Abstract
Plant shoots are characterized by indeterminate growth resulting from the action of a population of stem cells in the shoot apical meristem (SAM). Indeterminacy within the SAM is specified in part by the class I knox homeobox genes. The myb domain proteins rough sheath2 (RS2) and ASYMMETRIC LEAVES1 (AS1) from maize (Zea mays) and Arabidopsis thaliana, respectively, are required to establish determinacy during leaf development. These proteins are part of a cellular memory system that in response to a stem cell–derived signal keeps knox genes in an off state during organogenesis. Here, we show that RS2/AS1 can form conserved protein complexes through interaction with the DNA binding factor ASYMMETRIC LEAVES2, a predicted RNA binding protein (RIK, for RS2-Interacting KH protein), and a homologue of the chromatin-remodeling protein HIRA. Partial loss of HIRA function in Arabidopsis results in developmental defects comparable to those of as1 and causes reactivation of knox genes in developing leaves, demonstrating a direct role for HIRA in knox gene repression and the establishment of determinacy during leaf formation. Our data suggest that RS2/AS1 and HIRA mediate the epigenetic silencing of knox genes, possibly by modulating chromatin structure. Components of this process are conserved in animals, suggesting the possibility that a similar epigenetic mechanism maintains determinacy during both plant and animal development.
INTRODUCTION
Development in higher plants is a continuous process as organs emerge throughout the entire plant life cycle. Lateral organs, such as leaves and flowers, arise on the flank of the shoot apical meristem (SAM), which contains a population of self-renewing stem cells. These stem cells are maintained in a region of the SAM known as the central zone and divide to contribute daughter cells to the peripheral zone from which organ founder cells are recruited (for review, see Kidner et al., 2002). The reiterative process of organogenesis depends on a precise balance between stem cell maintenance and the differentiation of daughter cells. This balance is established through a negative feedback loop involving the homeodomain protein WUSCHEL (WUS), which specifies stem cell identity and positively regulates CLAVATA3 (CLV3), which in turn restricts the domain of WUS expression (Brand et al., 2000; Schoof et al., 2000).
Meristem maintenance also depends on the action of the class I knox genes. In Arabidopsis thaliana, SHOOT MERISTEMLESS (STM), BREVIPEDICELLUS (BP; also known as KNAT1), KNAT2, and KNAT6 make up the class I knox genes (Lincoln et al., 1994; Long et al., 1996; Semiarti et al., 2001). In maize (Zea mays), this homeobox gene family comprises nine genes, including knotted1 (kn1) (Kerstetter et al., 1994). Loss-of-function mutants in STM and kn1 lack a SAM, consistent with a role in meristem initiation and/or maintenance (Long et al., 1996; Vollbrecht et al., 2000). Mutations in BP alone have no meristem defect, but BP is conditionally redundant with STM (Byrne et al., 2002). A role in meristem function remains speculative for other class I knox genes. However, all class I knox genes are expressed in the SAM, and downregulation of knox expression is a key determinant that distinguishes stem cells and their derivatives in the SAM from lateral organ founder cells (Jackson et al., 1994; Long et al., 1996). Ectopic expression of knox genes in the developing leaf interferes with organ determination and results in the overproliferation of cells and a range of patterning defects (Smith et al., 1992; Sinha et al., 1993; Chuck et al., 1996). Therefore, knox genes have been implicated in prolonging cell division or maintaining an undifferentiated state.
The rough sheath2 (rs2), ASYMMETRIC LEAVES1 (AS1), and PHANTASTICA (PHAN) genes from maize, Arabidopsis, and Antirrhinum majus/tobacco (Nicotiana tabacum), respectively, are required to repress knox genes during leaf development (Schneeberger et al., 1998; Waites et al., 1998; Timmermans et al., 1999; Tsiantis et al., 1999; Byrne et al., 2000; Ori et al., 2000; McHale and Koning, 2004). These genes encode orthologous myb domain proteins and are expressed in a pattern complementary to the knox genes, in founder cells and lateral organ primordia. Recessive mutations in rs2 and as1 exhibit phenotypes that mimic the perturbations in cell determination and patterning that result from ectopic knox gene expression. However, the initial downregulation of knox genes that distinguishes founder cells from meristematic cells is preserved in rs2 and as1. Moreover, this initial downregulation in knox expression precedes the onset of rs2 expression, and STM is a negative regulator of AS1 (Timmermans et al., 1999; Tsiantis et al., 1999; Byrne et al., 2000). RS2 and AS1, therefore, are thought to repress knox gene expression after leaf initiation.
In rs2, reactivation of knox gene expression occurs only in a subset of rs2 null cells. KNOX proteins accumulate in stripes with sharp lateral boundaries, suggesting that rs2 null mutant leaves are clonal mosaics of knox+ and knox− sectors (Timmermans et al., 1999). The position of knox+ sectors varies among rs2 seedlings and does not correlate with normal developmental domains. Such variegated patterns of gene expression are typical of several classic epigenetic phenomena associated with a failure to stably maintain a repressive chromatin state in all cells of a lineage (Francis and Kingston, 2001). Therefore, we proposed that RS2 acts on knox genes as an epigenetic regulator (Timmermans et al., 1999). Several knox+ sectors affect multiple leaves in a pattern indicative of clonal sectors that originate in the meristematic cells of the SAM. RS2 thus acts in response to a factor/mark from the SAM to keep knox genes in an off state during organogenesis, thereby maintaining cellular differentiation.
Such cellular memory is a general feature of development, as the transcriptional output resulting from early patterning events needs to be stably maintained throughout many rounds of cell division. Loss of differentiated cell fates causes various abnormalities that can range from developmental defects to cancer. The Polycomb group proteins form one of the best-characterized cellular memory systems and are required to maintain the repression of developmental genes in animals and plants (Francis and Kingston, 2001; Hsieh et al., 2003). In this study, we show further evidence that RS2/AS1-mediated knox gene silencing involves a novel epigenetic mechanism. We show that these myb domain proteins can form conserved protein complexes through interaction with ASYMMETRIC LEAVES2 (AS2), a predicted RNA binding protein we named RS2-Interacting KH protein (RIK), and a homologue of the histone chaperone HIRA. In other species, HIRA is a chromatin-remodeling protein with known functions in heterochromatic and euchromatic gene silencing (Spector et al., 1997; Kaufman et al., 1998; Magnaghi et al., 1998; Sharp et al., 2001; Roberts et al., 2002). Mutational analysis revealed a direct role for HIRA in knox gene silencing. Partial loss of HIRA function in Arabidopsis results in developmental defects comparable to those of as1 and causes reactivation of several knox genes in developing leaves. Our data suggest that AS1, AS2, and HIRA act together to maintain knox gene silencing, likely by modulating chromatin structure. In addition, these results suggest that the process of maintaining determinacy may be partially conserved between plants and animals.
RESULTS
RS2 Interacts with Predicted DNA Binding and Chromatin-Modifying Proteins
To understand the mechanism by which rs2 maintains knox gene silencing, a yeast two-hybrid screen was performed to identify proteins that interact with RS2. A two-hybrid cDNA expression library was constructed from mRNA prepared from maize shoot apices including four to five leaf primordia that normally express rs2. A total of 3 × 106 independent clones were screened for their ability to interact with the non-myb domain region of RS2, which is highly conserved between RS2, AS1, and PHAN but does not display similarity to any other known proteins (Figure 1A). This region of RS2 includes the so-called C-terminal domain that can mediate homodimerization of RS2 (Theodoris et al., 2003). Accordingly, RS2 was one of the proteins identified in the screen (Table 1). In addition, a close homologue of the Arabidopsis protein AS2 was identified. AS2 is a member of the LBD family, which is characterized by the presence of a highly conserved N-terminal LOB domain that includes a Zn finger and leucine zipper–like motif and may mediate protein–protein and/or protein–DNA interactions (Iwakawa et al., 2002; Shuai et al., 2002). Phylogenetic analysis revealed that this maize homologue is more closely related to AS2 than any other Arabidopsis LBD family member (Figure 1C). The Arabidopsis AS2 protein has been shown to interact with AS1 both in yeast and in vitro (Xu et al., 2003) and is required to maintain the repression of knox genes in developing lateral organs (Ori et al., 2000; Semiarti et al., 2001; Lin et al., 2003). The fact that rs2 and as2 were detected in the two-hybrid screen verified that the library and screening conditions allowed the identification of biologically relevant interacting proteins. Other proteins identified in this screen include a DNA binding protein and two proteins with predicted roles in chromatin regulation (Table 1).
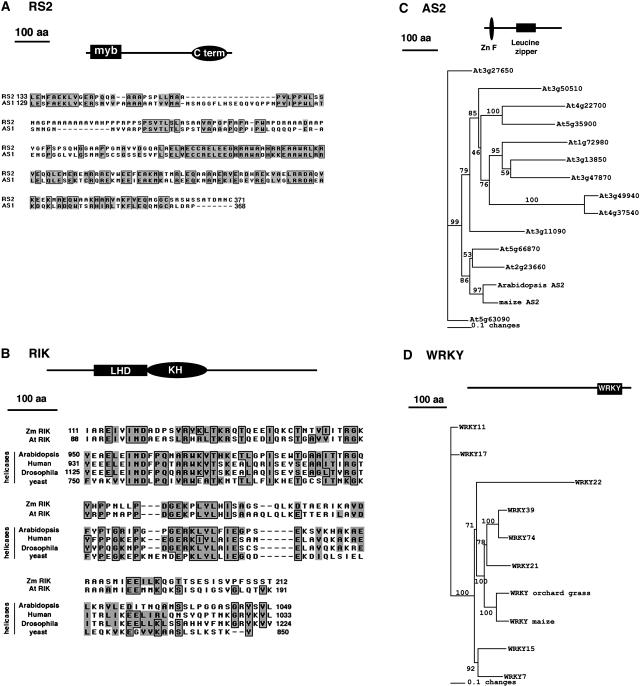
Figure 1.
RS2 and AS1 Have Closely Related Interaction Partners.
(A) RS2 and AS1 are orthologous myb domain transcription factors with a C-terminal domain (C term) necessary for dimerization (Theodoris et al., 2003). The alignment shows the homology between RS2 and AS1 in the region used for the two-hybrid screen and biochemical assays. Identical amino acids (aa) are boxed and shaded gray; similar amino acids are shaded only.
(B) RIK contains a KH domain and a motif similar to that found in helicases of several species (LHD, like helicase domain). Alignments of this LHD region from Arabidopsis and maize RIK with selected helicases highlight the presence of several highly conserved motifs in this domain.
(C) Maize AS2 contains a LOB domain (LBD) with a Zn finger and a leucine zipper motif. Phylogenetic analysis of the maize LBD protein identified in the two-hybrid screen and members of the Arabidopsis LBD family (Iwakawa et al., 2002; Shuai et al., 2002) indicates that the maize LBD protein is most closely related to Arabidopsis AS2.
(D) The maize WRKY1 protein identified in the two-hybrid screen contains a single C-terminal WRKY domain and is related to the class IId WRKY proteins from Arabidopsis (Eulgem et al., 2000). The phylogenetic analysis indicates that WRKY21, WRKY39, and WRKY74 are the nearest class IId Arabidopsis homologues and that maize WRKY1 is most closely related to a WRKY protein from orchardgrass involved with somatic embryogenesis.
Table 1.
Summary of Maize Proteins That Interact with RS2
| Interacting Proteins (AD) | RS2-BD | Number of Coloniesa | GenBank Accession Number |
|---|---|---|---|
| RS2 | ++ | 6 | AF143447 |
| AS2 | ++ | 1 | AY940681 |
| WRKY1 | ++ | 8 | AY940680 |
| RIK | ++ | 7 | AY940679 |
| HIRA | ++ | 2 | AY940678 |
A maize cDNA library was screened to identify proteins that interact with RS2 (see Methods). AD, GAL4 activation domain; BD, GAL4 DNA binding domain; ++, strong growth on 20 mM 3-AT.
The number of colonies identified for each protein.
The two-hybrid screen identified a class IId WRKY DNA binding factor (Eulgem et al., 2000). This WRKY protein (referred to as WRKY1) is most closely related to a WRKY protein from orchardgrass (Dactylis glomerata) that is involved in the development of somatic embryos from leaf tissue (Figure 1D) (Alexandrova and Conger, 2002). A direct connection between knox expression and somatic embryogenesis has not been demonstrated in orchardgrass, but knox misexpression in leaves of other species can lead to the formation of somatic shoots (Sinha et al., 1993; Chuck et al., 1996; Semiarti et al., 2001).
The two-hybrid screen also identified a K-homology (KH) RNA binding protein we named RIK that contains a second motif that is found near the C terminus of several RNA helicases from a variety of species; however, this motif has no described function (Figure 1B). The possible inclusion of RIK in a RS2 complex suggests that silencing at the knox loci may involve an RNA component.
Interestingly, RS2 was found to interact with HIRA (Table 1), a large (∼1000 amino acids) WD-repeat protein with close homologues in yeast and animals. HIRA proteins can direct changes in local chromatin organization and have a known role in heterochromatic and euchromatic gene silencing (Spector et al., 1997; Kaufman et al., 1998; Magnaghi et al., 1998; Roberts et al., 2002). The identification of HIRA as a protein that interacts with RS2 supports the idea that RS2-mediated silencing at the knox loci is an epigenetic phenomenon.
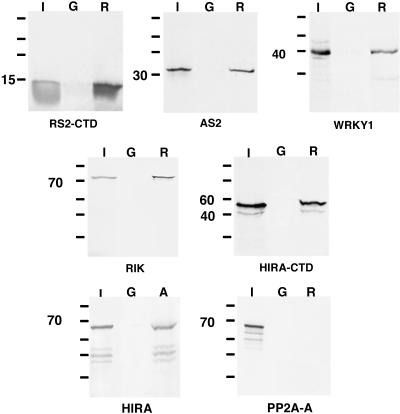
The RS2 protein interactions were confirmed biochemically and independent of the yeast two-hybrid system by glutathione S-transferase (GST) pull-down assays. A fusion protein between GST and the non-myb domain of RS2 (GST-RS2) was used to coprecipitate in vitro transcribed and translated radiolabeled proteins synthesized from each gene identified in the two-hybrid screen. To determine the specificity of the RS2-GST protein in this in vitro assay, we tested for binding of protein phosphatase regulatory subunit A. This protein was originally identified in the two-hybrid screen, but its interaction with RS2 could not be verified in subsequent more stringent analyses or in the GST-RS2 pull-down assay (Figure 2). RS2 has been shown previously to form dimers, and the Arabidopsis AS1 and AS2 proteins can interact in vitro (Theodoris et al., 2003; Xu et al., 2003). Accordingly, both RS2 and AS2 were specifically precipitated with GST-RS2 (Figure 2). Also WRKY1, RIK, and HIRA could be precipitated by GST-RS2 but not by GST alone. In the case of HIRA, the C-terminal region was sufficient for interaction with RS2. This region does not include the WD repeats, indicating that this protein–protein interaction domain is not necessary for RS2 binding.
Figure 2.
In Vitro GST Pull-Down Assays Identified Five RS2-Interacting Proteins.
Fusion proteins between GST and the non-myb domains of RS2 (R) or AS1 (A) were used to coprecipitate in vitro transcribed and translated proteins synthesized from the genes identified in the two-hybrid screen. The interacting protein being analyzed is indicated below each panel. For RS2 and HIRA, the C-terminal 124 amino acids and the C-terminal 450 amino acids, respectively (corresponding to the partial cDNAs obtained in the two-hybrid screen), were tested for their ability to interact with GST-RS2 and GST-AS1. Lanes I, 10% of the amount of each in vitro synthesized protein; lanes G, pull-down assay with GST alone; lanes R, pull-down assay with the GST-RS2 fusion protein; lane A, pull-down assay with the GST-AS1 fusion protein. Protein sizes are indicated in kilodaltons.
The RS2-Interacting Proteins Function in the Nuclei of Developing Leaf Primordia
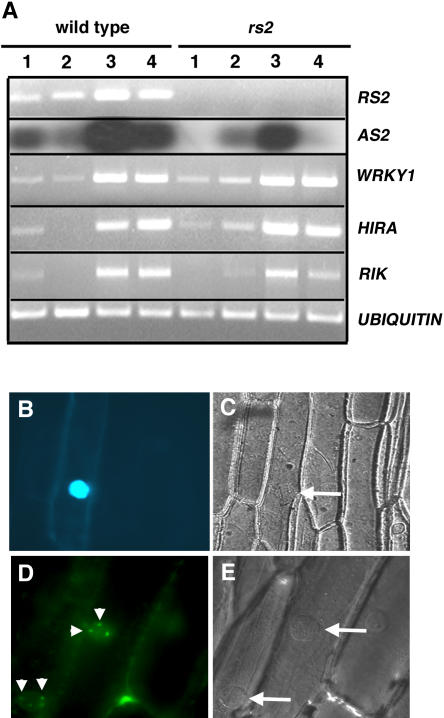
Proteins that act together with RS2 to maintain a silenced state at the knox loci should be expressed in developing leaf primordia. Expression of rs2 and the genes encoding the RS2-interacting proteins was analyzed at various stages of seedling leaf development by semiquantitative RT-PCR. rs2 is expressed most abundantly in young leaf primordia, but expression persists in the sheath and blade tissues of fully expanded seedling leaves (Figure 3A). The expression profile of maize as2 resembles that of rs2, although as2 transcripts are less abundant, consistent with the fact that as2 was recovered relatively infrequently in the two-hybrid screen (Table 1). The maize as2 expression pattern is also comparable to that of AS2 in Arabidopsis (Iwakawa et al., 2002; Lin et al., 2003). As was observed for rs2, transcript levels for wrky1, rik, and hira are more abundant in apices and young leaf primordia than in fully expanded leaf tissues. The overlapping expression patterns are consistent with the possibility that the proteins identified in the two-hybrid screen function together with RS2 in knox gene silencing.
Figure 3.
The RS2-Interacting Proteins Function in Nuclei of Developing Leaf Primordia.
(A) Semiquantitative RT-PCR was performed on RNA isolated from fully expanded blade tissue (lanes 1), sheath tissue (lanes 2), young leaf primordia (P5 to P8) (lanes 3), and shoot apices comprising the SAM and approximately four leaf primordia (lanes 4). RNA was isolated from 2-week-old wild-type and rs2 seedlings as indicated at top. The genes being analyzed are indicated at right.
(B) to (E) C-terminal fluorescence-tagged fusions to RIK ([B] and [C]) and HIRA ([D] and [E]) were transiently expressed in onion cells. Fluorescent images ([B] and [D]) were taken under UV light and show that RIK accumulates throughout the nucleus and that HIRA localizes to subnuclear foci (arrowheads in [D]). Images in (C) and (E) were taken using Nomarski optics and highlight the position of the nucleus within each cell (arrows).
Expression of the genes encoding the RS2-interacting proteins is maintained in rs2 null mutants, although differences in transcript abundance relative to the wild type were observed (Figure 3A). Expression of wrky1, rik, and hira appears to be slightly increased in sheath tissue of rs2 mutants, whereas as2 and rik are less abundantly expressed in rs2 apices compared with the wild type. Whether these changes in expression are significant with respect to the phenotype conferred by the rs2 mutant remains to be determined, but it should be noted that the phenotype conferred by rs2 results predominantly from misexpression of knox genes in developing sheath tissue. These results also indicate that RS2 is not essential for the expression of its interacting proteins.
Proteins involved in the regulation of gene expression should localize to the nucleus. RS2 and AS2 are known to function in the nucleus, whereas WRKY1 contains a well-characterized DNA binding domain and, therefore, is predicted to function in the nucleus as well (Eulgem et al., 2000; Iwakawa et al., 2002; Theodoris et al., 2003). To determine the subcellular localization of RIK and HIRA, C-terminal fusions of cyan fluorescent protein (CFP) to RIK and green fluorescent protein (GFP) to HIRA were transiently expressed in onion (Allium cepa) cells. Expression of CFP alone resulted in fluorescence in both the nucleus and the cytoplasm, as described previously (Kim et al., 2002). By contrast, fluorescence resulting from the RIK-CFP (Figure 3B) and HIRA-GFP (Figure 3D) fusion proteins was limited to the nuclei of expressing cells. Interestingly, transient expression of a HIRA-GFP in nondividing onion cells revealed that the maize HIRA homologue exhibits subnuclear localization (Figure 3D). Mammalian HIRA exhibits a variable subnuclear localization that is cell cycle and phosphorylation dependent (DeLucia et al., 2001). Phosphorylated HIRA in mitotically active cells is distributed diffusely throughout the nucleus, whereas in nondividing cells, HIRA is dephosphorylated and present in a punctate subnuclear pattern. In addition, HIRA promotes the formation of senescence-associated heterochromatin foci that are thought to repress proliferation-promoting genes (Zhang et al., 2005). Attempts to monitor the nuclear expression pattern of HIRA in dividing cells have been unsuccessful to date, probably because of the extreme sensitivity of plant cells to altered HIRA levels (see below). However, the transient expression data suggest that HIRA in plants may similarly recruit target genes into specialized transcriptionally silent heterochromatic foci.
Evolutionary Conservation of the RS2 Protein Interactions
The Arabidopsis AS1 gene encodes the functional orthologue of RS2. Recessive mutations in as1 also cause the ectopic expression of knox genes in developing leaf primordia (Byrne et al., 2000; Ori et al., 2000). However, in contrast with the variegated patterns of knox reactivation observed in rs2 null mutants, a BP:β-glucuronidase (GUS) reporter construct shows uniform misexpression at the base of each as1 leaf (Ori et al., 2000). To ascertain whether AS1- and RS2-mediated knox gene silencing involves similar protein complexes and mechanisms, Arabidopsis homologues of the RS2-interacting proteins were cloned and tested for their ability to interact with the non-myb domain of AS1 (Table 2). Consistent with earlier reports (Theodoris et al., 2003; Xu et al., 2003), AS1 could interact with itself and with AS2 in the yeast two-hybrid system. Phylogenetic and BLAST analyses identified three Arabidopsis WRKY proteins, WRKY21, -39, and -74, that are closely related to maize WRKY1 (Figure 1D). All three Arabidopsis homologues are expressed in vegetative tissues, but none interacted with AS1. Similarly, AS1 did not interact with maize WRKY1, whereas RS2 did interact with WRKY74 from Arabidopsis. This indicates that the function of these WRKY proteins is not conserved between maize and Arabidopsis.
Table 2.
Summary of Arabidopsis Homologues of RS2-Interacting Proteins to Bind AS1
| Arabidopsis Homologues (AD) | AS1-BD | GenBank Accession Number |
|---|---|---|
| AS1 | ++ | AF175996 |
| AS2 | ++ | AAL38032 |
| WRKY 21 | − | AF272747 |
| WRKY 39 | − | AF404860 |
| WRKY 74 | − | AF442398 |
| RIK | ++ | AY940684 |
The Arabidopsis homologues of the maize RS2-interacting proteins were fused to the GAL4 activation domain and the specified interactions were tested. AD, GAL4 activation domain; BD, GAL4 DNA binding domain; ++, strong growth on 20 mM 3-AT; −, no growth.
In Arabidopsis, RIK and HIRA each is encoded from a single gene. RT-PCR analysis showed that both genes are expressed in vegetative tissues. The interaction between RIK and AS1 was demonstrated in yeast (Table 2). However, expression of the Arabidopsis HIRA protein is lethal to yeast. Therefore, this interaction was tested in vitro by GST pull-down experiments. In maize, the C-terminal half of HIRA is sufficient for RS2 binding. An analogous region of Arabidopsis HIRA was transcribed and translated in vitro. This protein fragment can be precipitated by the GST-AS1 fusion protein but not by GST alone (Figure 2). RS2 and AS1 thus have the potential to form conserved protein complexes, suggesting that knox silencing and the establishment of determinacy in lateral organs may involve a similar epigenetic mechanism in maize and Arabidopsis.
HIRA Functions throughout Plant Development
The variegated pattern of knox gene reactivation in rs2 null mutants indicates that a subset of cells have lost the maintenance mechanism that stably propagates the repression of knox genes. The identification of HIRA as a component of the RS2 and AS1 complexes suggests a mechanism for the RS2/AS1-mediated epigenetic silencing of the knox loci and the maintenance of determinacy during organogenesis. HIRA is known not only to modulate chromatin structure during heterochromatic gene silencing but also to control the spatial and temporal expression of specific euchromatic genes (Spector et al., 1997; Magnaghi et al., 1998). Therefore, we determined a more detailed expression pattern for HIRA in Arabidopsis and determined its potential role in knox gene silencing and the specification of determinacy in developing lateral organs.
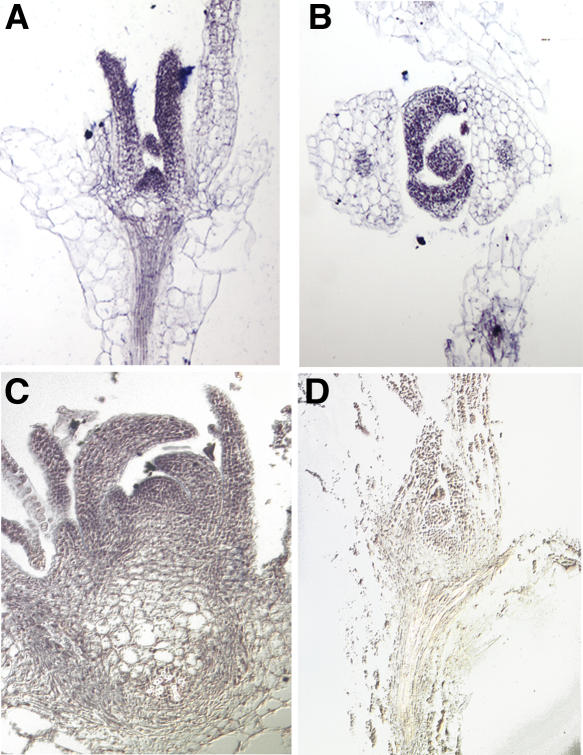
The expression pattern of HIRA during vegetative development in Arabidopsis was determined by in situ hybridization. HIRA is expressed strongly throughout the meristem and young leaf primordia (Figures 4A and 4B). In older leaves, its expression is reduced and becomes confined to the vascular bundles. HIRA transcripts were also detected in the vasculature of hypocotyls. This expression pattern is consistent with the HIRA expression profile observed in maize and suggests that HIRA may be expressed predominantly in tissues containing actively dividing cells. AS1 functions in developing leaf primordia; thus, this expression pattern is also consistent with the suggested role for HIRA in the AS1-mediated knox gene silencing. However, HIRA is expressed more widely than AS1 and includes the knox-expressing cells in the SAM, suggesting that HIRA has additional roles as well.
Figure 4.
HIRA Is Expressed in the Meristem and Developing Leaf Primordia.
(A) and (B) In situ hybridization with an antisense HIRA probe shows that HIRA is expressed throughout the meristem and young leaves of wild-type Arabidopsis seedlings. In older leaves, HIRA expression is restricted to the vascular bundles.
(C) In situ hybridization with an antisense HIRA probe shows that hira cosuppressed plants have an expression pattern comparable to that of wild-type plants.
(D) In situ hybridization with a sense HIRA probe in wild-type plants.
To identify the function of HIRA, we obtained two Arabidopsis lines from the SiGNaL collection (Alonso et al., 2003), each with a T-DNA insertion in the 5′ untranslated region (UTR) of HIRA. Homozygosity for either null allele results in embryo lethality (see Supplemental Figure 1 online). To determine whether HIRA is required during later stages of development, we generated transgenic lines that express a 35S:HIRA transgene to select for mutants that confer a weak hira phenotype as a result of posttranscriptional gene silencing. Surprisingly, only five independent 35S:HIRA transgenic lines were obtained upon screening of ∼20,000 T0 seeds. This unusually low frequency of viable transformants suggests that plants, like yeast, may be very sensitive to variations in normal HIRA expression. The five transgenic lines that were recovered developed vegetative and floral phenotypes (Figure 5). These phenotypes vary in severity between the lines but have been stably inherited for multiple generations. Semiquantitative RT-PCR of leaf tissues of the different 35S:HIRA lines revealed that HIRA transcript levels in each line are reduced compared with those in the wild type (Figure 6). However, in situ hybridization analysis suggests that their pattern of HIRA expression is unchanged (Figure 4C). The developmental defects of the 35S:HIRA plants thus seem to result from partial cosuppression of HIRA and may be reminiscent of a weak loss-of-function phenotype conferred by hira. To emphasize that the 35S:HIRA lines do not confer a gain-of-function phenotype, we hereafter refer to them as hira lines. The rosettes of the hira cosuppression plants are smaller and more compact compared with those of normal Columbia plants (Figures 5A and 5B). Their leaves have shortened petioles and appear pinched as a result of an upward curling of the leaf blade near the petiole and midvein. However, the margins of hira leaves are curled under. as1-1 plants in the Columbia background display similar defects, although the leaf-curling phenotype of as1-1 is generally more severe (Figure 5C). Leaves in the hira cosuppression lines are also asymmetric and frequently develop lobes in the proximal region of the blade. These lobes are often more pronounced than the lobes in as1-1 (Figures 5G and 5I). Like as1-1, the number and identity of the floral organs are unaffected in the hira lines, but the sepals, petals, and stamens are reduced in size such that the carpels become exposed prematurely (Figures 5D and 5F). The diverse phenotypes observed in the hira cosuppression lines and null mutants suggest that HIRA affects distinct developmental processes. However, the phenotypic similarity between as1-1 and the hira cosuppression lines and the observed physical interaction between AS1 and HIRA suggest that the knox loci may be among the HIRA targets.
Figure 5.
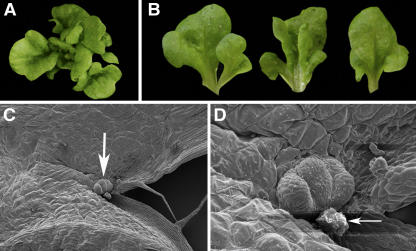
HIRA and AS1 Maintain Determinacy during Organogenesis.
Arabidopsis plants with reduced HIRA expression ([A] and [D]) develop vegetative and floral phenotypes reminiscent of as1 ([C] and [F]). Compared with the wild type ([B] and [E]), as1-1 and cosuppressed hira seedlings are more compact and develop leaves with shortened petioles and blades that are curled under (arrows in [A] and [C]). The sepals, petals, and stamens of cosuppressed hira and as1 flowers are reduced in size, exposing the carpel prematurely (arrows in [D] and [F]). Unlike wild-type leaves (H), cosuppressed hira leaves (G) and as1 leaves (I) are asymmetric and develop lobes (arrows). Expression from the BP:GUS transgene is limited to the meristem of wild-type seedlings (J), but in cosuppressed hira plants (K), GUS activity is extended into developing leaf primordia. In situ hybridization shows that the AS1 expression pattern in cosuppressed hira plants resembles that in wild-type plants; AS1 is expressed in incipient (arrow) and young leaf primordia but not in the meristem (asterisk) of hira cosuppressed plants (L).
Figure 6.
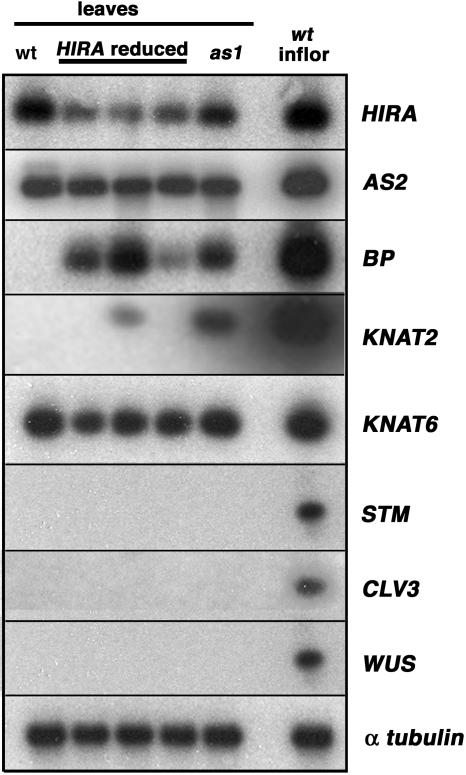
BP and KNAT2 Are Misexpressed in hira and as1 Leaves.
RT-PCR analysis of mature leaves from wild-type, as1, and cosuppressed hira plants indicates that HIRA transcript levels are reduced in cosuppressed hira plants compared with wild-type and as1 plants. AS2 expression is unaffected by a reduction in HIRA transcripts compared with that in wild-type plants. BP and KNAT2 are not expressed in wild-type leaves but are ectopically expressed in leaves of hira cosuppressed plants and as1. By contrast, KNAT6 is expressed in wild-type leaves, and this expression is unaffected in the hira cosuppressed plants and as1 mutants. Expression of STM, CLV3, and WUS is restricted to meristematic tissues in wild-type plants, hira cosuppressed plants, or as1 plants. RT-PCR analyses of α-tubulin transcripts and of RNA prepared from wild-type inflorescences were included as positive controls. These RT-PCR products were analyzed by DNA gel blot hybridization to verify that small amounts of STM, CLV3, and WUS were not misexpressed.
HIRA Maintains knox Gene Silencing in Developing Leaves
The as1-1 phenotype results, at least in part, from misexpression of BP and KNAT2 in developing leaves (Byrne et al., 2000; Ori et al., 2000; Semiarti et al., 2001). To determine whether misexpression of these knox genes also contributes to the phenotypes observed in hira cosuppression lines, we used semiquantitative RT-PCR to compare the expression of the knox genes in mature leaves of wild-type, as1-1, and hira cosuppression lines (Figure 6). As in as1-1 mutants, BP is misexpressed in hira leaves, although the level of BP misexpression varied between the three hira lines analyzed. knox misexpression in as1, as well as in the maize rs2 mutant, occurs predominantly near the base of the leaf (Timmermans et al., 1999; Ori et al., 2000). To determine the pattern of BP misexpression in hira cosuppressed leaves, a GUS reporter gene driven by the BP promoter was introduced into the hira lines. This BP:GUS reporter construct has been shown to contain the cis-acting sequences necessary for repression by AS1 and AS2 (Ori et al., 2000). Accordingly, BP:GUS expression in wild-type plants is limited to the meristem (Figure 5J). In hira cosuppressed plants, the GUS reporter is expressed in the meristem but is also misexpressed in developing leaves (Figure 5K). GUS activity was observed throughout young leaves but became gradually restricted to the petiole and primary veins during leaf development. This pattern of BP misexpression is comparable to that observed in as1 (Ori et al., 2000).
KNAT2 is expressed strongly in wild-type vegetative and inflorescence meristems but also at a low level in young leaves. In as1 mutant leaves, KNAT2 expression is upregulated (Byrne et al., 2000; Semiarti et al., 2001). KNAT2 was not expressed in the wild-type leaf samples analyzed in this study, possibly because these leaves were more mature than those used in previous studies. However, expression of KNAT2 was observed in leaves of as1-1 as well as in one of the hira cosuppression lines (Figure 6). By contrast, expression of the remaining two Arabidopsis class I knox genes, KNAT6 and STM, appears unaffected in the hira lines. KNAT6 is expressed during normal leaf development, but no appreciable differences in the level of KNAT6 expression were observed in any of the tissues analyzed (Figure 6). Expression of STM is normally limited to meristematic cells of the SAM, and this expression pattern is unchanged in as1 mutants (Byrne et al., 2000). Similarly, STM transcripts were undetectable in the leaves of hira cosuppressed plants. These observations indicate that the different knox genes may be under the control of distinct regulatory pathways. Thus, although reduced levels of AS1 and HIRA do not appreciably affect the expression of STM and KNAT6, these genes are required to maintain the repression of BP and KNAT2 during normal leaf development.
AS1 and HIRA could conceivably act linearly in a pathway regulating knox gene expression. If so, the phenotypic resemblance between weak hira cosuppressed plants and as1 null alleles would predict that as1 expression is lost or reduced significantly in the hira lines. However, in situ hybridization revealed that AS1 is expressed abundantly in incipient and young leaf primordia of hira seedlings (Figure 5L). This AS1 expression pattern is identical to that previously reported for wild-type plants (Byrne et al., 2000). Also, expression of AS2, which functions with AS1 in the silencing of knox genes, is unchanged in the hira cosuppression lines (Figure 6). Together, these results and the observation that AS1 and HIRA can physically interact suggest that these proteins act together in a complex that maintains determinacy by repressing BP and KNAT2 during normal leaf development.
HIRA shares homology with another chromatin-remodeling protein, the p60 subunit of chromatin assembly factor1 (CAF1) (Sherwood et al., 1993). In yeast, CAF1 and HIRA have distinct but overlapping functions (Kaufman et al., 1998), even though these proteins regulate gene expression in a DNA synthesis–dependent and –independent manner, respectively (Ray-Gallet et al., 2002; Tagami et al., 2004). In plants, FASCIATA1 (FAS1) and FAS2, which encode components of CAF1, affect meristem function by regulating the expression of WUS and CLV3 (Kaya et al., 2001). Meristem organization appears normal in hira cosuppressed plants (Figure 5L), and RT-PCR analysis revealed that WUS and CLV3 are not misexpressed in hira leaves (Figure 6). These data suggest that although HIRA and FAS2 are close homologues, these proteins likely regulate meristem function and organogenesis through independent pathways.
Misexpression of BP and KNAT2 in as1 and hira cosuppressed leaves is associated with phenotypes that are relatively mild compared with the phenotypes of leaves expressing 35S:BP, which are deeply lobed and develop ectopic stipules and meristems in the sinus regions between the lobes (Chuck et al., 1996). This suggests that silencing at the knox loci is partially maintained in both backgrounds. Although this is not surprising for the weak hira cosuppression lines, the as1-1 allele is amorphic (Byrne et al., 2000). To further evaluate how HIRA functions along with AS1 to regulate knox gene silencing, the 35S:HIRA transgene was crossed into the as1-1 background. Reduced HIRA expression in as1-1 gives rise to plants with severely lobed leaves, and some leaves appear to develop separate leaflets (Figures 7A and 7B). Scanning electron microscopy analysis of such double mutant leaves showed that ectopic meristems often develop at the junction of leaflets with the petiole (Figures 7C and 7D). Thus, hira as1-1 plants exhibit a more severe phenotype that is similar to the phenotype observed in 35S:BP lines. This synergistic interaction between hira and as1-1 suggests that, even though AS1 is unique in the Arabidopsis genome, AS1 function is partially redundant and other factors may meditate the recruitment of HIRA to the knox loci.
Figure 7.
Double Mutants Reveal a Synergistic Interaction between hira and as1.
Both the cosuppressed hira plants and as1 mutants develop curled-under leaves that have shorter petioles than wild-type leaves and are asymmetrically lobed (see Figures 5A to 5C).
(A) These phenotypes are enhanced in hira cosuppressed lines homozygous for as1.
(B) Detached mature leaves illustrate the severe lobing and the occurrence of apparent individual leaflets.
(C) Scanning electron microscopy analysis of leaf 1 from (B) reveals the presence of an ectopic meristem (arrow) at the position where the leaflet-like structure branches from the petiole.
(D) Higher magnification of the ectopic meristem in (C) shows the presence of overproliferated but undifferentiated cell masses (arrow) surrounding the ectopic meristem.
DISCUSSION
RS2/AS1 and HIRA Maintain Determinacy during Organogenesis
Indeterminacy within the SAM is specified in part by the knox homeobox genes. Downregulation of knox gene expression is a key step in leaf initiation, and silencing of these genes needs to be maintained for the progression of normal organogenesis. The maize and Arabidopsis myb domain proteins RS2 and AS1 have orthologous functions in the negative regulation of knox genes (Schneeberger et al., 1998; Timmermans et al., 1999; Tsiantis et al., 1999; Byrne et al., 2000; Ori et al., 2000). However, expression and genetic analyses indicate that these myb proteins act after the initial downregulation in knox expression to maintain knox gene silencing during subsequent leaf development. Moreover, the pattern of ectopic KNOX accumulation in rs2 leaves suggests that an epigenetic mechanism maintains silencing at the knox loci (Timmermans et al., 1999).
Consistent with an epigenetic mode of knox gene repression during leaf development, we have shown that RS2 and AS1 can interact with HIRA. The yeast and mammalian HIRA proteins are known to modulate chromatin structure, both during heterochromatic gene silencing and to control the spatial and temporal expression of specific euchromatic genes (Spector et al., 1997; Kaufman et al., 1998; Magnaghi et al., 1998; Roberts et al., 2002). Mutations in Arabidopsis that lead to reduced expression of HIRA cause developmental defects similar to those of as1 that are accompanied by misexpression of BP and KNAT2 in developing leaves. AS1 is expressed normally in hira cosuppressed seedlings, and HIRA transcripts can be detected at normal levels in as1. These observations substantiate the physical interaction between RS2/AS1 and HIRA and are consistent with the idea that these proteins are part of a cellular memory system that ensures the inheritance of a repressed transcriptional state at the knox loci, thereby maintaining determinacy throughout leaf development. Mutations in the chromatin-remodeling proteins PICKLE (PKL) and SERRATE (SE) have also been implicated in knox gene regulation (Ori et al., 2000). Although mutations in these genes can enhance the as1 phenotype, neither pkl nor se single mutants misexpress knox genes. Therefore, these genetic interactions likely reflect an additive effect resulting from a widespread loss of chromatin regulation that affects multiple leaf developmental pathways.
The precise mechanism by which HIRA maintains knox gene silencing throughout leaf development is not currently known. HIRA contributes to heterochromatic gene silencing in yeast by recruiting the histone deposition protein anti-silencing factor1 (Sharp et al., 2001). In mammals, HIRA was found to be uniquely responsible for replication-independent nucleosome assembly and exchange of the histone variant H3.3, which is typically associated with transcriptionally active genes (Ray-Gallet et al., 2002; Tagami et al., 2004). However, HIRA has also been shown to interact with histone deacetylase (Ahmad et al., 2003). Acetylation of histone H3 at Lys-9 and Lys-14 are well-characterized modifications associated with actively transcribed genes. Deacetylation of these histone residues is interrelated with their methylation, which leads to heterochromatin formation and silencing in plants, animals, and fungi (for review, see Matzke and Birchler, 2005). Thus, HIRA could potentially direct changes in local chromatin organization at the knox loci through the addition of new histones, by replacement of histone variants, or by altering the covalent modifications associated with histone tails.
RS2/AS1 May Mediate the Targeting of HIRA to the knox Loci
Loss of HIRA function in Arabidopsis leads to early embryonic lethality, suggesting that HIRA may control the chromatin state at genomic regions other than the knox loci. In the seedling, HIRA is expressed throughout the meristem and developing leaf primordia. Thus, additional factors likely mediate the targeting of HIRA to specific loci. In mouse, the function of HIRA in the neural crest is mediated through its interaction with the homeodomain protein PAX3 (Magnaghi et al., 1998). Similarly, the genetic and physical interactions between RS2/AS1 and HIRA suggest that these myb domain proteins may recruit HIRA to the knox loci during leaf development. However, the synergistic interaction between as1-1 and the weak hira cosuppressed lines indicates partial redundancy in AS1 function. Moreover, RS2/AS1 contain unusual myb domains, and attempts to demonstrate binding of these proteins to DNA in vitro have been unsuccessful (Theodoris et al., 2003). These observations suggest that RS2/AS1 require cofactors to recruit HIRA to the knox loci. AS2 would be an obvious candidate. AS2 interacts with RS2/AS1 in both Arabidopsis and maize, and AS2 contains Zn finger and leucine zipper–like motifs that are known to mediate protein–protein and protein–DNA interactions (Iwakawa et al., 2002; Shuai et al., 2002). In Arabidopsis, as2 has defects in leaf development that are comparable to those of as1 and the weakly cosuppressed hira lines, and all three genes are required to maintain the repression of BP and KNAT2 in developing organs. However, double mutant analysis showed that as2 is epistatic to as1 (Byrne et al., 2000). Therefore, factors other than AS2 and RS2/AS1 also mediate the targeting of HIRA to the knox loci. The epistatic interaction between as2 and as1 further indicates that RS2/AS1 function depends on AS2, thus presenting the possibility that AS2 contributes to the binding of RS2/AS1 at the knox loci.
Perhaps the RNA binding protein RIK acts together with RS2/AS1 in the recruitment of HIRA. The Polycomb-repressive complex-1 in animals also includes an RNA binding protein that is thought to have a role in directing this complex to target loci (Zhang et al., 2004). The interaction between RS2/AS1 and RIK suggests that regulatory RNAs may function in the repression of knox genes. RNA has been implicated in a wide range of gene-silencing phenomena and can mediate the sequence-dependent establishment of epigenetic marks, such as DNA and/or histone modifications (Wassenegger, 2000; Jeffery and Nakielny, 2004; Matzke and Birchler, 2005). Such epigenetic modifications are typically interpreted by transcription factors and chromatin-remodeling complexes to regulate gene expression. In support of this notion, the clonal sectors of knox reactivation in rs2 mutants suggest that a factor/mark is present at the knox genes in the meristematic cells of the SAM that gives rise to a metastable silenced state upon founder cell recruitment, which is reinforced through the action of RS2/AS1. Although the nature of this factor/mark remains elusive and may conceivably involve proteins other than those identified in the two-hybrid screen, RIK is expressed in the SAM as well as in developing leaves (T. Phelps-Durr and M. Timmermans, unpublished results).
Divergence in the Functions of RS2, AS1, and PHAN
RS2 and AS1 have nearly identical protein interaction partners, suggesting that the mechanism of knox gene silencing may be largely conserved between maize and Arabidopsis. However, knox genes are misexpressed ubiquitously at the base of all as1 leaves, whereas in rs2 leaf primordia, knox genes become reactivated in a variable variegated pattern (Timmermans et al., 1999; Ori et al., 2000; Byrne et al., 2002). This indicates that, despite the partial functional redundancy, Arabidopsis has a stricter requirement for AS1 to maintain the downregulation in knox expression established during founder cell recruitment. The initial repressed state could be inherently more stable in maize. Alternatively, additional proteins could contribute to maize knox gene silencing, albeit with less efficiency than RS2. The interaction between WRKY1 and RS2 may substantiate such divergence in knox gene regulation between maize and Arabidopsis. Although it is not clear whether WRKY1 functions in this process, the close homology with a somatic embryogenesis-associated WRKY protein from orchardgrass makes it conceivable that WRKY1 acts along with RS2 in knox gene regulation (Alexandrova and Conger, 2002).
Irrespective of the function of WRKY1, the fact that this interaction is not conserved in Arabidopsis reveals species-specific variation in the function of RS2/AS1. Differences in PHAN function also exist in Antirrhinum and tobacco (Waites et al., 1998; McHale and Koning, 2004). These RS2/AS1 orthologues are required for the proper regulation of knox gene expression, but they also have a well-defined role in the specification of adaxial fate. Such phenotypic differences may reflect functional differences in the targets of PHAN. More likely, however, they reflect differences in the regulatory functions of RS2, AS1, and PHAN. Although these proteins are nearly identical in the myb domain, other regions exhibit more extensive sequence variation. As a result, RS2, AS1, and PHAN may have coevolved with unique binding partners, such as WRKY1, that lead to divergence in complex activity or targeting. As in the APETALLA3 and PISTILLATA gene lineages, such functional divergence may have played a prominent role in the evolution of new morphologies (Lamb and Irish, 2003). In tomato (Lycopersicon esculentum) and other compound-leafed species, the regulatory relationship between PHAN and the knox genes has diverged, and this is thought to have contributed to the acquisition of compound leaf morphology (Kim et al., 2003).
Establishment of Determinacy in Animals May Involve a Similar Epigenetic Mechanism
Epigenetic mechanisms that coordinate the stable inheritance of early cell fate decisions are a fundamental feature of development. In plants and animals, several chromatin-remodeling proteins have been identified that ensure the stable inheritance of activated or repressed states at developmental loci. For instance, in Drosophila, such cellular memory at the HOX gene cluster is maintained by chromatin-remodeling factors belonging to the Polycomb group (Francis and Kingston, 2001). However, the epigenetic mechanism of homeobox gene silencing in plants is likely distinct from that in animals, because the Polycomb-repressive complex-1, which exerts long-term repression, is not conserved in plants (Hsieh et al., 2003). Our results indicate that RS2/AS1 and HIRA are part of a cellular memory system that maintains knox gene silencing and determinacy throughout organogenesis. We propose that RIK binds a meristem-derived regulatory RNA, which establishes a metastable silenced state at the knox loci. AS2 and RS2/AS1 act upon this initial repressed state and recruit HIRA and perhaps other chromatin-remodeling factors to the knox loci. These establish a stable repressive chromatin state that is faithfully inherited throughout leaf development. Thus, like Polycomb-mediated silencing, knox gene silencing may involve the successive recruitment of distinct proteins or protein complexes that create target specificity, establish a transient repressive state, and finally form a somatically stable silenced state.
Several observations suggest that a similar genetic pathway may regulate determinacy during animal development. Expression of the homeobox gene Msx1 is required for amphibian limb regeneration and can induce dedifferentiation of mouse muscle cells (Odelberg et al., 2000; Kumar et al., 2004). Noncoding RNAs play a role in the establishment of differentiated cell lineages from bone marrow and adult neural stem cells (Kuwabara et al., 2004; Tagoh et al., 2004). Moreover, both HIRA and RIK are conserved throughout the animal lineages, and HIRA may have related functions in mammalian development. The replication timing of HIRA is altered in patients with DiGeorge syndrome, a developmental disorder thought to arise from defects in the multipotent neural crest progenitor cells (D'Antoni et al., 2004). In mouse, HIRA physically interacts with PAX transcription factors that regulate stem cell pluripotency, and loss of HIRA function leads to neural crest–associated defects (Magnaghi et al., 1998; Chi and Epstein, 2002; Roberts et al., 2002). Thus, our results may provide a framework for the establishment and maintenance of determinacy in plants as well as animals.
METHODS
Yeast Two-Hybrid Screen
The non-myb domain region of RS2 (amino acids 133 to 370) was cloned into pGBT10, creating a fusion between RS2 and the GAL4 DNA binding domain (GDB) (Van Aelst, 1998). Poly(A)+ RNA was isolated from shoot apices of 2-week-old maize (Zea mays) seedlings using Trizol reagent (Gibco BRL) and PolyATract (Promega). Approximately 5 μg of poly(A)+ RNA was primed with oligo(dT) and converted into cDNA using the Stratagene cDNA synthesis kit and the manufacturer's suggested protocol. cDNAs were cloned downstream of the GAL4 activation domain into the EcoRI and XhoI sites of the pGADGH vector. The library consisted of ∼3 × 106 independent clones with average insert size of 1.6 kb, ranging between 0.5 and 3.5 kb. The library was transformed into Escherichia coli, clones were scraped from the agar plates, and DNA was prepared according to standard protocols. The two-hybrid screen was performed as described (Van Aelst, 1998). Approximately 107 GAD-cDNA clones were transformed into yeast strain PJ69-4A expressing the GDB-RS2 fusion protein. Transformants were selected for His prototrophy in the presence of 20 mM 3-amino-1,2,4-triazole (3-AT) and subsequently replica-plated and screened for adenine prototrophy. GAD-cDNA plasmids were isolated from His+ colonies by transforming crude yeast DNA extracts into the leuB− E. coli strain HB101 and selecting for Leu prototrophy. Interaction specificity was determined by retransforming the GAD plasmids into PJ69-4A carrying GDB-RS2, pGBT10, or a nonspecific GDB-RAS bait (Van Aelst, 1998) and selecting for His prototrophy in the presence of 0, 20, and 40 mM 3-AT. Further verification of positive interactions was performed by β-galactosidase filter assays. cDNA inserts of positive GAD plasmids were sequenced. Full-length cDNA for hira was isolated using 5′ rapid amplification of cDNA ends PCR (Roche) according to the manufacturer's protocol. ClustalW alignments between the RS2-interacting proteins and their Arabidopsis thaliana homologues were generated using MacVector 6.5.1 (Oxford Molecular Group), with a gap weight of 15.00 and a length weight of 0.30. Parsimony analyses were performed on conserved domains using PAUP4.0. Consensus trees and bootstrap values were determined after 1000 replicates. Full-length cDNA clones for AS2, the Arabidopsis homologues of RIK, and WRKY1 were obtained by RT-PCR and cloned into pGADGH to create GAD fusion proteins. Interactions between these fusion proteins and the non-myb domain of AS1 (amino acids 130 to 367) were determined as described above.
GST Pull-Down Assays
The non-myb domain of RS2 or AS1 was cloned into pGEX-6P (Amersham Biosciences), creating an N-terminal fusion with GST. Fusion proteins were expressed and isolated from BL21 E. coli cells as described (Smith and Johnson, 1988). cDNA clones identified in the yeast two-hybrid screen were transcribed and translated in vitro using the TNT T7 Quick for PCR DNA kit (Promega) according to the manufacturer's protocol. Fifty microliters of glutathione Sepharose 4B beads (Pharmacia Biotech) was incubated for 1 h at 4°C with 20 μg of GST, GST-RS2, or GST-AS1 protein in a total volume of 500 μL of 50 mM Hepes, pH 7.4, 10 mM EDTA, and 0.1% Nonidet P-40. After washing once, the beads were incubated with 20 μL of in vitro–translated protein for 2 h at 4°C in 500 μL of the same buffer. Beads were collected by centrifugation and washed three times in Hepes buffer containing 200 mM NaCl. Precipitated proteins were analyzed by SDS-PAGE.
RT-PCR Analysis
Total RNA was isolated using Trizol reagent according to the manufacturer's instructions (Gibco BRL). Approximately 2 μg of DNaseI-treated RNA was primed with oligo(dT) and converted into cDNA using moloney murine leukemia virus reverse transcriptase (New England Biolabs). The cDNA concentrations in different tissue samples were equalized semiquantitatively based on the amplification of ubiquitin in maize and α-tubulin in Arabidopsis. Subsequent PCRs were performed using standard procedures. The annealing temperature was 60°C, and each PCR product was analyzed after 30 cycles. The primers used for amplification of the various maize and Arabidopsis cDNAs are available upon request.
Subcellular Localizations
For the construction of fluorescent fusion proteins, the coding regions of hira and rik were amplified with primers that included appropriate restriction sites and were cloned upstream of plant enhanced GFP and CFP, respectively (gifts from Eric Lam, Rutgers University, New Brunswick, NJ). These fusions were inserted into an expression cassette containing the cauliflower mosaic virus 35S promoter and octopine synthase terminator. Approximately 1.5 μg of 1-μm gold particles (Bio-Rad) were coated with 0.15 μg of plasmid DNA according to the manufacturer's instructions and were bombarded into onion (Allium cepa) cells at 900 pounds per square inch with a PDS-100 helium biolistic particle delivery system (Bio-Rad). After overnight incubation in a humid chamber, onion epidermal tissues were mounted in water and viewed at ×20 and ×40 magnification with an Axioplan 2 fluorescent compound microscope (Zeiss).
Plant Materials
A full-length HIRA cDNA was cloned in the sense orientation behind the cauliflower mosaic virus 35S promoter in a T-DNA binary vector that confers kanamycin resistance to plants (PS119). This T-DNA was transformed into plants of the Columbia ecotype. The as1-1 allele results from a frameshift mutation in the non-myb domain of AS1 (Byrne et al., 2000). This allele was introgressed five times into the Columbia ecotype. T-DNA insertion lines for HIRA were obtained from SiGNaL (Alonso et al., 2003) and correspond to accessions SALK019573 and SALK143806. The positions of the T-DNA insertions were verified by PCR. Segregation analysis was also performed by PCR. The line expressing the BP:GUS reporter gene (Ori et al., 2000) was a gift from S. Hake (Plant Gene Expression Center, Albany, CA). All plants were grown under long-day conditions at 20°C.
In Situ Hybridizations
Ten-day-old seedlings were fixed and embedded as described previously (Jackson, 1991). Digoxigenin-labeled probes were prepared by in vitro transcription according to the manufacturer's protocol (Stratagene). The AS1-specific probe comprises amino acids 138 to 368 and part of the 3′ UTR. The HIRA-specific probe includes amino acids 613 to 1052 and part of the 3′ UTR. Both probes were used at a concentration of 0.5 ng·μl−1·kb−1. Tissue sections were pretreated, hybridized, and washed using published protocols (Jackson, 1991).
Phylogenetic Analysis
Phylogenetic analysis of LBD family members was based on the LOB domain (Shuai et al., 2002). Analysis of the WRKY proteins was based on the conserved N-terminal (amino acids 1 to 100, approximately) and WRKY domains (Eulgem et al., 2000). These regions were aligned with the ClustalW program available in MacVector. Phylogenetic trees were generated in the program PAUP4.0 using the Neighbor Joining method with a 1000 bootstrap output.
Accession Numbers
Accession numbers for the major genes discussed in this article are listed in Tables 1 and 2. GenBank accession numbers for the helicases shown in the alignment in Figure 1 are as follows: Arabidopsis, NP173516; human, NP055644; Drosophila, AAF48446; and yeast, NP009796. The Arabidopsis Genome Initiative locus identifiers for the Arabidopsis WRKY genes are as follows: WRKY11, At4g32550; WRKY17, At2g24570; WRKY22, At4g01250; WRKY15, At2g23320; and WRKY7, At4g24240. The accession number for the orchardgrass WRKY gene is AAG42147. The accession numbers corresponding to the remaining WRKY proteins in Figure 1D are presented in Table 1 or 2.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. HIRA Insertion Lines Are Embryo Lethal.
Supplemental Figure 2. Sequence Analysis of the LOB Domains from Selected LBD Family Members.
Supplemental Figure 3. Sequence Alignment of the Various WRKY Family Members.
Supplementary Material
Acknowledgments
We thank Sarah Hake for the BP:GUS reporter line and Tim Mulligan for excellence in plant care. We also thank Arooba Alam, Jason Pellegrino, and Josh Egan for the analysis of HIRA insertion lines and their role in developing the HIRA fluorescent fusion constructs. This work was supported by a National Science Foundation grant awarded to M.C.P.T. and a Perkin Fellowship for Women in Science awarded to M.C.P.T. and T.L.P.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Marja C.P. Timmermans (timmerma@cshl.edu).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.035477.
References
- Ahmad, A., Takami, Y., and Nakayama, T. (2003). WD dipeptide motifs and LXXLL motif of chicken HIRA are necessary for transcription repression and the latter motif is essential for interaction with histone deacetylase-2 in vivo. Biochem. Biophys. Res. Commun. 312, 1266–1272. [DOI] [PubMed] [Google Scholar]
- Alexandrova, K.S., and Conger, B.V. (2002). Isolation of two somatic embryogenesis-related genes from orchardgrass (Dactylis glomerata). Plant Sci. 162, 301–307. [Google Scholar]
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657. [DOI] [PubMed] [Google Scholar]
- Brand, U., Fletcher, J.C., Hobe, M., Meyerowitz, E.M., and Simon, R. (2000). Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289, 617–619. [DOI] [PubMed] [Google Scholar]
- Byrne, M.E., Barley, R., Curtis, M., Arroyo, J.M., Dunham, M., Hudson, A., and Martienssen, R.A. (2000). ASYMMETRIC LEAVES1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 408, 967–971. [DOI] [PubMed] [Google Scholar]
- Byrne, M.E., Simorowski, J., and Martienssen, R.A. (2002). ASYMMETRIC LEAVES1 reveals knox gene redundancy in Arabidopsis. Development 129, 1957–1965. [DOI] [PubMed] [Google Scholar]
- Chi, N., and Epstein, J.A. (2002). Getting your Pax straight: Pax proteins in development and disease. Trends Genet. 18, 41–47. [DOI] [PubMed] [Google Scholar]
- Chuck, G., Lincoln, C., and Hake, S. (1996). KNAT1 induces lobed leaves with ectopic meristems when overexpressed in Arabidopsis. Plant Cell 8, 1277–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Antoni, S., Mattina, T., Di Mare, P., Federico, C., Motta, S., and Saccone, S. (2004). Altered replication timing of the HIRA/Tuple1 locus in the DiGeorge and velocardiofacial syndromes. Gene 333, 111–119. [DOI] [PubMed] [Google Scholar]
- DeLucia, F., Lorain, S., Scamps, C., Galisson, F., Machold, J., and Lipinski, M. (2001). Subnuclear localization and mitotic phosphorylation of HIRA, the human homologue of Saccharomyces cerevisiae transcriptional regulators Hir1p/Hir2p. Biochem. J. 358, 447–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem, T., Rushton, P.J., Robatzek, S., and Somssich, I.E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206. [DOI] [PubMed] [Google Scholar]
- Francis, N.J., and Kingston, R.E. (2001). Mechanisms of transcriptional memory. Nat. Rev. Mol. Cell Biol. 2, 409–421. [DOI] [PubMed] [Google Scholar]
- Hsieh, T.F., Hakim, O., Ohad, N., and Fischer, R.L. (2003). From flour to flower: How Polycomb group proteins influence multiple aspects of plant development. Trends Plant Sci. 8, 439–445. [DOI] [PubMed] [Google Scholar]
- Iwakawa, H., Ueno, Y., Semiarti, E., Onouchi, H., Kojima, S., Tsukaya, H., Hasebe, M., Soma, T., Ikezaki, M., Machida, C., and Machida, Y. (2002). The ASYMMETRIC LEAVES1 gene of Arabidopsis thaliana, required for formation of symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant Cell Physiol. 43, 467–478. [DOI] [PubMed] [Google Scholar]
- Jackson, D. (1991). In situ hybridization in plants. In Molecular Plant Pathology: A Practical Approach, D.J. Bowles, S.J. Gurr, and M. McPherson, eds (Oxford, UK: Oxford University Press), pp. 163–174.
- Jackson, D., Veit, B., and Hake, S. (1994). Expression of maize KNOTTED-1 related homeobox genes in the shoot apical meristem predicts patterns of morphogenesis in the vegetative shoot. Development 120, 405–413. [Google Scholar]
- Jeffery, L., and Nakielny, S. (2004). Components of the DNA methylation system of chromatin control are RNA binding proteins. J. Biol. Chem. 279, 49479–49487. [DOI] [PubMed] [Google Scholar]
- Kaufman, P.D., Cohen, J.L., and Osley, M.A. (1998). Hir proteins are required for position-dependent gene silencing in Saccharomyces cerevisiae in the absence of chromatin assembly factor I. Mol. Cell. Biol. 18, 4793–4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaya, H., Shibahara, K.I., Taoka, K.I., Iwabuchi, M., Stillman, B., and Araki, T. (2001). FASCIATA genes for chromatin assembly factor-1 in Arabidopsis maintain the cellular organization of apical meristems. Cell 104, 131–142. [DOI] [PubMed] [Google Scholar]
- Kerstetter, R., Vollbrecht, E., Lowe, B., Veit, B., Yamaguchi, J., and Hake, S. (1994). Sequence analysis and expression patterns divide the maize knotted1-like homeobox genes into two classes. Plant Cell 6, 1877–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidner, C., Timmermans, M., Byrne, M., and Martienssen, R. (2002). Developmental genetics of the angiosperm leaf. Adv. Bot. Res. 38, 191–234. [Google Scholar]
- Kim, J.Y., Yuan, Z., Cilia, M., Khalfan-Jagani, Z., and Jackson, D. (2002). Intercellular trafficking of a KNOTTED1 green fluorescent protein fusion in the leaf and shoot meristem of Arabidopsis. Proc. Natl. Acad. Sci. USA 99, 4103–4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, M., McCormick, S., Timmermans, M., and Sinha, N. (2003). The expression domain of PHANTASTICA determines leaflet placement in compound leaves. Nature 424, 438–443. [DOI] [PubMed] [Google Scholar]
- Kumar, A., Velloso, C.P., Imokawa, Y., and Brockes, J.P. (2004). The regenerative plasticity of isolated urodele myofibers and its dependence on MSX1. PLoS Biol. 2, E218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwabara, T., Hsieh, J., Nakashima, K., Taira, K., and Gage, F.H. (2004). A small modulatory dsRNA specifies the fate of adult neural stem cells. Cell 116, 779–793. [DOI] [PubMed] [Google Scholar]
- Lamb, R.S., and Irish, V.F. (2003). Functional divergence within the APETALA3/PISTILLATA floral homeotic gene lineages. Proc. Natl. Acad. Sci. USA 100, 6558–6563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, W.C., Shuai, B., and Springer, P.S. (2003). The Arabidopsis LATERAL ORGAN BOUNDARIES-domain gene ASYMMETRIC LEAVES2 functions in the repression of KNOX gene expression and in adaxial-abaxial patterning. Plant Cell 15, 2241–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln, C., Long, J., Yamaguchi, J., Serikawa, K., and Hake, S. (1994). A knotted1-like homeobox gene in Arabidopsis is expressed in the vegetative meristem and dramatically alters leaf morphology when overexpressed in transgenic plants. Plant Cell 6, 1859–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, J.A., Moan, E.I., Medford, J.I., and Barton, M.K. (1996). A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379, 66–69. [DOI] [PubMed] [Google Scholar]
- Magnaghi, P., Roberts, C., Lorain, S., Lipinski, M., and Scambler, P.J. (1998). HIRA, a mammalian homologue of Saccharomyces cerevisiae transcriptional co-repressors, interacts with Pax3. Nat. Genet. 20, 74–77. [DOI] [PubMed] [Google Scholar]
- Matzke, M.A., and Birchler, J.A. (2005). RNAi-mediated pathways in the nucleus. Nat. Rev. Genet. 6, 24–35. [DOI] [PubMed] [Google Scholar]
- McHale, N.A., and Koning, R.E. (2004). PHANTASTICA regulates development of the adaxial mesophyll in Nicotiana leaves. Plant Cell 16, 1251–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odelberg, S.J., Kollhoff, A., and Keating, M.T. (2000). Dedifferentiation of mammalian myotubes induced by msx1. Cell 103, 1099–1109. [DOI] [PubMed] [Google Scholar]
- Ori, N., Eshed, Y., Chuck, G., Bowman, J.L., and Hake, S. (2000). Mechanisms that control knox gene expression in the Arabidopsis shoot. Development 127, 5523–5532. [DOI] [PubMed] [Google Scholar]
- Ray-Gallet, D., Quivy, J., Scamps, C., Martini, E., Lipinski, M., and Almouzni, G. (2002). HIRA is critical for a nucleosome assembly pathway independent of DNA synthesis. Mol. Cell 9, 1091–1100. [DOI] [PubMed] [Google Scholar]
- Roberts, C., Sutherland, H.F., Farmer, H., Kimber, W., Halford, S., Carey, A., Brickman, J.M., Wynshaw-Boris, A., and Scambler, P.J. (2002). Targeted mutagenesis of the Hira gene results in gastrulation defects and patterning abnormalities of mesoendodermal derivatives prior to early embryonic lethality. Mol. Cell. Biol. 22, 2318–2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneeberger, R., Tsiantis, M., Freeling, M., and Langdale, J.A. (1998). The rough sheath2 gene negatively regulates homeobox gene expression during maize leaf development. Development 125, 2857–2865. [DOI] [PubMed] [Google Scholar]
- Schoof, H., Lenhard, M., Haecker, A., Mayer, K.F., Jurgens, G., and Laux, T. (2000). The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100, 635–644. [DOI] [PubMed] [Google Scholar]
- Semiarti, E., Ueno, Y., Tsukaya, H., Iwakawa, H., Machida, C., and Machida, Y. (2001). The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem-related homeobox genes in leaves. Development 128, 1771–1783. [DOI] [PubMed] [Google Scholar]
- Sharp, J.A., Fouts, E.T., Krawitz, D.C., and Kaufman, P.D. (2001). Yeast histone deposition protein Asf1p requires Hir proteins and PCNA for heterochromatic silencing. Curr. Biol. 11, 463–473. [DOI] [PubMed] [Google Scholar]
- Sherwood, P.W., Tsang, S.V.-M., and Osley, M.A. (1993). Characterization of HIR1 and HIR2, two genes required for regulation of histone gene transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 13, 28–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuai, B., Reynaga-Pena, C.G., and Springer, P.S. (2002). The LATERAL ORGAN BOUNDARIES gene defines a novel, plant-specific gene family. Plant Physiol. 129, 747–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha, N.R., Williams, R.E., and Hake, S. (1993). Overexpression of the maize homeobox gene, knotted-1, causes a switch from determinate to indeterminate cell fates. Genes Dev. 7, 787–795. [DOI] [PubMed] [Google Scholar]
- Smith, D.B., and Johnson, K.S. (1988). Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene 67, 31–40. [DOI] [PubMed] [Google Scholar]
- Smith, L.G., Greene, B., Veit, B., and Hake, S. (1992). A dominant mutation in the maize homeobox gene, knotted-1, causes its ectopic expression in leaf cells with altered fates. Development 116, 21–30. [DOI] [PubMed] [Google Scholar]
- Spector, M.S., Raff, A., DeSilva, H., Lee, K., and Osley, M.A. (1997). Hir1p and Hir2p function as transcriptional corepressors to regulate histone gene transcription in the Saccharomyces cerevisiae cell cycle. Mol. Cell. Biol. 17, 545–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagami, H., Ray-Gallet, D., Almouzni, G., and Nakatani, Y. (2004). Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 116, 51–61. [DOI] [PubMed] [Google Scholar]
- Tagoh, H., Melnik, S., Lefevre, P., Chong, S., Riggs, A.D., and Bonifer, C. (2004). Dynamic reorganization of chromatin structure and selective DNA demethylation prior to stable enhancer complex formation during differentiation of primary hematopoietic cells in vitro. Blood 103, 2950–2955. [DOI] [PubMed] [Google Scholar]
- Theodoris, G., Inada, N., and Freeling, M. (2003). Conservation and molecular dissection of ROUGH SHEATH2 and ASYMMETRIC LEAVES1 function in leaf development. Proc. Natl. Acad. Sci. USA 100, 6837–6842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmermans, M.C., Hudson, A., Becraft, P.W., and Nelson, T. (1999). ROUGH SHEATH2: A Myb protein that represses knox homeobox genes in maize lateral organ primordia. Science 284, 151–153. [DOI] [PubMed] [Google Scholar]
- Tsiantis, M., Schneeberger, R., Golz, J.F., Freeling, M., and Langdale, J.A. (1999). The maize rough sheath2 gene and leaf development programs in monocot and dicot plants. Science 284, 154–156. [DOI] [PubMed] [Google Scholar]
- Van Aelst, L. (1998). Two-hybrid analysis of Ras-Raf interactions. Methods Mol. Biol. 84, 201–222. [DOI] [PubMed] [Google Scholar]
- Vollbrecht, E., Reiser, L., and Hake, S. (2000). Shoot meristem size is dependent on inbred background and presence of the maize homeobox gene, knotted1. Development 127, 3161–3172. [DOI] [PubMed] [Google Scholar]
- Waites, R., Selvadurai, H.R.N., Oliver, I.R., and Hudson, A. (1998). The PHANTASTICA gene encodes a MYB transcription factor involved in growth and dorsoventrality of lateral organs in Antirrhinum. Cell 93, 779–789. [DOI] [PubMed] [Google Scholar]
- Wassenegger, M. (2000). RNA-directed DNA methylation. Plant Mol. Biol. 43, 203–220. [DOI] [PubMed] [Google Scholar]
- Xu, L., Xu, Y., Dong, A., Sun, Y., Pi, L., Xu, Y., and Huang, H. (2003). Novel as1 and as2 defects in leaf adaxial-abaxial polarity reveal the requirement for ASYMMETRIC LEAVES1 and 2 and ERECTA functions in specifying adaxial identity. Development 130, 4097–4107. [DOI] [PubMed] [Google Scholar]
- Zhang, J., Christoforou, A., Aravind, L., Emmons, S.W., van den Heuvel, S., and Haber, D.A. (2004). The C. elegans polycomb gene SOP-2 encodes an RNA binding protein. Mol. Cell 14, 841–847. [DOI] [PubMed] [Google Scholar]
- Zhang, R., et al. (2005). Formation of MacroH2A-containing senescence-associated heterochromatin foci and senescence driven by ASF1a and HIRA. Dev. Cell 8, 19–30. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.