Lolle and colleagues observed frequent true reversion of hothead (hth) mutations of Arabidopsis thaliana, whereby up to 10% of the progeny of self-pollinated homozygous hth/hth mutants carried a grandparental HTH allele (Lolle et al., 2005). Instability was not limited to the HTH gene: hth mutants displayed conversion of genome-wide polymorphisms to the genotype of a recent ancestor. The possibility that a mutator phenotype caused indiscriminate changes, thereby increasing the reversion rate of hth, was discounted because reverted HTH alleles showed no additional nucleotide changes, whereas 50 to 100 changes per sequenced allele would be expected from the ∼5% hth reversion rate. These genetic instabilities were interpreted as a memory of a past genotypic state. A cache of double-stranded RNA from the previous genotype was proposed, which, at times of stress corrects mutated sites in a template-directed manner. The existence of an alternate reserve genome appears consistent with the data and would necessitate a revision of our theories of heredity. Two recent commentaries on this discovery suggested that, instead of an RNA cache, the hth-correcting mechanism depends on ectopic gene conversion (Chaudhury, 2005) or on an elusive DNA cache (Ray, 2005). Thus, all explanations offered until now invoke novel genetic phenomena. We offer an alternative explanation based on established genetic and evolutionary principles. HTH could be a metabolic enzyme whose impairment causes the accumulation of toxic and mutagenic compounds that act at random DNA sites. The observed high reversion rate of hth could result from selection of rare revertant cells, such as pollen and meristem cells. The frequency of changes at other genomic sites would be overestimated by restriction enzyme analysis of DNA from chimeric tissue. Consider the following information.
HTH deficiency could result in accumulation of secondary metabolites that are toxic and mutagenic. The HTH gene encodes a product whose closest characterized relative is an MBC oxidoreductase that hydrolyzes the aromatic mandelonitrile in stone fruits (Zheng and Poulton, 1995) (Figure 1A). Arabidopsis has complex metabolic pathways for the production of glucosinolates (Wittstock and Halkier, 2002; Brown et al., 2003), some of which are aromatic and resemble mandelonitrile (Figure 1), and aromatic glucosinolates and their derivatives are mutagenic (Musk et al., 1995; Kassie et al., 1999; Kassie and Knasmuller, 2000; Canistro et al., 2004). The sticky surface properties of the HTH phenotype, which suggest alterations of cuticle, cell wall, and perhaps membranes, might also be an effect of the toxic mutator.
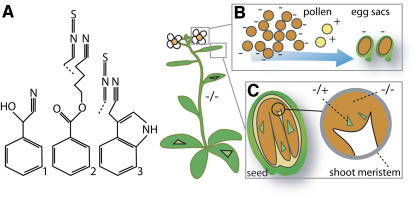
Figure 1.
Selection and Chimerism.
(A) Mandelonitrile (1), the substrate of the HTH homolog mandelonitrile lyase, and nitriles and isothiocyanates (2 and 3) produced by hydrolysis of seed glucosinolates. Nitriles or isothiocyanates and their derivatives could be the toxic and mutagenic compounds that accumulate in Arabidopsis hth mutants (−/−) in the toxic mutator hypothesis.
(B) Rare revertant pollen (+) outcompetes mutant pollen (−) during self-fertilization, resulting in frequent +/− heterozygotes.
(C) Revertant sectors are formed in the seed, meristem, and adult plant body.
If an alternative genetic reservoir exists and it restores mutations to the previous genotype, it should be revised gradually so that its memory should fade with cell divisions. Yet, data that addresses the reversion rate are available in a previous article by the same group and are inconsistent with the above expectation (Lolle et al., 1998). In complementation tests between hth alleles, wild-type plants were observed among the progeny of trans-heterozygotes and were thought to result from intragenic recombination. These were almost certainly revertants and not recombinants. The hth-10/hth-8 trans-heterozygote produced 30 wild types among 1858 individuals, whereas hth-10/hth-4 produced only mutant progeny (n = 1629) (Lolle et al., 1998; Krolikowski et al., 2003). The same alleles, hth-10 and hth-4, were reported to revert in the 2005 article and to do so through multiple generations (Lolle et al., 2005). Thus, the rate of reversion increased from 1998 to 2005 instead of decreasing.
Selection at multiple stages of development could bias the measured reversion rate. For example, several thousand pollen compete for 20 to 40 eggs. Rare HTH pollen could outcompete the preponderant hth pollen (containing the toxic metabolite), resulting in the enrichment of hth/HTH zygotes because the reversion rate was much higher for pollen than for eggs (Figure 1B). Our selection hypothesis appears inconsistent with the reported 3:1 progeny ratio from selfed heterozygotes because selection in favor of HTH pollen should decrease the fraction of hth/hth progeny, but HTH could have a paternal effect where an hth/HTH heterozygous anther provides its hth pollen with sufficient HTH enzyme to avoid a crisis. An hth/hth anther would provide no enzyme. Selection favoring revertant cells could also act in the shoot apical meristem, in the flower meristem, and in the anther primordia. In meristem tissues, selection could be influenced by the defective cellular surface properties of hth mutants that lead to postgenital organ fusion (Lolle et al., 1998). Selection could also act on seed, decreasing germination of hth/hth zygotes in favor of HTH/hth individuals.
The precise reversion of hth mutations without the appearance of secondary mutations is consistent with a template-driven repair but also with the alternative hypothesis of mutagenesis and selection. As explained above, any significant mutation rate coupled to selection can yield frequent revertants, and the probability of finding secondary mutations at HTH revertant alleles is proportional to the mutation rate. In fact, the highest mutation rate compatible with survival is likely to be significantly lower than the rates that would generate frequent concurrent mutations on the HTH gene. We know from TILLING experiments the genomic consequences of moderately intense mutagenesis: in Arabidopsis, it produces an average of one mutation for every 170 kb (Greene et al., 2003). Increasing the mutagenesis rate has progressively deleterious consequences on viability (S.H. Reynolds, B. Till, and L. Comai, unpublished results), and we estimate that a mutation rate 10 times higher (an average of one mutation every 17 kb) may not be compatible with growth of this diploid organism. Even this higher mutation density would be unlikely to result in frequent additional mutations in the gene fragment surveyed by Lolle et al. Furthermore, selection would tend to favor early revertants because they are less likely to have accumulated deleterious mutations in the genome. This, in turn, would favor revertants with little to no secondary mutations. Revertants would also have a lower mutation rate, making the rate of postreversion mutations lower than that of prereversion mutations. Therefore, it would be unlikely that any random mutagenic treatment in a living diploid organism could produce frequent concurrent mutations on the HTH gene.
If mutagenesis is random, why are second site intragenic suppressors not found? Different proteins display different responses to selection for mutant reversion. With some proteins, precise reversion of deleterious mutations is the predominant response when strong selection for restoration of protein activities is applied (Rossiter et al., 1990; Coombs et al., 1994; Gee et al., 1994; Wang et al., 2004). It is possible that the HTH protein may belong to this category and that its activity may be difficult to restore by secondary intragenic changes.
A seemingly compelling argument against selection is the appearance of revertants at presumably unselected polymorphic loci at the rate of 1 to 4%. This measured rate could be higher than the real per nucleotide reversion rate for two reasons. The genotype at these sites was scored by analyzing restriction endonuclease digests. Thus, multiple nucleotide mutations can change a cut site to a no-cut site. Changes from no-cut to cut must entail a true reversion. Since both were scored as equivalent changes, it is possible that changes of the first type were preponderant, and the true reversion rate per nucleotide could be lower than 1 to 4%. A second factor that could inflate the observed reversion rate is chimerism (Figure 1C). This possibility, resulting from somatic DNA changes, is acknowledged by the authors and supported by the higher rate of DNA changes measured in developing seeds (Lolle et al., 2005). DNA from chimeric tissue can be scored as heterozygous by PCR analysis, but somatic rather than germinal change would imply a lower mutation rate than the reported rate.
Our mutator hypothesis may appear complicated, but it comports with the data. Aromatic glucosinolates are abundant in Arabidopsis seeds and flowers (Wittstock and Halkier, 2002; Brown et al., 2003), and a block in their metabolism could lead to toxicity and mutagenicity (Musk et al., 1995; Kassie et al., 1999; Kassie and Knasmuller, 2000; Canistro et al., 2004) and to genomic instability (Henikoff, 2005). The delayed and progressive onset of reversion could arise from suppressor mutations that ameliorate the toxic effect. Such mutations could affect DNA repair proteins (Karran and Bignami, 1994), increasing the mutation rate. Testing the toxic mutator hypothesis would be simple: sequencing of heritable polymorphic changes observed at various loci should reveal whether the changes are template directed (i.e., reflect the ancestral genotype) or random and without reference to the ancestry. The mutator potential of hth mutants could be assessed by TILLING (Till et al., 2003) and should be associated with novel mutant phenotypes. Selection in favor of HTH pollen could be tested in pollen mixing experiments. Suppressor mutations at other loci could be distinguished from effects derived from the HTH locus by mapping.
In conclusion, there are two components to this hypothesis for hth reversion: a mechanism that generates variation (pollen and seed of hth mutants are steeped in a self-made mutagen) and a mechanism that amplifies the variation (selection for rare reverted cells increases the observed number of reversion events). This and other explanations (Chaudhury, 2005; Ray, 2005) offered for this phenomenon should help in designing experiments that elucidate its nature.
Acknowledgments
We are thankful to several colleagues for comments and particularly to Arnold Bendich for encouragement and editorial suggestions.
References
- Brown, P.D., Tokuhisa, J.G., Reichelt, M., and Gershenzon, J. (2003). Variation of glucosinolate accumulation among different organs and developmental stages of Arabidopsis thaliana. Phytochemistry 62, 471–481. [DOI] [PubMed] [Google Scholar]
- Canistro, D., Croce, C.D., Iori, R., Barillari, J., Bronzetti, G., Poi, G., Cini, M., Caltavuturo, L., Perocco, P., and Paolini, M. (2004). Genetic and metabolic effects of gluconasturtiin, a glucosinolate derived from cruciferae. Mutat. Res. 545, 23–35. [DOI] [PubMed] [Google Scholar]
- Chaudhury, A. (2005). Hothead healer and extragenomic information. Nature 437, E1–E2. [DOI] [PubMed] [Google Scholar]
- Coombs, K.M., Mak, S.C., and Petrycky-Cox, L.D. (1994). Studies of the major reovirus core protein sigma 2: Reversion of the assembly-defective mutant tsC447 is an intragenic process and involves back mutation of Asp-383 to Asn. J. Virol. 68, 177–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gee, P., Maron, D.M., and Ames, B.N. (1994). Detection and classification of mutagens: A set of base-specific Salmonella tester strains. Proc. Natl. Acad. Sci. USA 91, 11606–11610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene, E.A., Codomo, C.A., Taylor, N.E., Henikoff, J.G., Till, B.J., Reynolds, S.H., Enns, L.C., Burtner, C., Johnson, J.E., Odden, A.R., Comai, L., and Henikoff, S. (2003). Spectrum of chemically induced mutations from a large-scale reverse-genetic screen in Arabidopsis. Genetics 164, 731–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff, S. (2005). Rapid changes in plant genomes. Plant Cell 17, 2852–2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karran, P., and Bignami, M. (1994). DNA damage tolerance, mismatch repair and genome instability. Bioessays 16, 833–839. [DOI] [PubMed] [Google Scholar]
- Kassie, F., and Knasmuller, S. (2000). Genotoxic effects of allyl isothiocyanate (AITC) and phenethyl isothiocyanate (PEITC). Chem. Biol. Interact. 127, 163–180. [DOI] [PubMed] [Google Scholar]
- Kassie, F., Pool-Zobel, B., Parzefall, W., and Knasmuller, S. (1999). Genotoxic effects of benzyl isothiocyanate, a natural chemopreventive agent. Mutagenesis 14, 595–604. [DOI] [PubMed] [Google Scholar]
- Krolikowski, K.A., Victor, J.L., Wagler, T.N., Lolle, S.J., and Pruitt, R.E. (2003). Isolation and characterization of the Arabidopsis organ fusion gene HOTHEAD. Plant J. 35, 501–511. [DOI] [PubMed] [Google Scholar]
- Lolle, S.J., Hsu, W., and Pruitt, R.E. (1998). Genetic analysis of organ fusion in Arabidopsis thaliana. Genetics 149, 607–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lolle, S.J., Victor, J.L., Young, J.M., and Pruitt, R.E. (2005). Genome-wide non-mendelian inheritance of extra-genomic information in Arabidopsis. Nature 434, 505–509. [DOI] [PubMed] [Google Scholar]
- Musk, S.R., Smith, T.K., and Johnson, I.T. (1995). On the cytotoxicity and genotoxicity of allyl and phenethyl isothiocyanates and their parent glucosinolates sinigrin and gluconasturtiin. Mutat. Res. 348, 19–23. [DOI] [PubMed] [Google Scholar]
- Ray, A. (2005). RNA cache or genome trash? Nature 437, E1–E2. [DOI] [PubMed] [Google Scholar]
- Rossiter, B.J., Muzny, D.M., Hampson, I., Caskey, C.T., and Fox, M. (1990). Induced reversion of a spontaneous point mutation within the Chinese hamster HPRT gene to the wild-type sequence. Mutagenesis 5, 605–608. [DOI] [PubMed] [Google Scholar]
- Till, B.J., et al. (2003). Large-scale discovery of induced point mutations with high-throughput TILLING. Genome Res. 13, 524–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H.H., Yu, H.H., and Wong, S.M. (2004). Mutation of Phe50 to Ser50 in the 126/183-kDa proteins of Odontoglossum ringspot virus abolishes virus replication but can be complemented and restored by exact reversion. J. Gen. Virol. 85, 2447–2457. [DOI] [PubMed] [Google Scholar]
- Wittstock, U., and Halkier, B.A. (2002). Glucosinolate research in the Arabidopsis era. Trends Plant Sci. 7, 263–270. [DOI] [PubMed] [Google Scholar]
- Zheng, L., and Poulton, J.E. (1995). Temporal and spatial expression of amygdalin hydrolase and (R)-(+)-mandelonitrile lyase in black cherry seeds. Plant Physiol. 109, 31–39. [DOI] [PMC free article] [PubMed] [Google Scholar]