Abstract
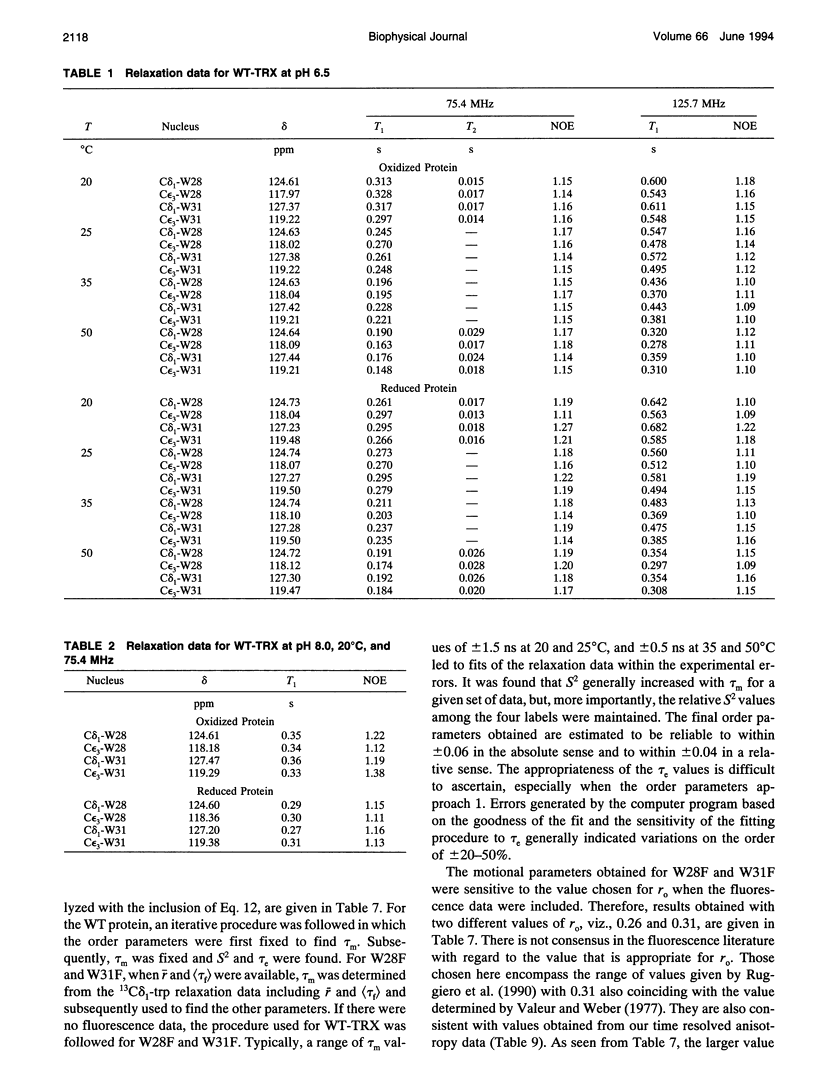
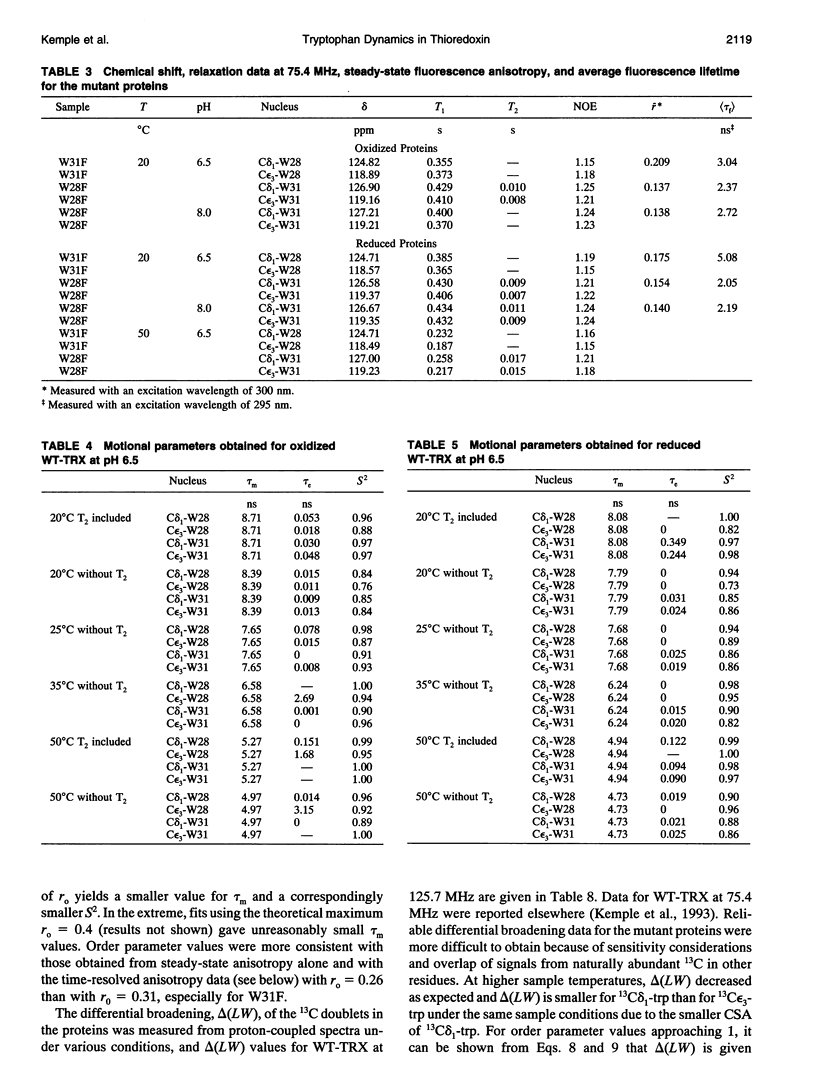
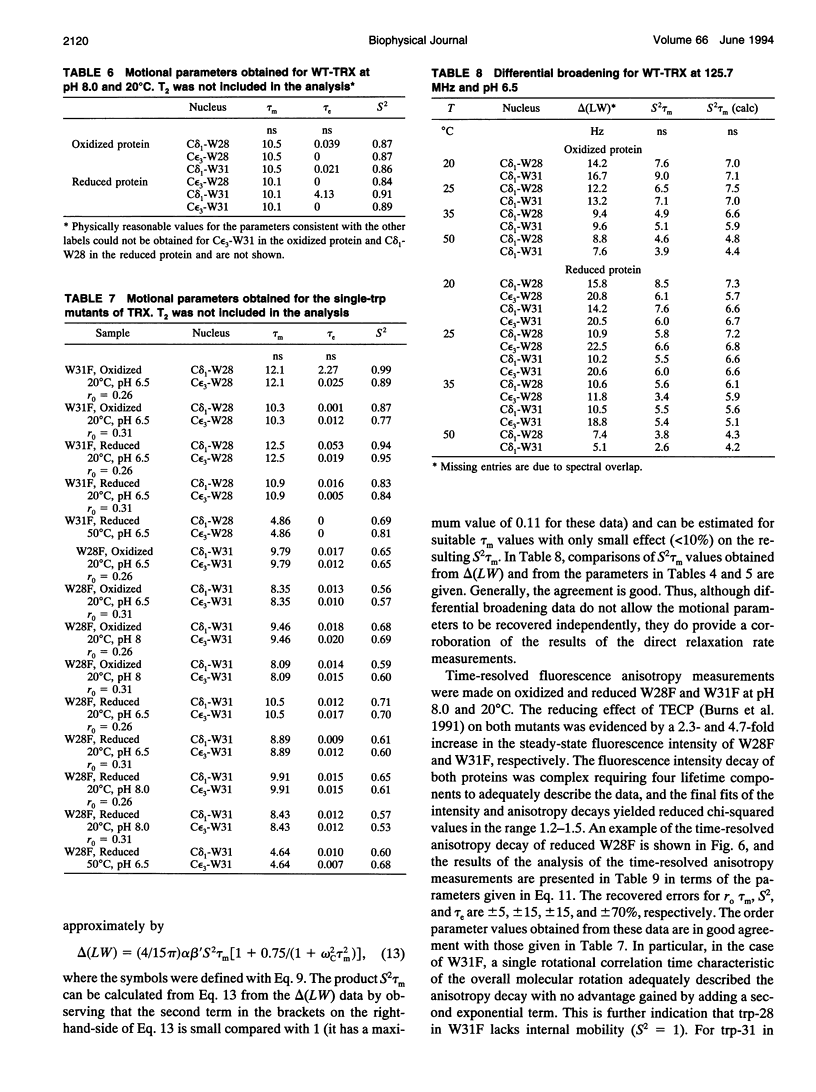
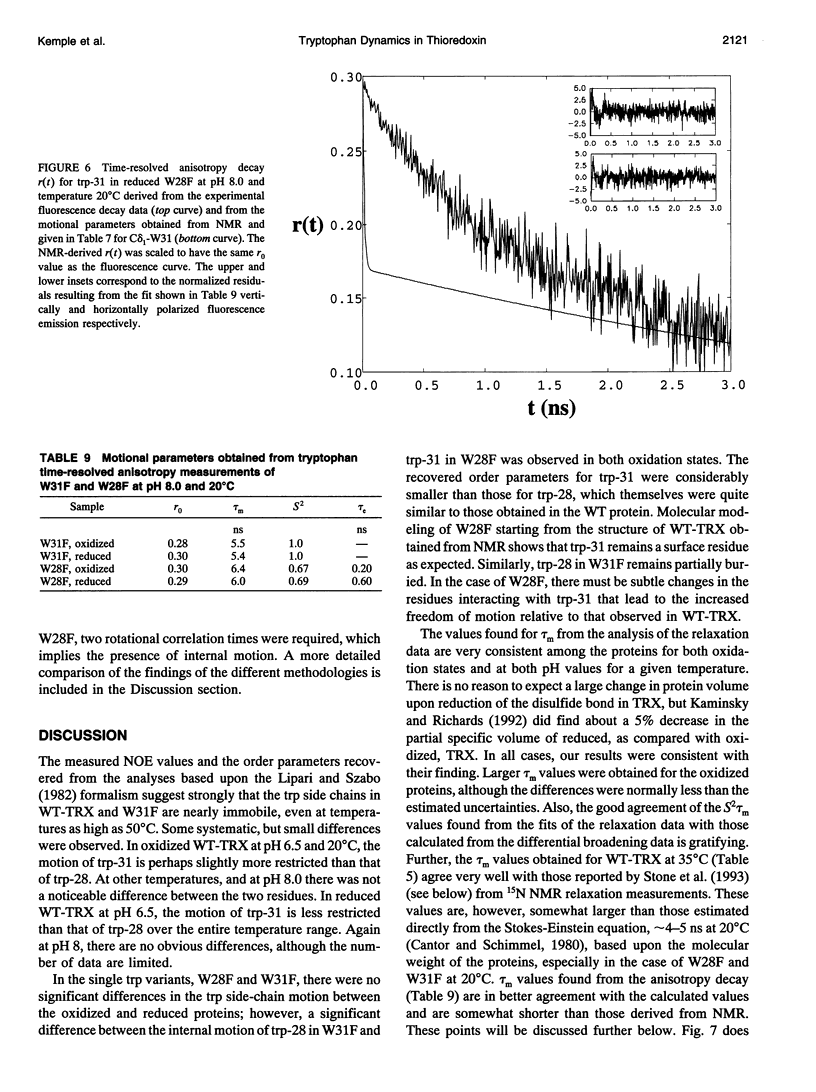
The rotational motion of tryptophan side chains in oxidized and reduced wild-type (WT) Escherichia coli thioredoxin and in two single-tryptophan variants of E. coli thioredoxin was studied in solution in the temperature range 20-50 degrees C from 13C-NMR relaxation rate measurements at 75.4 and 125.7 MHz and at 20 degrees C from steady-state and time-resolved trp fluorescence anisotropy measurements. Tryptophan enriched with 13C at the delta 1 and epsilon 3 sites of the indole ring was incorporated into WT thioredoxin and into two single-trp mutants, W31F and W28F, in which trp-28 or trp-31 of WT thioredoxin was replaced, respectively, with phenylalanine. The NMR relaxation data were interpreted using the Lipari and Szabo "model-free" approach (G. Lipari and A. Szabo. 1982. J. Amer. Chem. Soc. 104:4546-4559) with trp steady-state anisotropy data included for the variants at 20 degrees C. Values for the correlation time for the overall rotational motion (tau m) from NMR of oxidized and reduced WT thioredoxin at 35 degrees C agree well with those given by Stone et al. (Stone, M. J., K. Chandrasekhar, A. Holmgren, P. E. Wright, and H. J. Dyson. 1993. Biochemistry. 32:426-435) from 15N NMR relaxation rates, and the dependence of tau m on viscosity and temperature was in accord with the Stokes-Einstein relationship. Order parameters (S2) near 1 were obtained for the trp side chains in the WT proteins even at 50 degrees C. A slight increase in the amplitude of motion (decrease in S2) of trp-31, which is near the protein surface, but not of trp-28, which is partially buried in the protein matrix, was observed in reduced relative to oxidized WT thioredoxin. For trp-28 in W31F, order parameters near 1 (S2 > or = 0.8) at 20 degrees C were found, whereas trp-31 in W28F yielded the smallest order parameters (S2 approximately 0.6) of any of the cases. Analysis of time-resolved anisotropy decays in W28F and W31F yielded S2 values in good agreement with NMR, but gave tau m values about 60% smaller. Generally, values of tau e, the effective correlation time for the internal motion, were < or = 60 ps from NMR, whereas somewhat longer times were obtained from fluorescence. The ability of NMR and fluorescence techniques to detect subnanosecond motions in proteins reliably is examined.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Axelsen P. H., Gratton E., Prendergast F. G. Experimentally verifying molecular dynamics simulations through fluorescence anisotropy measurements. Biochemistry. 1991 Feb 5;30(5):1173–1179. doi: 10.1021/bi00219a002. [DOI] [PubMed] [Google Scholar]
- Axelsen P. H., Prendergast F. G. Molecular dynamics of tryptophan in ribonuclease-T1. II. Correlations with fluorescence. Biophys J. 1989 Jul;56(1):43–66. doi: 10.1016/S0006-3495(89)82651-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbato G., Ikura M., Kay L. E., Pastor R. W., Bax A. Backbone dynamics of calmodulin studied by 15N relaxation using inverse detected two-dimensional NMR spectroscopy: the central helix is flexible. Biochemistry. 1992 Jun 16;31(23):5269–5278. doi: 10.1021/bi00138a005. [DOI] [PubMed] [Google Scholar]
- Cheng J. W., Lepre C. A., Chambers S. P., Fulghum J. R., Thomson J. A., Moore J. M. 15N NMR relaxation studies of the FK506 binding protein: backbone dynamics of the uncomplexed receptor. Biochemistry. 1993 Sep 7;32(35):9000–9010. doi: 10.1021/bi00086a004. [DOI] [PubMed] [Google Scholar]
- Clore G. M., Driscoll P. C., Wingfield P. T., Gronenborn A. M. Analysis of the backbone dynamics of interleukin-1 beta using two-dimensional inverse detected heteronuclear 15N-1H NMR spectroscopy. Biochemistry. 1990 Aug 14;29(32):7387–7401. doi: 10.1021/bi00484a006. [DOI] [PubMed] [Google Scholar]
- Drapeau G. R., Brammar W. J., Yanofsky C. Amino acid replacements of the glutamic acid residue at position 48 in the tryptophan synthetase A protein of Escherichia coli. J Mol Biol. 1968 Jul 28;35(2):357–367. doi: 10.1016/s0022-2836(68)80030-0. [DOI] [PubMed] [Google Scholar]
- Dyson H. J., Gippert G. P., Case D. A., Holmgren A., Wright P. E. Three-dimensional solution structure of the reduced form of Escherichia coli thioredoxin determined by nuclear magnetic resonance spectroscopy. Biochemistry. 1990 May 1;29(17):4129–4136. doi: 10.1021/bi00469a016. [DOI] [PubMed] [Google Scholar]
- Dyson H. J., Holmgren A., Wright P. E. Assignment of the proton NMR spectrum of reduced and oxidized thioredoxin: sequence-specific assignments, secondary structure, and global fold. Biochemistry. 1989 Aug 22;28(17):7074–7087. doi: 10.1021/bi00443a044. [DOI] [PubMed] [Google Scholar]
- Eftink M. R., Ghiron C. A. Exposure of tryptophanyl residues and protein dynamics. Biochemistry. 1977 Dec 13;16(25):5546–5551. doi: 10.1021/bi00644a024. [DOI] [PubMed] [Google Scholar]
- Eftink M. R., Ghiron C. A., Kautz R. A., Fox R. O. Fluorescence lifetime studies with staphylococcal nuclease and its site-directed mutant. Test of the hypothesis that proline isomerism is the basis for nonexponential decays. Biophys J. 1989 Mar;55(3):575–579. doi: 10.1016/S0006-3495(89)82851-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elofsson A., Nilsson L. How consistent are molecular dynamics simulations? Comparing structure and dynamics in reduced and oxidized Escherichia coli thioredoxin. J Mol Biol. 1993 Oct 20;233(4):766–780. doi: 10.1006/jmbi.1993.1551. [DOI] [PubMed] [Google Scholar]
- Elofsson A., Rigler R., Nilsson L., Roslund J., Krause G., Holmgren A. Motion of aromatic side chains, picosecond fluorescence, and internal energy transfer in Escherichia coli thioredoxin studied by site-directed mutagenesis, time-resolved fluorescence spectroscopy, and molecular dynamics simulations. Biochemistry. 1991 Oct 8;30(40):9648–9656. doi: 10.1021/bi00104a012. [DOI] [PubMed] [Google Scholar]
- Holmgren A., Sjöberg B. M. Immunochemistry of thioredoxin. I. Preparation and cross-reactivity of antibodies against thioredoxin from Escherichia coli and bacteriophage T4. J Biol Chem. 1972 Jul 10;247(13):4160–4164. [PubMed] [Google Scholar]
- Holmgren A., Söderberg B. O., Eklund H., Brändén C. I. Three-dimensional structure of Escherichia coli thioredoxin-S2 to 2.8 A resolution. Proc Natl Acad Sci U S A. 1975 Jun;72(6):2305–2309. doi: 10.1073/pnas.72.6.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmgren A. Thioredoxin and glutaredoxin systems. J Biol Chem. 1989 Aug 25;264(24):13963–13966. [PubMed] [Google Scholar]
- Holmgren A. Thioredoxin. Annu Rev Biochem. 1985;54:237–271. doi: 10.1146/annurev.bi.54.070185.001321. [DOI] [PubMed] [Google Scholar]
- Holmgren A. Tryptophan fluorescence study of conformational transitions of the oxidized and reduced form of thioredoxin. J Biol Chem. 1972 Apr 10;247(7):1992–1998. [PubMed] [Google Scholar]
- Kaminsky S. M., Richards F. M. Reduction of thioredoxin significantly decreases its partial specific volume and adiabatic compressibility. Protein Sci. 1992 Jan;1(1):22–30. doi: 10.1002/pro.5560010104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katti S. K., LeMaster D. M., Eklund H. Crystal structure of thioredoxin from Escherichia coli at 1.68 A resolution. J Mol Biol. 1990 Mar 5;212(1):167–184. doi: 10.1016/0022-2836(90)90313-B. [DOI] [PubMed] [Google Scholar]
- Kinosita K., Jr, Kawato S., Ikegami A. A theory of fluorescence polarization decay in membranes. Biophys J. 1977 Dec;20(3):289–305. doi: 10.1016/S0006-3495(77)85550-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakowicz J. R., Maliwal B. P., Cherek H., Balter A. Rotational freedom of tryptophan residues in proteins and peptides. Biochemistry. 1983 Apr 12;22(8):1741–1752. doi: 10.1021/bi00277a001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langsetmo K., Fuchs J., Woodward C. Escherichia coli thioredoxin folds into two compact forms of different stability to urea denaturation. Biochemistry. 1989 Apr 18;28(8):3211–3220. doi: 10.1021/bi00434a015. [DOI] [PubMed] [Google Scholar]
- LeMaster D. M., Richards F. M. NMR sequential assignment of Escherichia coli thioredoxin utilizing random fractional deuteriation. Biochemistry. 1988 Jan 12;27(1):142–150. doi: 10.1021/bi00401a022. [DOI] [PubMed] [Google Scholar]
- Ludescher R. D., Volwerk J. J., de Haas G. H., Hudson B. S. Complex photophysics of the single tryptophan of porcine pancreatic phospholipase A2, its zymogen, and an enzyme/micelle complex. Biochemistry. 1985 Dec 3;24(25):7240–7249. doi: 10.1021/bi00346a033. [DOI] [PubMed] [Google Scholar]
- Matthews S. J., Jandu S. K., Leatherbarrow R. J. 13C NMR study of the effects of mutation on the tryptophan dynamics in chymotrypsin inhibitor 2: correlations with structure and stability. Biochemistry. 1993 Jan 19;32(2):657–662. doi: 10.1021/bi00053a034. [DOI] [PubMed] [Google Scholar]
- Mérola F., Rigler R., Holmgren A., Brochon J. C. Picosecond tryptophan fluorescence of thioredoxin: evidence for discrete species in slow exchange. Biochemistry. 1989 Apr 18;28(8):3383–3398. doi: 10.1021/bi00434a038. [DOI] [PubMed] [Google Scholar]
- Peng J. W., Wagner G. Mapping of the spectral densities of N-H bond motions in eglin c using heteronuclear relaxation experiments. Biochemistry. 1992 Sep 15;31(36):8571–8586. doi: 10.1021/bi00151a027. [DOI] [PubMed] [Google Scholar]
- Rance M., Sørensen O. W., Bodenhausen G., Wagner G., Ernst R. R., Wüthrich K. Improved spectral resolution in cosy 1H NMR spectra of proteins via double quantum filtering. Biochem Biophys Res Commun. 1983 Dec 16;117(2):479–485. doi: 10.1016/0006-291x(83)91225-1. [DOI] [PubMed] [Google Scholar]
- Redfield C., Boyd J., Smith L. J., Smith R. A., Dobson C. M. Loop mobility in a four-helix-bundle protein: 15N NMR relaxation measurements on human interleukin-4. Biochemistry. 1992 Nov 3;31(43):10431–10437. doi: 10.1021/bi00158a003. [DOI] [PubMed] [Google Scholar]
- Reutimann H., Straub B., Luisi P. L., Holmgren A. A conformational study of thioredoxin and its tryptic fragments. J Biol Chem. 1981 Jul 10;256(13):6796–6803. [PubMed] [Google Scholar]
- Ross J. B., Schmidt C. J., Brand L. Time-resolved fluorescence of the two tryptophans in horse liver alcohol dehydrogenase. Biochemistry. 1981 Jul 21;20(15):4369–4377. doi: 10.1021/bi00518a021. [DOI] [PubMed] [Google Scholar]
- Schneider D. M., Dellwo M. J., Wand A. J. Fast internal main-chain dynamics of human ubiquitin. Biochemistry. 1992 Apr 14;31(14):3645–3652. doi: 10.1021/bi00129a013. [DOI] [PubMed] [Google Scholar]
- Stone M. J., Chandrasekhar K., Holmgren A., Wright P. E., Dyson H. J. Comparison of backbone and tryptophan side-chain dynamics of reduced and oxidized Escherichia coli thioredoxin using 15N NMR relaxation measurements. Biochemistry. 1993 Jan 19;32(2):426–435. doi: 10.1021/bi00053a007. [DOI] [PubMed] [Google Scholar]
- Valeur B., Weber G. Resolution of the fluorescence excitation spectrum of indole into the 1La and 1Lb excitation bands. Photochem Photobiol. 1977 May;25(5):441–444. doi: 10.1111/j.1751-1097.1977.tb09168.x. [DOI] [PubMed] [Google Scholar]
- Weaver A. J., Kemple M. D., Brauner J. W., Mendelsohn R., Prendergast F. G. Fluorescence, CD, attenuated total reflectance (ATR) FTIR, and 13C NMR characterization of the structure and dynamics of synthetic melittin and melittin analogues in lipid environments. Biochemistry. 1992 Feb 11;31(5):1301–1313. doi: 10.1021/bi00120a005. [DOI] [PubMed] [Google Scholar]
- Weaver A. J., Kemple M. D., Prendergast F. G. Fluorescence and 13C NMR determination of side-chain and backbone dynamics of synthetic melittin and melittin analogues in isotropic solvents. Biochemistry. 1989 Oct 17;28(21):8624–8639. doi: 10.1021/bi00447a053. [DOI] [PubMed] [Google Scholar]
- Weaver A. J., Kemple M. D., Prendergast F. G. Tryptophan sidechain dynamics in hydrophobic oligopeptides determined by use of 13C nuclear magnetic resonance spectroscopy. Biophys J. 1988 Jul;54(1):1–15. doi: 10.1016/S0006-3495(88)82925-4. [DOI] [PMC free article] [PubMed] [Google Scholar]



