Abstract
Expression of the viral silencing suppressor P1/HC-Pro in plants causes severe developmental anomalies accompanied by defects in both short interfering RNA (siRNA) and microRNA (miRNA) pathways. P1/HC-Pro transgenic lines fail to accumulate the siRNAs that mediate RNA silencing and are impaired in both miRNA processing and function, accumulating abnormally high levels of miRNA/miRNA* processing intermediates as well as miRNA target messages. Both miRNA and RNA silencing pathways require participation of DICER-LIKE (DCL) ribonuclease III-like enzymes. Here, we investigate the effects of overexpressing DCL1, one of four Dicers in Arabidopsis thaliana, on P1/HC-Pro–induced defects in development and small RNA metabolism. Expression of a DCL1 cDNA transgene (35S:DCL1) produced a mild gain-of-function phenotype and largely rescued dcl1 mutant phenotypes. The 35S:DCL1 plants were competent for virus-induced RNA silencing but were impaired in transgene-induced RNA silencing and in the accumulation of some miRNAs. Ectopic DCL1 largely alleviated developmental anomalies in P1/HC-Pro plants but did not correct the P1/HC-Pro–associated defects in small RNA pathways. The ability of P1/HC-Pro plants to suppress RNA silencing and the levels of miRNAs, miRNA*s, and miRNA target messages in these plants were essentially unaffected by ectopic DCL1. These data suggest that P1/HC-Pro defects in development do not result from general impairments in small RNA pathways and raise the possibility that DCL1 participates in processes in addition to miRNA biogenesis.
INTRODUCTION
Small regulatory RNAs play crucial roles in a variety of genetic regulatory processes in a broad range of eukaryotes, including plants (recently reviewed in Hunter and Poethig, 2003; Baulcombe, 2004; Dugas and Bartel, 2004; Mallory and Vaucheret, 2004). One class of small regulatory RNAs, the short interfering RNAs (siRNAs), are produced in the RNA silencing pathway and serve as a defense against invading nucleic acids, such as viruses, transposons, and transgenes (Vance and Vaucheret, 2001). Such invaders trigger silencing by producing long fully double-stranded RNAs (dsRNA) that are cleaved into siRNAs by a ribonuclease III-like enzyme called Dicer. The siRNAs then act as guides in an endonuclease complex called RNA-induced silencing complex (RISC) that destroys any RNA having high homology to the dsRNA trigger. Consistent with the idea that RNA silencing is an antiviral mechanism, many plant viruses encode proteins that interfere with the process at one or more steps (reviewed in Roth et al., 2004; Silhavy and Burgyan, 2004).
Another class of small regulatory RNAs, the microRNAs (miRNAs), is produced from endogenous cellular genes and has been implicated as key developmental regulators. They arise from nonprotein encoding precursor RNAs at one locus and act in trans to negatively regulate the expression of a target mRNA from another locus. Similar to siRNAs, miRNAs are produced by the activity of a Dicer and then are incorporated into a RISC-like complex to act as guides to direct the complex to specific mRNA targets (reviewed in Ambros, 2004; Bartel, 2004; Dugas and Bartel, 2004; Mallory and Vaucheret, 2004). In contrast with the majority of animal miRNAs, most plant miRNAs direct the cleavage of target mRNAs (Llave et al., 2002; Kasschau et al., 2003; Palatnik et al., 2003; Xie et al., 2003), though at least some act primarily by inhibiting translation of the target mRNA (Aukerman and Sakai, 2003; Chen, 2004). Interestingly, miRNA-directed pathways in plants share a number of common biochemical features and genetic requirements with those directed by siRNAs. The integration of these two pathways can be seen in the biogenesis of a third class of small RNAs termed trans-acting siRNAs. These small RNAs arise after miRNA-directed cleavage of a precursor RNA that is subsequently copied into dsRNA by the activity of an RNA-dependent RNA polymerase that is also required for production of the siRNAs that mediate sense transgene–induced RNA silencing (Peragine et al., 2004; Vazquez et al., 2004; Allen et al., 2005; Williams et al., 2005). Perhaps reflecting the shared features of the small RNA pathways, many of the plant viral proteins that suppress RNA silencing also affect miRNA biogenesis and/or function and alter plant development (Mallory et al., 2002b; Kasschau et al., 2003; Chapman et al., 2004; Dunoyer et al., 2004; Roth et al., 2004; Silhavy and Burgyan, 2004).
The Dicers that mediate small RNA biogenesis are complex multifunctional proteins that contain an RNA helicase domain and usually a PAZ RNA binding domain in addition to the ribonuclease and dsRNA binding domains found in canonical RNase IIIs (Figure 3A; Schauer et al., 2002; Song et al., 2003). In addition to cleaving substrates to produce small RNAs, Dicers play a role in early steps in miRNA precursor processing (Kurihara and Watanabe, 2004) and are required for the assembly of small RNAs into the RISC complex (Lee et al., 2004; Pham et al., 2004). Many animals have a single Dicer that mediates all the required processing events for both siRNAs and miRNAs (Grishok et al., 2001; Ketting et al., 2001; Knight and Bass, 2001), whereas Drosophila melanogaster has two Dicers that are functionally diverged, one required primarily for siRNA production and the other primarily for miRNA production (Lee et al., 2004; Pham et al., 2004; Tijsterman and Plasterk, 2004). The two Dicers in the fungi Neurospora crassa are redundant for transgene-induced RNA silencing (Catalanotto et al., 2004), whereas only one of the two Dicers in the fungus Magnaporthe oryzae is required for this process (Kadotani et al., 2004).
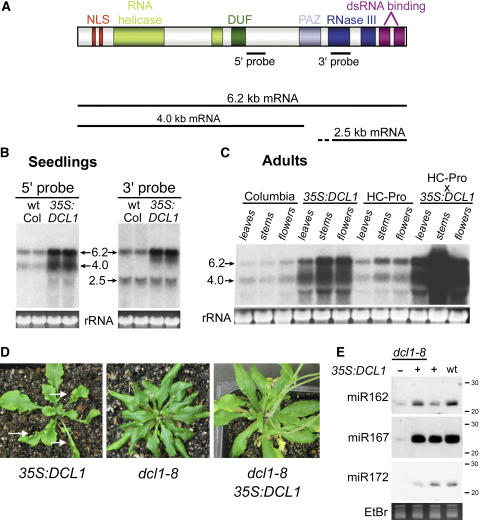
Figure 3.
Overexpression of DCL1 mRNA Largely Complements the dcl1-8 Mutation.
(A) The predicted domains of the DCL1 protein from Schauer et al. (2002). NLS is the nuclear localization signal, and DUF refers to domain of unknown function 283. The location of 5′ and 3′ probes used in (B) and (C) and the sequence content of the major DCL1 transcripts are indicated. The dashed line at the beginning of the 2.5-kb RNA represents the known heterogeneity in the 5′ end of this transcript containing intron 14 sequences.
(B) 35S:DCL1 seedlings accumulate high levels of DCL1 mRNA. RNA gel blot analysis of RNA isolated from 10-d seedlings of wild-type plants or plants homozygous for 35S:DCL1 transgene (line 12), as indicated, using the 5′ or 3′ hybridization probes indicated in (A). Arrows indicate the location of the 6.2-kb full-length DCL1 transcript as well as the 4.0- and 2.5-kb smaller DCL1 transcripts. Ethidium bromide staining of 25S rRNA is shown as a loading control.
(C) Adult 35S:DCL1 plants accumulate high levels of DCL1 mRNA. RNA gel blot analysis of RNA from rosette leaves, stems, or flowers of adult wild-type plants, 35S:DCL1 transgenic plants, P1/HC-Pro transgenic plants, and P1/HC-Pro × 35S:DCL1 plants, as indicated, using the 5′ hybridization probe indicated in (A). The location of the 6.2-kb full-length and the 4.0-kb DCL1 transcripts is indicated. Ethidium bromide staining of 25S rRNA is shown as a loading control.
(D) Rosette leaf morphology is affected by altered levels of DCL1 activity. 35S:DCL1 Arabidopsis plants sometimes show abaxialized (curled up) rosette leaves as shown in the left picture, in contrast with dcl1-8 mutant rosette leaves, which are adaxialized (curled under) as shown in the middle picture. The dcl1-8 adaxialized rosette leaf phenotype is largely complemented in plants expressing the 35S:DCL1 transgene (right picture). Arrows indicate adaxialized leaves.
(E) RNA gel blot showing the levels of the indicated miRNAs in rosette leaves of young dcl1-8 rosette leaves in the absence (−) or presence (+) of the 35S:DCL1 transgene as compared with those in the equivalent tissues of 35S:DCL1 plants or wild-type plants. The location of molecular weight RNA markers of 20 and 30 nucleotides are indicated to the right of the figure. Ethidium bromide (EtBr) staining of the predominant RNA species in the low molecular weight fraction is shown as a loading control.
The Arabidopsis thaliana genome contains four putative DICER-LIKE genes (DCL1-DCL4; Schauer et al., 2002), and the exact roles of these enzymes are still being determined. Their function in small RNA biogenesis has been addressed via mutant analysis, using the weak loss-of-function dcl1-7 and dcl1-9 alleles and the null dcl2-1, dcl3-1, and dcl4-1 insertion alleles (dcl1 null alleles are embryo lethal) (Lang et al., 1994; Schwartz et al., 1994; Ray et al., 1996; Jacobsen et al., 1999; Golden et al., 2002; Xie et al., 2004, 2005; Gasciolli et al., 2005; Yoshikawa et al., 2005). The weak dcl1 alleles are defective in at least two steps of miRNA precursor processing (Kurihara and Watanabe, 2004), accumulate reduced levels of most mature miRNAs (Park et al., 2002; Reinhart et al., 2002), and have severe developmental defects. By contrast, dcl2-1, dcl3-1, and dcl4-1 null alleles have no reported defects in miRNA accumulation and exhibit mild or no developmental anomalies (Xie et al., 2004; Gasciolli et al., 2005). The roles of the other Arabidopsis DCL genes in RNA silencing are less well understood. The dcl2-1 null allele causes a transient decrease in siRNA accumulation in response to infection with Turnip crinkle virus but not to either Turnip mosaic virus (TuMV) or Cucumber mosaic virus (Xie et al., 2004), suggesting that it plays a role in at least some types of virus-induced RNA silencing. DCL3 has been implicated in production of a class of larger small RNAs that mediate chromatin modifications associated with transcriptional gene silencing (Chan et al., 2004; Xie et al., 2004). DCL4 is required for the accumulation of trans-acting siRNAs (Gasciolli et al., 2005; Xie et al., 2005; Yoshikawa et al., 2005). However, the Dicer(s) that produces the siRNAs that direct transgene-induced RNA silencing has not yet been identified. While dcl1-9 mutant plants were shown to be competent for silencing induced by a hairpin transgene (Finnegan et al., 2003), this finding does not eliminate a role for DCL1 in transgene-induced RNA silencing since a null allele could not be used. The other DCL genes have not yet been tested to determine if they play a role in transgene-induced RNA silencing.
P1/HC-Pro (for proteinase1/helper component-proteinase) is a plant viral protein that blocks both virus-induced RNA silencing and transgene-induced RNA silencing (Anandalakshmi et al., 1998; Brigneti et al., 1998; Kasschau and Carrington, 1998; Llave et al., 2000; Mallory et al., 2001; Dunoyer et al., 2004) but not transcriptional gene silencing (Marathe et al., 2000; Mette et al., 2001). In tobacco (Nicotiana tabacum), suppression of sense transgene–induced silencing by HC-Pro completely eliminates detectable levels of 21- to 24-nucleotide siRNAs (Mallory et al., 2001). The suppression of inverted repeat transgene silencing by HC-Pro also blocks the accumulation of the 21- to 24-nucleotide siRNA but allows accumulation of longer 25- 26-nucleotide small RNAs as well as long dsRNAs (Mallory et al., 2002b). Similarly, long dsRNA accumulates in transgenic Arabidopsis lines suppressed by HC-Pro for hairpin transgene-induced silencing (Dunoyer et al., 2004). The reduction in short siRNAs, which are the products of Dicer, accompanied by the simultaneous accumulation of long dsRNA, which is the substrate of Dicer, suggests that HC-Pro alters the activity of the enzyme that produces siRNAs from long dsRNA substrates.
Expression of P1/HC-Pro also impairs miRNA biogenesis and function. In contrast with siRNAs, miRNA levels are enhanced in P1/HC-Pro transgenic lines (Mallory et al., 2002b; Kasschau et al., 2003; Chapman et al., 2004; Dunoyer et al., 2004). Paradoxically, some miRNA target messages are also increased, suggesting that these miRNAs are not functional (Kasschau et al., 2003; Dunoyer et al., 2004). P1/HC-Pro lines also accumulate miRNA* species, small RNAs largely complementary to the mature miRNA sequence that are thought to arise as a labile intermediate in the processing of the miRNA precursor (Chapman et al., 2004; Dunoyer et al., 2004). These results suggest that P1/HC-Pro impairs strand separation of an intermediate miRNA/miRNA* duplex and/or the subsequent transfer of the miRNA strand into the RISC complex, thereby inhibiting miRNA-directed target cleavage. Because assembly of miRNA into a functional RISC requires Dicer activity in at least some organisms (Lee et al., 2004; Pham et al., 2004), these observations are consistent with the hypothesis that P1/HC-Pro interferes with miRNA-directed mRNA degradation at a Dicer-mediated step.
The developmental phenotypes of P1/HC-Pro plants have been ascribed to the inhibition of miRNA-directed target degradation (Kasschau et al., 2003; Xie et al., 2003; Chapman et al., 2004; Dunoyer et al., 2004). Supporting this view is the observation that mutations in genes involved in miRNA biogenesis (such as DCL1, HUA ENHANCER1, HYPONASTIC LEAVES1, and ARGONAUTE1) partially phenocopy P1/HC-Pro lines (Ray et al., 1996; Bohmert et al., 1998; Jacobsen et al., 1999; Lu and Fedoroff, 2000; Chen et al., 2002; Golden et al., 2002). However, because P1/HC-Pro transgenic Arabidopsis lines are additionally impaired in RNA silencing (Dunoyer et al., 2004; this article), it remains possible that RNA silencing also plays an important, yet unappreciated, role in the regulation of developmental pathways. Alternatively, some of the effects of P1/HC-Pro on development may reflect an influence on factors that function independently of small RNA metabolism.
Here, we test the hypothesis that P1/HC-Pro interferes with small RNA metabolism and normal development by altering the activity of one or more Dicers. Ectopic overexpression of DCL1 would be expected to overcome P1/HC-Pro–mediated defects that arise due to interference with DCL1 function and perhaps to compensate for P1/HC-Pro interference with the other Dicers as well. Indeed, we find that overexpression of DCL1 is sufficient to relieve the developmental anomalies in P1/HC-Pro plants but surprisingly does not relieve the impairments in the biogenesis of small RNAs or those in small RNA-directed RNA degradation pathways in those plants. These results suggest that developmental defects in the P1/HC-Pro plants arise due to aberrant Dicer activity and raise the possibility that Dicers play an important role in development that is independent of target RNA degradation via small RNA pathways.
RESULTS
Effect of P1/HC-Pro Expression on Phenotype in Arabidopsis
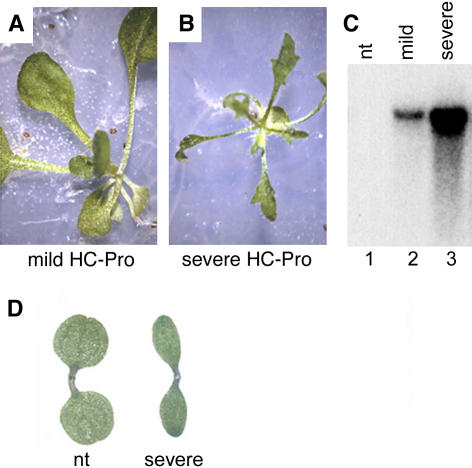
Developmental abnormalities have been reported previously in both tobacco and Arabidopsis transgenic lines expressing P1/HC-Pro, and these have been attributed to defects in small RNA pathways (Anandalakshmi et al., 2000; Mallory et al., 2002a; Kasschau et al., 2003; Chapman et al., 2004; Dunoyer et al., 2004). To further investigate the nature of the defects caused by P1/HC-Pro, we generated Arabidopsis transgenic lines expressing the 5′ proximal region of the TuMV genome encoding the P1 and HC-Pro proteins. Analysis of TuMV P1/HC-Pro primary transformants indicated a range of severity in developmental phenotypes, which are first detectable after germination, with a positive correlation between the degree of phenotypic abnormality and the level of P1/HC-Pro expression (Figure 1). Thus, the primary transformants with a severe phenotype accumulated higher levels of P1/HC-Pro mRNA than transformants with a much weaker phenotype (Figures 1A to 1C). The most severely affected P1/HC-Pro transgenic lines had narrow cotyledons and thus could be distinguished from the wild type at germination (Figure 1D).
Figure 1.
The Severity of P1/HC-Pro Phenotype in Primary Transformants Correlates with P1/HC-Pro mRNA Accumulation.
(A) The mild phenotype exhibited by a subset of P1/HC-Pro primary transformants in Arabidopsis.
(B) The severe phenotype exhibited by the majority of P1/HC-Pro primary transformants in Arabidopsis.
(C) RNA gel blot analysis showing the levels of P1/HC-Pro mRNA in total RNA from a pool of individual primary transformants. The level of P1/HC-Pro mRNA is higher in the pool of plants displaying a severe phenotype as shown in (B) (lane 3) than that in the pool of plants displaying a mild phenotype as shown in (A) (lane 2). Lane 1 shows the absence of P1/HC-Pro mRNA in vector only–transformed Arabidopsis plants.
(D) Severely phenotypic P1/HC-Pro Arabidopsis plants have narrow cotyledons as compared with those of nontransformed (nt) seedlings.
The Arabidopsis P1/HC-Pro line chosen for further experiments had severe developmental abnormalities in leaves and flowers similar to those previously reported by others (Kasschau et al., 2003; Dunoyer et al., 2004). Additional defects in branching pattern and vasculature were also noted in the P1/HC-Pro transgenic plants (Figures 2A to 2F). Reduced fertility was also noted in the P1/HC-Pro transgenic line and was associated with defects in both male and female floral organs (Figures 2G to 2J). The P1/HC-Pro transgenic line used in all our experiments was hemizygous for the transgene, as plants homozygous for the transgene were never recovered despite extensive efforts to do so (data not shown).
Figure 2.
Developmental Phenotypes in P1/HC-Pro Transgenic Arabidopsis Plants.
(A) and (B) Branching abnormalities associated with the P1/HC-Pro transgene. The typical monopodial branching pattern of wild-type inflorescences (A), unlike the primitive branching pattern in P1/HC-Pro inflorescences (B).
(C) to (F) Vascular patterning abnormalities in P1/HC-Pro transgenic Arabidopsis flowers. A sepal from a nontransgenic flower (C), as compared with a sepal from a P1/HC-Pro transgenic flower (D), showing irregular edges and reduced interconnections in the vascular system. A petal from a nontransgenic flower (E), as contrasted with a petal from a P1/HC-Pro transgenic flower (F), showing reduced interconnections in the vascular system.
(G) to (J) Reduced fertility in P1/HC-Pro transgenic Arabidopsis is associated with defects in both male and female floral organs. The microsporangia (in yellow), the sites of pollen grain generation and maturation in the anther, of a nontransgenic stamen (G). The microsporangia in the anther of a P1/HC-Pro transgenic stamen (H), which are generally smaller and bound by more connective tissue in P1/HC-Pro transgenic plants than in nontransgenic plants. A pistil of a P1/HC-Pro transgenic flower (right), showing the reduced number of ovules compared with a nontransgenic pistil (left) (I). The papillae on the pistil of a P1/HC-Pro transgenic flower (right) are longer and less dense than those of nontransgenic plants (left) (J).
Ectopic Expression of DCL1 cDNA
If the P1/HC-Pro–associated defects in development are due to defects in Dicer-mediated small RNA pathways, then overexpression of DCL1 might suppress some or all aspects of the P1/HC-Pro phenotype. In order to test this hypothesis, we generated plant lines that ectopically overexpress DCL1 by transformation of Arabidopsis with an expression cassette containing the full-length 6.2-kb DCL1 cDNA under control of the constitutive cauliflower mosaic virus 35S promoter (35S:DCL1). Three of the twelve primary transformants that contained a single transgene locus (lines 2, 9, and 12) were found to overexpress the predicted 6.2-kb DCL1 transcript in both seedling and adult stages (Figure 3B and 3C; data not shown). Interestingly, accumulation of the 6.2 kb DCL1 mRNA was consistently higher in stems and flowers than in rosette leaves of adult plants in both lines (Figure 3C; data not shown). Two previously identified smaller DCL1 transcripts were also detected in the 35S:DCL1 plants (Figure 3A): a 4.0-kb RNA containing 5′ DCL1 sequences and a 2.5-kb RNA containing primarily 3′ DCL1 sequences as well as variable amounts of intron 14 sequences (Jacobsen et al., 1999; Golden et al., 2002; Xie et al., 2003). It is thought that these smaller transcripts arise due to aberrant splicing of the 14th intron of DCL1 (Xie et al., 2003). Interestingly, the 2.5-kb transcript was present at wild-type levels, as expected, but the 4.0-kb transcript accumulated in parallel with the 6.2-kb mRNA and was more heterogeneous than in the wild type (Figures 3B and 3C). These results suggest that the 4.0-kb RNA transcript in the 35S:DCL1 plant does not arise due to aberrant splicing but is a product of premature termination of the 35S:DCL1 transgene or the breakdown of DCL1 mRNA. Expression of P1/HC-Pro increased the steady state level of the 6.2- and 4.0-kb DCL1 transcripts by threefold to fivefold in both wild-type and 35S:DCL1 plants (Figure 3C).
Although 35S:DCL1 lines overexpressed the DCL1 mRNA, it was possible that the lines would fail to accumulate DCL1 protein because the DCL1 mRNA is targeted by miR162, which, in theory, could repress translation of the DCL1 mRNA. To test if expression of the 35S:DCL1 transgene produced a protein with DCL1 activity, the 35S:DCL1 (line 12) transgene locus was crossed into the dcl1-8 mutant background. Homozygous dcl1-8 mutants show the same developmental abnormalities as the two other partial loss-of-function dcl1 alleles (dcl1-7 and dcl1-9), including delayed flowering, as seen by an increase in the number of rosette leaves at bolting (Table 1), adaxialized (curled under) rosette leaves (Figure 3D, Table 2), female sterility, and severe reductions in miRNA accumulation (Figure 3E) (Lang et al., 1994; Ray et al., 1996; Jacobsen et al., 1999; Golden et al., 2002; Park et al., 2002; Reinhart et al., 2002; Xie et al., 2004). The dcl1-8 defects in leaf morphology and flowering time were largely complemented in homozygous dcl1-8 plants carrying the 35S:DCL1 transgene (Figure 3D, Table 1); moreover, fertility was partially restored. Furthermore, the accumulation of each of three tested miRNAs was returned to wild-type or near wild-type level in these plants (Figure 3E). The ability of the 35S:DCL1 transgene to complement dcl1-8 mutant phenotypes indicates that the transgenic locus produces functional protein capable of DCL1 activities.
Table 1.
Effects of 35S:DCL1 on Flowering Time and Leaf Number at Flowering
| Strain | Transgene | Na | Lightb | Flowering time (Days)c | Leaf Numberc |
|---|---|---|---|---|---|
| Co-gld | – | 68 | 18L, 6D | 25.3 ± 1.8 | 7.6 ± 0.7 |
| Line 2 | 35S:DCL1 | 72 | 18L, 6D | 25.7 ± 2.1e | 6.4 ± 1.0f |
| Line 9 | 35S:DCL1 | 72 | 18L, 6D | 24.2 ± 1.7f | 4.8 ± 0.7f |
| Line 12 | 35S:DCL1 | 72 | 18L, 6D | 23.3 ± 1.8f | 5.3 ± 0.6f |
| Cod | – | 19 | 24L | ND | 12.5 ± 1.7g |
| Dcl1+ | – | 21 | 24L | ND | 12.8 ± 2.3eg |
| Dcl1+ | 35S:DCL1 | 31 | 24L | ND | 13.4 ± 1.8eg |
| dcl1-8 | 35S:DCL1 | 6 | 24L | ND | 16.3 ± 1.9fg |
| dcl1-8 | – | 5 | 24L | ND | 30.2 ± 3.2f |
Number of plants scored per strain.
18L, 6D, growth in 18 h light/6 h dark; 24L, growth in constant light.
Mean ± sd; ND, not determined.
Co-gl, Columbia glabrous1; Co, Columbia.
Not significantly different from wild-type plants using Fisher-Behrens procedure (for means with unequal population variances; Campbell, 1989).
d value significant at the 99% level compared with wild-type plants using Fisher-Behrens.
d value significant at the 99% level compared with homozygous dcl1-8 plants using Fisher-Behrens.
Table 2.
Effects of the Level of DCL1 Gene Activity on the Patterning of Lateral Organsa
| Strain | Phenotypic Class | Nb | % Fused | % Adaxialized | % Abaxialized |
|---|---|---|---|---|---|
| Co-glc | Wild type | 373 | 0.0 | 0.0 | 0.0 |
| F2 of dcl1-7/+ | Dcl1+ | 312 | 0.0 | 0.0 | 0.0 |
| (Co-gl) | Dcl1− | 188 | 0.0 | 29.3d | 0.0 |
| F2 of dcl1-8/+ | Dcl1+ | 307 | 0.0 | 0.0 | 0.0 |
| (Co-gl) | Dcl1− | 280 | 0.0 | 23.8d | 0.0 |
| F2 of dcl1-7/+ | Dcl1+ | 316 | 1.0 | 0.0 | 0.0 |
| (Ler)c | Dcl1− | 219 | 0.5 | 14.6 | 0.0 |
| F2 of dcl1-8/+ | Dcl1+ | 312 | 0.0 | 0.0 | 0.0 |
| (Ler) | Dcl1− | 201 | 1.4 | 14.1 | 0.0 |
| F2 of dcl1-9/+ | Dcl1+ | 212 | 0.0 | 0.5 | 0.0 |
| (Ler) | Dcl1− | 105 | 0.0 | 30.5d | 0.0 |
| Co-gl | Wild type | 520 | 0.0 | 0.0 | 0.6 |
| Line 2 (Co)c | 35S:DCL1 | 458 | 0.2 | 2.0 | 1.1 |
| Line 9 (Co) | 35S:DCL1 | 345 | 0.3 | 0.0 | 30.4d |
| Line 12 (Co) | 35S:DCL1 | 382 | 0.0 | 0.0 | 10.7e |
| Null hypothesisf | 4311 | 0.2 | 3.6 | 4.8 |
Data were analyzed using the p × q table and a one-sided significance test with 45° of freedom (Campbell, 1989).
Number of leaves scored per strain.
Co-gl, Columbia glabrous1; Ler, Landsberg erecta; Co, Columbia.
χ2 value significant at the 99.9% level.
χ2 value significant at the 95% level.
Based on the frequency of each phenotypic class in the total population (Campbell, 1989).
To determine the effect of the 35S:DCL1 transgene in the wild-type DCL1 background, developmental progression was examined in these lines. Ectopic DCL1 in the wild-type DCL1 background had only subtle effects on plant development. Plants homozygous for the 35S:DCL1 transgene had a slight acceleration of flowering time (Table 1) and an expansion of the abaxial fates of rosette leaves (Figure 3D, Table 2) and were fully fertile. No obvious developmental phenotypes were observed in plants hemizygous for the 35S:DCL1 transgene (data not shown). We can eliminate the possibility that the developmental phenotypes are due to transcriptional or RNA silencing of the DCL1 gene or to feedback repression due to enhanced levels of miR162 because abundant full-length DCL1 transcripts are detected in the 35S:DCL1 lines (Figures 3B and 3C) and because the 35S:DCL1 transgene is able to complement defects in the dcl1-8 mutant (Figures 3D and 3E). As the observed phenotype of the 35S:DCL1 plants is the opposite of the loss-of-function dcl1 alleles (early versus late flowering; abaxialized versus adaxialized leaves), we can conclude that the 35S:DCL1 transgene is functioning equivalently to a true gain-of-function allele of DCL1.
35S:DCL1 Suppresses the Developmental Phenotype of P1/HC-Pro Plants
To investigate the possibility that P1/HC-Pro phenotypes are due to altered Dicer activity, we examined the effect of ectopic overexpression of DCL1 on the morphology of plants expressing P1/HC-Pro in the F1 offspring of a cross between the homozygous 35S:DCL1 and the dominant hemizygous P1/HC-Pro plants. Surprisingly, plants that were hemizygous for both transgenes had morphologically normal fertile flowers, unlike the nearly sterile flowers produced by plants hemizygous for P1/HC-Pro (Figure 4A). Additionally, the rosette leaves of the double hemizygous plants displayed only a modest lobing as compared with the severely serrated leaves of the P1/HC-Pro line (Figure 4A). The weakening of the P1/HC-Pro phenotype by 35S:DCL1 is not due to either transcriptional or RNA silencing of the P1/HC-Pro transgene because the level of P1/HC-Pro mRNA is similar in P1/HC-Pro and P1/HC-Pro × 35S:DCL1 plants (Figure 4B). Thus, ectopic expression of DCL1 suppresses the majority of the morphological defects observed in P1/HC-Pro plants, supplying further evidence for the model that P1/HC-Pro affects normal plant development by altering the activity of a Dicer.
Figure 4.
The 35S:DCL1 Locus Largely Rescues Morphological Features of P1/HC-Pro–Expressing Arabidopsis Plants.
(A) Ectopic DCL1 rescues developmental defects in P1/HC-Pro transgenic plants. The morphology of the rosette leaves and flowers is shown for wild-type (Columbia), homozygous 35S:DCL1, hemizygous P1/HC-Pro, and doubly hemizygous P1/HC-Pro × 35S:DCL1 lines. The heavily serrated leaves and sterile flowers of the P1/HC-Pro plants are almost completely suppressed in the P1/HC-Pro × 35S:DCL1 lines.
(B) P1/HC-Pro mRNA is expressed in P1/HC-Pro × 35S:DCL1offspring. RNA gel blot analysis showing levels of P1/HC-Pro mRNA in the leaves, stems, and flowers of P1/HC-Pro transgenic plants and the equivalent tissues from P1/HC-Pro × 35S:DCL1 plants, as indicated. Ethidium bromide staining of 25S rRNA is shown as a loading control.
35S:DCL1 Does Not Alter the Accumulation of miRNA or miRNA*s in P1/HC-Pro Plants
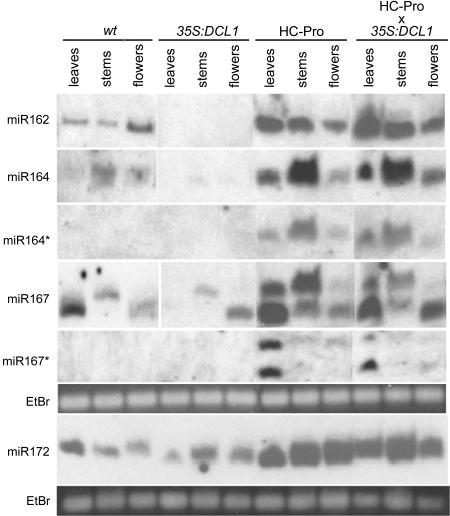
The developmental abnormalities of P1/HC-Pro plants have previously been ascribed to impairments in miRNA biogenesis and function (Kasschau et al., 2003; Chapman et al., 2004). This is because both miRNAs and miRNA* species accumulate to high levels in P1/HC-Pro plants, suggesting that HC-Pro interferes with the assembly of the mature miRNA strand into the RISC complex. If the P1/HC-Pro effects on miRNA biogenesis are mediated through a DCL protein, then ectopic DCL1 might be expected to correct the anomalous accumulation of miRNA* and/or miRNA in the P1/HC-Pro line. We therefore examined the accumulation of four miRNAs and two miRNA*s in leaves, stems, and flowers of 35S:DCL1, P1/HC-Pro, and P1/HC-Pro × 35S:DCL1 plants. Surprisingly, the accumulation of a subset of miRNAs was impaired in 35S:DCL1 plants in a tissue-specific and developmental manner. The accumulation of two of the four miRNAs was decreased ∼10-fold in both stem and flower tissues (Figure 5). In addition, levels of three of the four miRNAs examined were dramatically reduced in the rosette leaves of plants in the late stages of flowering (Figure 5), though little or no reduction was noted in the rosette leaves of plants immediately after bolting (Figure 3E). By contrast, levels of all four miRNAs were increased severalfold in the P1/HC-Pro plants as compared with wild-type plants (Figure 5). Furthermore, the accumulation of miRNA* species, which are nearly undetectable in wild-type and 35S:DCL1 plants, was increased to an even greater extent, consistent with previously reported results (Figure 5; Chapman et al., 2004; Dunoyer et al., 2004). Interestingly, the levels of both miRNAs and miRNA* species in the P1/HC-Pro × 35S:DCL1 plants were almost indistinguishable from those in the P1/HC-Pro plants (Figure 5). Thus, even though the severe developmental anomalies in the P1/HC-Pro line were alleviated in the 35S:DCL1 background, the anomalies in accumulation of miRNAs and miRNA*s were not. These results suggest that the alleviation of P1/HC-Pro phenotype by ectopic DCL1 is not mediated by a general correction of the abnormal accumulation of miRNAs and miRNA*s in these plants.
Figure 5.
The Accumulation of miRNAs in 35S:DCL1, P1/HC-Pro, and P1/HC-Pro × 35S:DCL1 Plants.
The accumulation of the indicated miRNAs and miRNA*s was determined from RNA gel blot analysis of low molecular weight RNA. RNA was isolated from the indicated tissues of adult plants at a late stage of flowering. The same RNA samples were fractionated on two gels. Probes to miR162, miR164, miR164*, miR167, and miR167* were hybridized to a blot derived from one gel, whereas the probe to miR172 was hybridized to a blot derived from the other gel. Ethidium bromide (EtBr) staining of the predominant RNA species in the low molecular weight fraction is shown as a loading control.
35S:DCL1 Does Not Alter the Accumulation of miRNA Targets in P1/HC-Pro Plants
Despite the increased miRNA accumulation in P1/HC-Pro plants, the levels of some, but not all, miRNA target mRNAs are elevated in these plants, suggesting a P1/HC-Pro–mediated impairment in miRNA function (Kasschau et al., 2003; Dunoyer et al., 2004). Thus, even though 35S:DCL1 does not alter the accumulation of miRNAs or miRNA* species in P1/HC-Pro plants, it is still possible that the 35S:DCL1 transgene could suppress the P1/HC-Pro phenotype by restoring miRNA-directed target degradation. To test this hypothesis, the accumulation of selected miRNA target mRNAs was examined in P1/HC-Pro plants, with and without 35S:DCL1. Two of the three target mRNAs examined, AUXIN RESPONSE FACTOR8 (ARF8; At5g37020; miR167 target) and AP2-LIKE (At2g28550; miR172 target), were previously shown to undergo miRNA-directed cleavage that was inhibited by P1/HC-Pro in some tissues (Kasschau et al., 2003). The third, encoding a NAC domain protein (At5g61430), is a target of miR164 as shown by the detection of cleavage products (Mallory et al., 2004). Consistent with earlier results, the levels of some target messages were higher in plants expressing P1/HC-Pro than in the equivalent tissues of wild-type controls (for example, the ARF8 and NAC domain protein mRNAs; Figure 6). However, we did not observe an increase in accumulation of AP2-LIKE mRNA in P1/HC-Pro plants as previously reported (Kasschau et al., 2003), possibly because of differences in the particular tissues and developmental stages examined. The effect of 35S:DCL1 alone on miRNA target accumulation was inconclusive because the observed changes were small and inconsistent; although an inverse correlation between the level of a particular miRNA and its corresponding target could sometimes be noted (for example, a decrease in miR164 in stems is correlated with an increase in the NAC domain protein target mRNA in stems; Figures 5 and 6). By contrast, the effect of ectopic DCL1 on miRNA target messages in the P1/HC-Pro background was clear in every case. The levels of all three miRNA targets in the P1/HC-Pro × 35S:DCL1 plants were the same as in the P1/HC-Pro plants (Figure 6). It seems likely, therefore, that the developmental abnormalities of P1/HC-Pro plants and the phenotypic rescue by ectopic DCL1 are not mediated by a general miRNA-directed effect on target mRNA stability or at least not by effects mediated by the specific miRNAs examined here.
Figure 6.
The Accumulation of miRNA Target mRNAs in 35S:DCL1, P1/HC-Pro, and P1/HC-Pro × 35S:DCL1 Plants.
The accumulation of the indicated mRNA targets was determined from RNA gel blot analysis of high molecular weight RNA isolated from the indicated plants. Ethidium bromide staining of 25S rRNA is shown as a loading control.
35S:DCL1 Does Not Rescue either Sense Transgene–Induced Silencing or Virus-Induced RNA Silencing in P1/HC-Pro Plants
To investigate if 35S:DCL1 could prevent P1/HC-Pro–mediated suppression of RNA silencing in addition to correcting P1/HC-Pro–mediated developmental anomalies, we looked at the effect of 35S:DCL1 on P1/HC-Pro suppression of both sense transgene–induced silencing and virus-induced RNA silencing (VIGS). For the sense transgene–induced silencing experiments, P1/HC-Pro, 35S:DCL1, and P1/HC-Pro × 35S:DCL1 plants were crossed to the well-characterized L1 line, which contains a silenced sense transgene encoding β-glucuronidase (GUS). L1 plants initiate silencing at ∼2 to 3 weeks after germination as evidenced by a dramatic decrease in GUS mRNA and the simultaneous appearance of high levels of GUS siRNAs (Figure 7A, lanes 5 and 6). By contrast, L1 plants expressing P1/HC-Pro fail to silence and thus had high levels of full-length GUS mRNA and undetectable levels of GUS siRNAs at the same developmental stage (Figure 7A, lane 1). Overexpression of DCL1 did not rescue L1 silencing in the P1/HC-Pro line: plants with all three loci had high levels of GUS mRNA and undetectable levels of GUS siRNAs (Figure 7A, lanes 3 and 4). Unexpectedly, the 35S:DCL1 plants were also partially suppressed for silencing of the L1 transgene in the absence of a P1/HC-Pro transgene (Figure 7A, lane 2). Analysis of F2 offspring indicated that suppression of L1 silencing occurred whether the plants were homozygous or hemizygous for either the L1 or the 35S:DCL1 locus (data not shown). These results raise the possibility that DCL1 plays a role in at least some kinds of RNA silencing and strongly suggest that the alleviation of the P1/HC-Pro developmental phenotype by 35S:DCL1 does not reflect a rescue of sense transgene–induced RNA silencing.
Figure 7.
35S:DCL1 Does Not Rescue P1/HC-Pro Suppression of either Sense Transgene–Induced or Virus-Induced RNA Silencing.
(A) Sense transgene–induced RNA silencing was monitored by RNA gel blot analysis of high molecular weight RNA to detect GUS mRNA (top blot) and of low molecular weight RNA to detect GUS siRNAs (bottom blot). The RNAs in lanes 1 to 5 are derived from plants hemizygous for the L1 GUS-silenced locus. The presence or absence of the P1/HC-Pro and 35S:DCL1 transgenes is indicated above the blot. The RNA in lane 2 is from a single plant hemizygous for both 35S:DCL1 and L1 transgenes and is representative of four separate individuals that were assayed. The RNAs in lanes 3 and 4 are from two different individual plants hemizygous for all three transgenes. Lanes 5 and 6 show RNA from control plants lacking both P1/HC-Pro and 35S:DCL1 transgenes and either hemizygous (lane 5) or homozygous (lane 6) for the L1 locus.
(B) VIGS was assayed by RNA gel blot analysis of high molecular weight RNA to detect CH42 mRNA (top blot) and of low molecular weight RNA to detect CH42 siRNA (bottom blot). RNA was isolated from tissue pooled from 10 plants at 21 d after bombardment with the CH42 silencing vector. The presence or absence of the P1/HC-Pro and 35S:DCL1 transgenes is indicated above the blot. Ethidium bromide (EtBr) staining of 25S rRNA and the predominant RNA species in the low molecular weight fraction are shown as loading controls in (A) and (B). The molecular weight of the indicated small RNA species in (A) and (B) was inferred from the migration of DNA oligonucleotides that were subsequently standardized to RNA markers.
To determine if ectopic DCL1 prevents P1/HC-Pro suppression of VIGS, we bombarded wild-type, P1/HC-Pro, 35S:DCL1, and P1/HC-Pro × 35S:DCL1 plants with a Cabbage leaf curl geminivirus (CbLCV) vector carrying a portion of the endogenous gene CHLORATA42 (CH42), which encodes a component of the magnesium chelatase complex (Turnage et al., 2002). Silencing of this gene causes the inability to accumulate chlorophyll and results in pronounced yellowing or chlorosis of leaf tissue. Bombardment with the CH42 vector DNA resulted in viral infection in all inoculated plants as shown by the presence of symptoms and viral DNA (data not shown). When infected with the CH42 vector, both wild-type and 35S:DCL1 plants developed the severe chlorosis characteristic of VIGS of CH42, had nearly undetectable levels of CH42 mRNA, and accumulated CH42 siRNAs (Figure 7B, lanes 5 and 6). By contrast, both P1/HC-Pro and P1/HC-Pro × 35S:DCL1 plants infected with the CH42 vector had levels of CH42 mRNA equivalent to those in uninfected controls and showed no signs of chlorosis, indicating that P1/HC-Pro blocked VIGS even when DCL1 was overexpressed (Figure 7B, lanes 7 and 8). Interestingly, the siRNAs that accumulate in response to VIGS in this system were primarily of the larger variety previously associated with chromatin modifications rather than with RNA degradation (Mallory et al., 2002b; Chan et al., 2004; Xie et al., 2004). Moreover, the level of these larger small RNAs was not reduced in either P1/HC-Pro plants or in the P1/HC-Pro × 35S:DCL1 plants even though VIGS was suppressed as assayed by accumulation of the targeted CH42 mRNA. Overall, these results suggest that it is unlikely that the alleviation of P1/HC-Pro developmental phenotypes caused by ectopic 35S:DCL1 is due to rescue of an endogenous pathway related to either VIGS or sense transgene–induced RNA silencing.
DISCUSSION
The plant viral protein P1/HC-Pro has dramatic affects on RNA silencing and miRNA pathways and normal progression through development. The effect of P1/HC-Pro on the miRNA pathway is manifest as an increase in the levels of miRNA and miRNA*s and an increase in the levels of some miRNA target mRNAs in some plant tissues (Mallory et al., 2002b; Kasschau et al., 2003; Chapman et al., 2004; Dunoyer et al., 2004). Suppression of sense transgene–induced RNA silencing by P1/HC-Pro is associated with the elimination of siRNAs and the accumulation of target mRNA (Llave et al., 2000; Mallory et al., 2001). Both defects in small RNA metabolism are consistent with the hypothesis that P1/HC-Pro alters the activity of DCL enzymes, which produce small RNAs and are essential for their assembly into RISC. If P1/HC-Pro interferes with one or more DCL proteins, then additional copies of a Dicer might restore Dicer activity and allow the plant to overcome the effects of P1/HC-Pro on both development and small RNA metabolism. To test this hypothesis, we ectopically expressed Arabidopsis DCL1, a Dicer required for miRNA biogenesis, under the control of a constitutive promoter to see if it could rescue any aspect of the P1/HC-Pro phenotype.
Ectopic expression of DCL1 largely alleviated the severe developmental anomalies that characterize P1/HC-Pro transgenic Arabidopsis, resulting in plants with a near wild-type phenotype. To determine if the rescue of developmental anomalies by 35S:DCL1 correlated with the alleviation of any of the known defects in small RNA processes induced by P1/HC-Pro, we examined RNA silencing and miRNA pathways in P1/HC-Pro plants with and without the 35S:DCL1 transgene. Surprisingly, we found no significant differences with respect to small RNA metabolism in these experiments: the VIGS and sense transgene silencing phenotypes for both P1/HC-Pro plants and P1/HC-Pro × 35S:DCL1 plants were essentially identical. Furthermore, the levels of miRNAs, miRNA*s, and miRNA target messages in P1/HC-Pro × 35S:DCL1 plants were similar to those in P1/HC-Pro controls. Thus, the rescue of P1/HC-Pro developmental anomalies by ectopic DCL1 was not coupled to a general correction of P1/HC-Pro–mediated defects in either RNA silencing or miRNA pathways.
The simplest interpretation of these results is that P1/HC-Pro–mediated developmental defects, but not the defects in small RNA metabolism, are caused by interference with a DCL1 activity that can be rescued by increased expression of the gene. Alternatively, the mechanism of P1/HC-Pro action might be via interference with one or more of the other Arabidopsis DCL enzymes (DCL2, 3, or 4), and ectopic DCL1 can compensate for the effect of these enzymes on development but not for the other effects on small RNA-directed RNA degradation pathways. A further possibility is that P1/HC-Pro may not directly interfere with any aspect of Dicer function but rather with some other activity for which overexpression of DCL1 can compensate, perhaps by enhancing the efficiency of a downstream step in the pathways controlling development.
Current models attribute the developmental anomalies in P1/HC-Pro plants to an impairment in miRNA precursor processing that allows accumulation of a normally labile miRNA/miRNA* intermediate, thereby preventing assembly of a functional RISC and blocking miRNA-directed target degradation (Chapman et al., 2004). These models would predict that correction of defects in development would be accompanied by a correction of the aberrant accumulation of miRNA, miRNA*s, and miRNA target mRNAs. However, our results clearly show that the P1/HC-Pro developmental anomalies can be rescued without a concomitant rescue of these miRNA pathway defects. How then might 35S:DCL1 relieve the effects of P1/HC-Pro on development? Since our studies included a limited number of miRNAs, it remains possible that the observed alleviation of P1/HC-Pro phenotype by ectopic DCL1 is due to an effect on a subset of miRNAs rather than a general correction of the biogenesis and function of all miRNAs. An alternative hypothesis is that DCL1 and perhaps one or more of the other Arabidopsis DCL enzymes have a function in addition to their known roles in small RNA metabolism. In general, RNase IIIs cleave highly base-paired or fully dsRNAs and are important in the processing of rRNAs (Venema and Tollervey, 1999) and small nucleolar RNAs (Tycowski and Steitz, 2001) and in regulating the level of some pre-mRNAs (Danin-Kreiselman et al., 2003). Based on these known roles of canonical RNase IIIs, one possibility is that Dicers are involved in RNA processing events that directly regulate the accumulation of pre-mRNAs, small nuclear RNAs, or small nucleolar RNAs that play important roles in development.
An unexpected finding of our studies is that the 35S:DCL1 plants are not uniformly enhanced for known DCL1 functions. While the 35S:DCL1 transgene can largely complement a dcl1 mutation (indicating that the transgene produces a functional DCL1 protein), its phenotype in wild-type plants suggests that some DCL1 activities are enhanced, whereas others are impaired. The developmental phenotype in the homozygous DCL1 overexpressing plants is opposite those of dcl1 partial loss-of-function mutants with respect to flowering time and adaxialized/abaxialized patterning of leaves, suggesting enhanced DCL1 function. However, the accumulation of at least some miRNAs is decreased in the 35S:DCL1 plants in the wild-type DCL1 background, suggesting decreased DCL1 activity with regard to miRNA biogenesis. It is not unreasonable to think that overexpressing a Dicer might result in either increased or decreased activity depending upon the process being examined. For example, if a Dicer function was dependent on assembly with two or more other factors, then overexpression might titrate out these other factors, resulting in the formation of inactive complexes that contain only a subset of the required components. Similarly, if such factors are shared with other Dicers, then overexpression might also interfere with the activities of these other Dicers. By contrast, overexpression ought to enhance DCL1 activities that are not dependent on limiting amounts of cofactors by simply providing more enzyme. Using this rationale, the finding that both miRNA accumulation and sense transgene silencing are impaired in the 35S:DCL1 plants suggests that both processes depend on a Dicer complex that is not properly assembled when DCL1 is overexpressed. By contrast, an increase in a DCL1 activity that does not require cofactors may account for the rescue of P1/HC-Pro–induced developmental anomalies. Overall, the relatively mild developmental phenotypes of both the 35S:DCL1 and the P1/HC-Pro × 35S:DCL1 plants suggest that some degree of impairment in both RNA silencing and miRNA pathways can be tolerated in Arabidopsis without severe developmental consequences.
METHODS
Plant Lines and Transformation
35S:DCL1 lines were generated by transforming Arabidopsis thaliana (Columbia ecotype) with the 6.2-kb DCL1 sequence containing the 5′ and 3′ untranslated regions in binary vector pAL144 (Lloyd et al., 1992; kind gift of A. Lloyd). Plants expressing TuMV P1/HC-Pro were made by transforming Arabidopsis (Columbia ecotype) with binary vector PCX305 containing the P1/HC-Pro coding region from TuMV isolate 1 (kind gift of T. Pirone). A translation termination signal was inserted to produce the authentic C terminus of HC-Pro, which is generated by the autocatalytic activity of HC-Pro when the polyprotein is expressed from the virus. Both the DCL1 and the P1/HC-Pro cDNAs were cloned under control of the cauliflower mosaic virus 35S promoter. Line L1 (23b1) was the kind gift of H. Vaucheret and has been previously described (Elmayan et al., 1998).
Genotyping to Identify Homozygous dcl1-8 Plants Containing the 35S:DCL1 Transgene
The genotype of F2 plants segregating for both the dcl1-8 mutation and the 35S:DCL1 transgene was determined by a PCR-based assay using DCL1-specific primers (5′-CACGCTGTCAAGAAACATCCATAC-3′ and 5′-GCAGCTTTGTCATACTCGACGAC-3′). Using these primers, a PCR product of 340 bp is amplified from the wild-type DCL1 gene and the dcl1-8 mutant gene; however, the product amplified from the 35S:DCL1 transgene is only 262 bp because it lacks the introns that are present in the endogenous gene. The single nucleotide change causing the dcl1-8 mutation fortuitously also disrupts the recognition sequence for the restriction endonuclease SnaBI, and this polymorphism was used to distinguish the dcl1-8 mutant from the wild-type DCL1 gene (Golden et al., 2002; S. Schauer, D. Merchant, and A. Ray, unpublished data). DNA amplified from a wild-type plant produces 229- and 111-bp bands when digested with SnaBI, while DNA amplified from a homozygous dcl1-8 mutant is not cut by SnaBI, producing the original 340-bp PCR fragment. The presence of all three DNA fragments indicates that the sampled plant was heterozygous for the dcl1-8 mutation. The presence of the 35S:DCL1 transgene in the sampled plant DNA is indicated by the production of 151- and 111-bp fragments upon cleavage of the 262-bp transgene-specific PCR fragment with SnaBI.
VIGS Assay
For the VIGS assay, Arabidopsis plants were bombarded at the four- to six-leaf stage using the CbLCV-based vector system for silencing of the CH42 gene encoding magnesium cheletase that was kindly provided by D. Robertson and has been previously described (Turnage et al., 2002). Each cartridge contained 0.125 mg of 0.6-μm gold particles coated with a mixture of 1 μg each of the CbLCV A and B component vectors as described (Turnage et al., 2002). Plants were bombarded twice/cartridge using a Bio-Rad PDS 100 Helium system at a pressure of 80 p.s.i.
RNA Isolation and Hybridization
Plant material was frozen in liquid nitrogen, ground to a powder, and immediately vortexed in a preheated (80°C) mixture of ∼5 volumes/g tissue of extraction buffer (100 mM Tris-HCl, pH 8.0, 100 mM LiCl, 10 mM EDTA, and 1% sodium dodecyl sulfate) and an equal volume of phenol. Two volumes of chloroform/volume of phenol were added, the solution was mixed intermittently for 5 min, and phases were separated by centrifugation. An equal volume of 4 M LiCl and 10 mM EDTA, pH 7.0, was added to the aqueous phase and incubated overnight at 4°C. High molecular weight RNA was recovered by centrifugation at 12,000g for 15 min. The supernatant, containing DNA and low molecular weight RNAs, was made 10% in polyethylene glycol 8000 and 0.5 M NaCl and incubated for 30 min on ice to precipitate the DNA. After centrifugation to remove DNA, low molecular weight RNA was concentrated by ethanol precipitation.
RNA gel blot analysis of high and low molecular weight RNA were performed exactly as described (Vance, 1991; Mallory et al., 2002b) using Ambion ULTRAhyb hybridization solution at 45°C for DNA probes and 68°C for RNA probes. The nucleotide coordinates of hybridization probes (nucleotide numbers derived from mRNA sequences) were as follows: DCL1 5′ probe (At1g01040), nucleotides 3171 to 3410; DCL1 3′ probe (At1g01040), nucleotides 4577 to 4829; CH42 (At4g18480), nucleotides 1241 to 1557; NAC domain protein (At5g614330), nucleotides 870 to 1284; ARF8 (At5g37020), nucleotides 1988 to 2395; AP2-LIKE (At2g28550), nucleotides 11 to 452. GUS probes were derived from the 3′ 700 nucleotides of the GUS coding sequence. The P1/HC-Pro probe was derived from the TuMV P1/HC-Pro coding region. Probes were generated using PCR and either labeled directly using Ambion DECAprime II kit or transcribed with T7 RNA polymerase using Ambion MAXIscript after addition of a T7 promoter with an Ambion Lig'n Scribe kit. Specific miRNA probes were prepared by end-labeling antisense oligonucleotides with T4 polynucleotide kinase (New England Biolabs). Probes for miRNA* species 164 and 167 were an equal mix of end-labeled antisense oligonucleotides to the predicted miRNA* sequence of the two precursor genes (miR164a and b; miR167a and b) (Reinhart et al., 2002).
Accession Numbers
Sequence data from this article can be found in the EMBL/GenBank data libraries under accession numbers At1g01040 (DCL1), At4g18480 (CH42), At5g614330 (NAC domain protein), At5g37020 (ARF8), and At2g28550 (AP2-LIKE).
Acknowledgments
We thank Gail Pruss and Teresa A. Golden for critical review of the manuscript, Herve Vaucheret for providing the L1 Arabidopsis line, Dominique Robertson for providing the CbLCV vector constructs, and Tom Pirone for providing TuMV viral RNA. The research was supported by grants to VBV from the US Department of Agriculture, Competitive Grants Program, by a grant to V.B.V. and L.H.B. from the National Institutes of Health and by grants to A.R. from the National Science Foundation.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) are: Lewis H. Bowman (bowman@biol.sc.edu) and Vicki B. Vance (vance@biol.sc.edu).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.036608.
References
- Allen, E., Xie, Z., Gustafson, A.M., and Carrington, J.C. (2005). MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121, 207–221. [DOI] [PubMed] [Google Scholar]
- Ambros, V. (2004). The functions of animal microRNAs. Nature 16, 350–355. [DOI] [PubMed] [Google Scholar]
- Anandalakshmi, R., Marathe, R., Ge, X., Herr, J.M., Jr., Mau, C., Mallory, A., Pruss, G., Bowman, L., and Vance, V.B. (2000). A calmodulin-related protein that suppresses posttranscriptional gene silencing in plants. Science 290, 142–144. [DOI] [PubMed] [Google Scholar]
- Anandalakshmi, R., Pruss, G.J., Ge, X., Marathe, R., Mallory, A.C., Smith, T.H., and Vance, V.B. (1998). A viral suppressor of gene silencing in plants. Proc. Natl. Acad. Sci. USA 95, 13079–13084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aukerman, M.J., and Sakai, H. (2003). Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 11, 2730–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel, D.P. (2004). MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 23, 281–297. [DOI] [PubMed] [Google Scholar]
- Baulcombe, D. (2004). RNA silencing in plants. Nature 431, 356–363. [DOI] [PubMed] [Google Scholar]
- Bohmert, K., Camus, I., Bellini, C., Bouchez, D., Caboche, M., and Benning, C. (1998). AGO1 defines a novel locus of Arabidopsis controlling leaf development. EMBO J. 17, 170–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brigneti, G., Voinnet, O., Li, W.X., Ji, L.H., Ding, S.W., and Baulcombe, D.C. (1998). Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J. 17, 6739–6746. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Campbell, R.C. (1989). Statistics for Biologists, 3rd ed. (Cambridge, UK: Cambridge University Press).
- Catalanotto, C., Pallotta, M., ReFalo, P., Sachs, M.S., Vayssie, L., Macino, G., and Cogoni, C. (2004). Redundancy of the two dicer genes in transgene-induced posttranscriptional gene silencing in Neurospora crassa. Mol. Cell Biol. 24, 2536–2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, S.W., Zilberman, D., Xie, Z., Johansen, L.K., Carrington, J.C., and Jacobsen, S.E. (2004). RNA silencing genes control de novo DNA methylation. Science 303, 1336. [DOI] [PubMed] [Google Scholar]
- Chapman, E.J., Prokhnevsky, A.I., Gopinath, K., Dolja, V.V., and Carrington, J.C. (2004). Viral RNA silencing suppressors inhibit the microRNA pathway at an intermediate step. Genes Dev. 18, 1179–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X. (2004). A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303, 2022–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X., Liu, J., Cheng, Y., and Jia, D. (2002). HEN1 functions pleiotropically in Arabidopsis development and acts in C function in the flower. Development 129, 1085–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danin-Kreiselman, M., Lee, C.Y., and Chanfreau, G. (2003). RNAse III-mediated degradation of unspliced pre-mRNAs and lariat introns. Mol. Cell 11, 1279–1289. [DOI] [PubMed] [Google Scholar]
- Dugas, D.V., and Bartel, B. (2004). MicroRNA regulation of gene expression in plants. Curr. Opin. Plant Biol. 7, 512–520. [DOI] [PubMed] [Google Scholar]
- Dunoyer, P., Lecellier, C.H., Parizotto, E.A., Himber, C., and Voinnet, O. (2004). Probing the microRNA and small interfering RNA pathways with virus-encoded suppressors of RNA silencing. Plant Cell 16, 1235–1250. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Elmayan, T., Balzergue, S., Beon, F., Bourdon, V., Daubremet, J., Guenet, Y., Mourrain, P., Palauqui, J.C., Vernhettes, S., Vialle, T., Wostrikoff, K., and Vaucheret, H. (1998). Arabidopsis mutants impaired in cosuppression. Plant Cell 10, 1747–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan, E.J., Margis, R., and Waterhouse, P.M. (2003). Posttranscriptional gene silencing is not compromised in the Arabidopsis CARPEL FACTORY (DICER-LIKE1) mutant, a homolog of Dicer-1 from Drosophila. Curr. Biol. 13, 236–240. [DOI] [PubMed] [Google Scholar]
- Gasciolli, V., Mallory, A.C., Bartel, D.P., and Vaucheret, H. (2005). Partially redundant functions of Arabidopsis Dicer-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr. Biol. 15, 1494–1500. [DOI] [PubMed] [Google Scholar]
- Golden, T.A., Schauer, S.E., Lang, J.D., Pien, S., Mushegian, A.R., Grossniklaus, U., Meinke, D.W., and Ray, A. (2002). SHORT INTEGUMENTS1/SUSPENSOR1/CARPEL FACTORY, a Dicer homolog, is a maternal effect gene required for embryo development in Arabidopsis. Plant Physiol. 130, 808–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishok, A., Pasquinelli, A.E., Conte, D., Li, N., Parrish, S., Ha, I., Baillie, D.L., Fire, A., Ruvkun, G., and Mello, C.C. (2001). Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106, 23–34. [DOI] [PubMed] [Google Scholar]
- Hunter, C., and Poethig, R.S. (2003). miSSING LINKS: miRNAs and plant development. Curr. Opin. Genet. Dev. 13, 372–378. [DOI] [PubMed] [Google Scholar]
- Jacobsen, S.E., Running, M.P., and Meyerowitz, E.M. (1999). Disruption of an RNA helicase/RNAse III gene in Arabidopsis causes unregulated cell division in floral meristems. Development 126, 5231–5243. [DOI] [PubMed] [Google Scholar]
- Kadotani, N., Nakayashiki, H., Tosa, Y., and Mayama, S. (2004). One of the two dicer-like proteins in the filamentous fungi Magnaporthe oryzae genome is responsible for hairpin RNA-triggered RNA silencing and related siRNA accumulation. J. Biol. Chem. 279, 44467–44474. [DOI] [PubMed] [Google Scholar]
- Kasschau, K.D., and Carrington, J.C. (1998). A counterdefensive strategy of plant viruses: Suppression of posttranscriptional gene silencing. Cell 95, 461–470. [DOI] [PubMed] [Google Scholar]
- Kasschau, K.D., Xie, Z., Allen, E., Llave, C., Chapman, E.J., Krizan, K.A., and Carrington, J.C. (2003). P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Dev. Cell 4, 205–217. [DOI] [PubMed] [Google Scholar]
- Ketting, R.F., Fischer, S.E., Bernstein, E., Sijen, T., Hannon, G.J., and Plasterk, R.H. (2001). Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 15, 2654–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight, S.W., and Bass, B.L. (2001). A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science 293, 2269–2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara, Y., and Watanabe, Y. (2004). Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc. Natl. Acad. Sci. USA 101, 12753–12758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang, J.D., Ray, S., and Ray, A. (1994). sin1, a mutation affecting female fertility in Arabidopsis, interacts with mod1, its recessive modifier. Genetics 137, 1101–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y.S., Nakahara, K., Pham, J.W., Kim, K., He, Z., Sontheimer, E.J., and Carthew, R.W. (2004). Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell 117, 69–81. [DOI] [PubMed] [Google Scholar]
- Llave, C., Kasschau, K.D., and Carrington, J.C. (2000). Virus-encoded suppressor of posttranscriptional gene silencing targets a maintenance step in the silencing pathway. Proc. Natl. Acad. Sci. USA 97, 13401–13406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave, C., Xie, Z., Kasschau, K.D., and Carrington, J.C. (2002). Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297, 2053–2056. [DOI] [PubMed] [Google Scholar]
- Lloyd, A.M., Walbot, V., and Davis, R.W. (1992). Arabidopsis and Nicotiana anthocyanin production activated by maize regulators R and C1. Science 258, 1773–1775. [DOI] [PubMed] [Google Scholar]
- Lu, C., and Fedoroff, N. (2000). A mutation in the Arabidopsis HYL1 gene encoding a dsRNA binding protein affects responses to abscisic acid, auxin, and cytokinin. Plant Cell 12, 2351–2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory, A.C., Dugas, D.V., Bartel, D.P., and Bartel, B. (2004). MicroRNA regulation of NAC-domain targets is required for proper formation and separation of adjacent embryonic, vegetative, and floral organs. Curr. Biol. 14, 1035–1046. [DOI] [PubMed] [Google Scholar]
- Mallory, A.C., Ely, L., Smith, T.H., Marathe, R., Anandalakshmi, R., Fagard, M., Vaucheret, H., Pruss, G., Bowman, L., and Vance, V.B. (2001). HC-Pro suppression of transgene silencing eliminates the small RNAs but not transgene methylation or the mobile signal. Plant Cell 13, 571–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory, A.C., Parks, G., Endres, M.W., Baulcombe, D., Bowman, L.H., Pruss, G.J., and Vance, V.B. (2002. a). The amplicon-plus system for high-level expression of transgenes in plants. Nat. Biotechnol. 20, 622–625. [DOI] [PubMed] [Google Scholar]
- Mallory, A.C., Reinhart, B.J., Bartel, D., Vance, V.B., and Bowman, L.H. (2002. b). A viral suppressor of RNA silencing differentially regulates the accumulation of short interfering RNAs and micro-RNAs in tobacco. Proc. Natl. Acad. Sci. USA 99, 15228–15233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory, A.C., and Vaucheret, H. (2004). MicroRNAs: Something important between the genes. Curr. Opin. Plant Biol. 7, 120–125. [DOI] [PubMed] [Google Scholar]
- Marathe, R., Anandalakshmi, R., Smith, T.H., Pruss, G.J., and Vance, V.B. (2000). RNA viruses as inducers, suppressors and targets of post-transcriptional gene silencing. Plant Mol. Biol. 43, 295–306. [DOI] [PubMed] [Google Scholar]
- Mette, M.F., Matzke, A.J., and Matzke, M.A. (2001). Resistance of RNA-mediated TGS to HC-Pro, a viral suppressor of PTGS, suggests alternative pathways for dsRNA processing. Curr. Biol. 11, 1119–1123. [DOI] [PubMed] [Google Scholar]
- Palatnik, J.F., Allen, E., Wu, X., Schommer, C., Schwab, R., Carrington, J.C., and Weigel, D. (2003). Control of leaf morphogenesis by microRNAs. Nature 425, 257–263. [DOI] [PubMed] [Google Scholar]
- Park, W., Li, J., Song, R., Messing, J., and Chen, X. (2002). CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 12, 1484–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peragine, A., Yoshikawa, M., Wu, G., Albrecht, H.L., and Poethig, R.S. (2004). SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 18, 2368–2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham, J.W., Pellino, J.L., Lee, Y.S., Carthew, R.W., and Sontheimer, E.J. (2004). A Dicer-2-dependent 80s complex cleaves targeted mRNAs during RNAi in Drosophila. Cell 117, 83–94. [DOI] [PubMed] [Google Scholar]
- Ray, A., Lang, J.D., Golden, T., and Ray, S. (1996). SHORT INTEGUMENT (SIN1), a gene required for ovule development in Arabidopsis, also controls flowering time. Development 122, 2631–2638. [DOI] [PubMed] [Google Scholar]
- Reinhart, B.J., Weinstein, E.G., Rhoades, M.W., Bartel, B., and Bartel, D.P. (2002). MicroRNAs in plants. Genes Dev. 16, 1616–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth, B.M., Pruss, G.J., and Vance, V.B. (2004). Plant viral suppressors of RNA silencing. Virus Res. 102, 97–108. [DOI] [PubMed] [Google Scholar]
- Schauer, S.E., Jacobsen, S.E., Meinke, D.W., and Ray, A. (2002). DICER-LIKE1: Blind men and elephants in Arabidopsis development. Trends Plant Sci. 7, 487–491. [DOI] [PubMed] [Google Scholar]
- Schwartz, B.W., Yeung, E.C., and Meinke, D.W. (1994). Disruption of morphogenesis and transformation of the suspensor in abnormal suspensor mutants of Arabidopsis. Development 120, 3235–3245. [DOI] [PubMed] [Google Scholar]
- Silhavy, D., and Burgyan, J. (2004). Effects and side-effects of viral RNA silencing suppressors on short RNAs. Trends Plant Sci. 9, 76–83. [DOI] [PubMed] [Google Scholar]
- Song, J.J., Liu, J., Tolia, N.H., Schneiderman, J., Smith, S.K., Martienssen, R.A., Hannon, G.J., and Joshua-Tor, L. (2003). The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes. Nat. Struct. Biol. 10, 1026–1032. [DOI] [PubMed] [Google Scholar]
- Tijsterman, M., and Plasterk, R.H. (2004). Dicers at RISC; the mechanism of RNAi. Cell 117, 1–3. [DOI] [PubMed] [Google Scholar]
- Turnage, M.A., Muangsan, N., Peele, C.G., and Robertson, D. (2002). Geminivirus-based vectors for gene silencing in Arabidopsis. Plant J. 30, 107–114. [DOI] [PubMed] [Google Scholar]
- Tycowski, K.T., and Steitz, J.A. (2001). Non-coding snoRNA host genes in Drosophila: Expression strategies for modification guide snoRNAs. Eur. J. Cell Biol. 80, 119–125. [DOI] [PubMed] [Google Scholar]
- Vance, V., and Vaucheret, H. (2001). RNA silencing in plants—Defense and counterdefense. Science 292, 2277–2280. [DOI] [PubMed] [Google Scholar]
- Vance, V.B. (1991). Replication of potato virus X RNA is altered in coinfections with potato virus Y. Virology 182, 486–494. [DOI] [PubMed] [Google Scholar]
- Vazquez, F., Vaucheret, H., Rajagopalan, R., Lepers, C., Gasciolli, V., Mallory, A.C., Hilbert, J.L., Bartel, D.P., and Crete, P. (2004). Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol. Cell 16, 69–79. [DOI] [PubMed] [Google Scholar]
- Venema, J., and Tollervey, D. (1999). Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 33, 261–311. [DOI] [PubMed] [Google Scholar]
- Williams, L., Carles, C.C., Osmont, K.S., and Fletcher, J.C. (2005). A database analysis method identifies an endogenous trans-acting short-interfering RNA that targets the Arabidopsis ARF2, ARF3, and ARF4 genes. Proc. Natl. Acad. Sci. USA 102, 9703–9708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Z., Allen, E., Wilken, A., and Carrington, J.C. (2005). DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 102, 12984–12989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Z., Johansen, L.K., Gustafson, A.M., Kasschau, K.D., Lellis, A.D., Zilberman, D., Jacobsen, S.E., and Carrington, J.C. (2004). Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2, 642–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Z., Kasschau, K.D., and Carrington, J.C. (2003). Negative feedback regulation of Dicer-Like1 in Arabidopsis by microRNA-guided mRNA degradation. Curr. Biol. 13, 784–789. [DOI] [PubMed] [Google Scholar]
- Yoshikawa, M., Peragine, A., Park, M.Y., and Poethig, R.S. (2005). A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 19, 2164–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]