholar]
- 4.Hara, Y., T. Kaneko, A. Yoshimura, and I. Kato. 1996. Serum rheumatoid factor induced by intraperitoneal administration of periodontopathic bacterial lipopolysaccharide in mice. J. Periodont. Res. 31: 502–507. [DOI] [PubMed] [Google Scholar]
- 5.Hayashi, J., I. Saito, I. Ishikawa, and N. Miyasaka. 1994. Effects of cytokines and periodontopathic bacteria on the leukocyte function-associated antigen 1/intercellular adhesion molecule 1 pathway in gingival fibroblasts in adult periodontitis. Infect. Immun. 62: 5205–5212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heine, H., C. J. Kirschning, E. Lien, B. G. Monks, M. Rothe, and D. T. Golenbock. 1999. Cutting edge: cells that carry a null allele for Toll-like receptor 2 are capable of responding to endotoxin. J. Immunol. 162: 6971–6975. [PubMed] [Google Scholar]
- 7.Hirschfeld, M., C. J. Kirschning, R. Schwandner, H. Wesche, J. H. Weis, R. M. Wooten, and J. J. Weis. 1999. Cutting edge: inflammatory signaling by Borrelia burgdorferi lipoproteins is mediated by Toll-like receptor 2. J. Immunol. 163: 2382–2386. [PubMed] [Google Scholar]
- 8.Hirschfeld, M., J. J. Weis, V. Toshchakov, C. A. Salkowski, M. J. Cody, D. C. Ward, N. Qureshi, S. M. Michalek, and S. N. Vogel. 2001. Signaling by Toll-like receptor 2 and 4 agonists results in differential gene expression in murine macrophages. Infect. Immun. 69: 1477–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Howells, G. L. 1995. Cytokine networks in destructive periodontal disease. Oral Dis. 1: 266–270. [DOI] [PubMed] [Google Scholar]
- 10.Jenkinson, H. F., and D. Dymock. 1999. The microbiology of periodontal disease. Dent. Update 26: 191–197. [DOI] [PubMed] [Google Scholar]
- 11.Kamma, J. J., M. Nakou, and F. A. Manti. 1994. Microbiota of rapidly progressive periodontitis lesions in association with clinical parameters. J. Periodontol. 65: 1073–1078. [DOI] [PubMed] [Google Scholar]
- 12.Kamma, J. J., M. Nakou, and F. A. Manti. 1995. Predominant microflora of severe, moderate and minimal periodontal lesions in young adults with rapidly progressive periodontitis. J. Periodont. Res. 30: 66–72. [DOI] [PubMed] [Google Scholar]
- 13.Kirikae, T., T. Nitta, F. Kirikae, Y. Suda, S. Kusumoto, N. Qureshi, and M. Nakano. 1999. Lipopolysaccharides (LPS) of oral black-pigmented bacteria induce tumor necrosis factor production by LPS-refractory C3H/HeJ macrophages in a way different from that of Salmonella LPS. Infect. Immun. 67: 1736–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kirschning, C. J., H. Wesche, T. Merrill Ayres, and M. Rothe. 1998. Human Toll-like receptor 2 confers responsiveness to bacterial lipopolysaccharide. J. Exp. Med. 188: 2091–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koga, T., T. Nishihara, T. Fujiwara, T. Nisizawa, N. Okahashi, T. Noguchi, and S. Hamada. 1985. Biochemical and immunobiological properties of lipopolysaccharide (LPS) from Bacteroides gingivalis and comparison with LPS from Escherichia coli. Infect. Immun. 47: 638–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumada, H., Y. Haishima, T. Umemoto, and K. Tanamoto. 1995. Structural study on the free lipid A isolated from lipopolysaccharide of Porphyromonas gingivalis. J. Bacteriol. 177: 2098–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lamont, R. J., and H. F. Jenkinson. 1998. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol. Mol. Biol. Rev. 62: 1244–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lien, E., T. K. Means, H. Heine, A. Yoshimura, S. Kusumoto, K. Fukase, M. J. Fenton, M. Oikawa, N. Qureshi, B. Monks, R. W. Finberg, R. R. Ingalls, and D. T. Golenbock. 2000. Toll-like receptor 4 imparts ligand-specific recognition of bacterial lipopolysaccharide. J. Clin. Investig. 105: 497–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lien, E., T. J. Sellati, A. Yoshimura, T. H. Flo, G. Rawadi, R. W. Finberg, J. D. Carroll, T. Espevik, R. R. Ingalls, J. D. Radolf, and D. T. Golenbock. 1999. Toll-like receptor 2 functions as a pattern recognition receptor for diverse bacterial products. J. Biol. Chem. 274: 33419–33425. [DOI] [PubMed] [Google Scholar]
- 20.Manthey, C. L., P. Y. Perera, B. E. Henricson, T. A. Hamilton, N. Qureshi, and S. N. Vogel. 1994. Endotoxin-induced early gene expression in C3H/HeJ (Lpsd) macrophages. J. Immunol. 153: 2653–2663. [PubMed] [Google Scholar]
- 21.Means, T. K., E. Lien, A. Yoshimura, S. Wang, D. T. Golenbock, and M. J. Fenton. 1999. The CD14 ligands lipoarabinomannan and lipopolysaccharide differ in their requirement for Toll-like receptors. J. Immunol. 163: 6748–6755. [PubMed] [Google Scholar]
- 22.Medzhitov, R., P. Preston-Hurlburt, and C. A. Janeway, Jr. 1997. A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature 388: 394–397. [DOI] [PubMed] [Google Scholar]
- 23.Ogawa, T. 1993. Chemical structure of lipid A from Porphyromonas (Bacteroides) gingivalis lipopolysaccharide. FEBS Lett. 332: 197–201. [DOI] [PubMed] [Google Scholar]
- 24.Ogawa, T., Y. Asai, H. Yamamoto, Y. Taiji, T. Jinno, T. Kodama, S. Niwata, H. Shimauchi, and K. Ochiai. 2000. Immunobiological activities of a chemically synthesized lipid A of Porphyromonas gingivalis. FEMS Immunol. Med. Microbiol. 28: 273–281. [DOI] [PubMed] [Google Scholar]
- 25.Ogawa, T., and H. Uchida. 1996. Differential induction of IL-1β and IL-6 production by the nontoxic lipid A from Porphyromonas gingivalis in comparison with synthetic Escherichia coli lipid A in human peripheral blood mononuclear cells. FEMS Immunol. Med. Microbiol. 14: 1–13. [DOI] [PubMed] [Google Scholar]
- 26.Ogawa, T., H. Uchida, and K. Amino. 1994. Immunobiological activities of chemically defined lipid A from lipopolysaccharides of Porphyromonas gingivalis. Microbiology 140: 1209–1216. [DOI] [PubMed] [Google Scholar]
- 27.Page, R. C. 1991. The role of inflammatory mediators in the pathogenesis of periodontal disease. J. Periodont. Res. 26: 230–242. [DOI] [PubMed] [Google Scholar]
- 28.Poltorak, A., X. He, I. Smirnova, M. Y. Liu, C. V. Huffel, X. Du, D. Birdwell, E. Alejos, M. Silva, C. Galanos, M. Freudenberg, P. Ricciardi-Castagnoli, B. Layton, and B. Beutler. 1998. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science 282: 2085–2088. [DOI] [PubMed] [Google Scholar]
- 29.Poltorak, A., P. Ricciardi-Castagnoli, S. Citterio, and B. Beutler. 2000. Physical contact between lipopolysaccharide and Toll-like receptor 4 revealed by genetic complementation. Proc. Natl. Acad. Sci. USA 97: 2163–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schromm, A. B., E. Lien, P. Henneke, J. C. Chow, A. Yoshimura, H. Heine, E. Latz, B. G. Monks, D. A. Schwartz, K. Miyake, and D. T. Golenbock. 2001. Molecular genetic analysis of an endotoxin nonresponder mutant cell line. A point mutation in a conserved region of MD-2 abolishes endotoxin-induced signaling. J. Exp. Med. 194: 79–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwandner, R., R. Dziarski, H. Wesche, M. Rothe, and C. J. Kirschning. 1999. Peptidoglycan- and lipoteichoic acid-induced cell activation is mediated by Toll-like receptor 2. J. Biol. Chem. 274: 17406–17409. [DOI] [PubMed] [Google Scholar]
- 32.Tabeta, K., K. Yamazaki, S. Akashi, K. Miyake, H. Kumada, T. Umemoto, and H. Yoshie. 2000. Toll-like receptors confer responsiveness to lipopolysaccharide from Porphyromonas gingivalis in human gingival fibroblasts. Infect. Immun. 68: 3731–3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takeuchi, O., K. Hoshino, T. Kawai, H. Sanjo, H. Takada, T. Ogawa, K. Takeda, and S. Akira. 1999. Differential roles of TLR2 and TLR4 in recognition of Gram-negative and Gram-positive bacterial cell wall components. Immunity 11: 443–451. [DOI] [PubMed] [Google Scholar]
- 34.Tanamoto, K., S. Azumi, Y. Haishima, H. Kumada, and T. Umemoto. 1997. The lipid A moiety of Porphyromonas gingivalis lipopolysaccharide specifically mediates the activation of C3H/HeJ mice. J. Immunol. 158: 4430–4436. [PubMed] [Google Scholar]
- 35.Tani, Y., M. Tani, and I. Kato. 1997. Extracellular 37-kDa antigenic protein from Actinobacillus actinomycetemcomitans induces TNF-α, IL-1β, and IL-6 in murine macrophages. J. Dent. Res. 76: 1538–1547. [DOI] [PubMed] [Google Scholar]
- 36.Tapping, R. I., S. Akashi, K. Miyake, P. J. Godowski, and P. S. Tobias. 2000. Toll-like receptor 4, but not Toll-like receptor 2, is a signaling receptor for Escherichia and Salmonella lipopolysaccharides. J. Immunol. 165: 5780–5787. [DOI] [PubMed] [Google Scholar]
- 37.Wang, P. L., Y. Azuma, M. Shinohara, and K. Ohura. 2000. Toll-like receptor 4-mediated signal pathway induced by Porphyromonas gingivalis lipopolysaccharide in human gingival fibroblasts. Biochem. Biophys. Res. Commun. 273: 1161–1167. [DOI] [PubMed] [Google Scholar]
- 38.Werts, C., R. I. Tapping, J. C. Mathison, T. H. Chuang, V. Kravchenko, I. Saint Girons, D. A. Haake, P. J. Godowski, F. Hayashi, A. Ozinsky, D. M. Underhill, C. J. Kirschning, H. Wagner, A. Aderem, P. S. Tobias, and R. J. Ulevitch. 2001. Leptospiral lipopolysaccharide activates cells through a TLR2-dependent mechanism. Nat. Immunol. 2: 346–352. [DOI] [PubMed] [Google Scholar]
- 39.Yang, R. B., M. R. Mark, A. Gray, A. Huang, M. H. Xie, M. Zhang, A. Goddard, W. I. Wood, A. L. Gurney, and P. J. Godowski. 1998. Toll-like receptor-2 mediates lipopolysaccharide-induced cellular signalling. Nature 395: 284–288. [DOI] [PubMed] [Google Scholar]
- 40.Yoshimura, A., Y. Hara, T. Kaneko, and I. Kato. 1997. Secretion of IL-1β, TNF-α, IL-8 and IL-1ra by human polymorphonuclear leukocytes in response to lipopolysaccharides from periodontopathic bacteria. J. Periodont. Res. 32: 279–286. [DOI] [PubMed] [Google Scholar]
- 41.Yoshimura, A., E. Lien, R. R. Ingalls, E. Tuomanen, R. Dziarski, and D. Golenbock. 1999. Cutting edge: recognition of Gram-positive bacterial cell wall components by the innate immune system occurs via Toll-like receptor 2. J. Immunol. 163: 1–5. [PubMed] [Google Scholar]