Abstract
Promoter alterations in the csgD gene of Escherichia coli O157:H7 strains ATCC 43894 and ATCC 43895 are associated with variations in curli expression and the ability to bind Congo red dye. Red variants of each strain were more invasive for cultured HEp-2 cells than were white variants. An ATCC 43895 red variant was more virulent than a white variant in a mouse model. However, there were no differences in Shiga toxin production between red and white variants.
Many strains of Escherichia coli and Salmonella spp. produce surface appendages referred to as curli fibers (thin aggregative fimbriae) in an environmentally regulated, rpoS-dependent manner (3, 14, 15). E. coli curli fibers bind Congo red dye and a variety of human serum and tissue proteins and are important in biofilm formation (7, 12, 15, 16, 20, 24). Curli expression and recognition may trigger cytokine induction during E. coli sepsis (1). Curli fiber genesis requires the products of two divergently transcribed operons (7). The csgBA operon encodes the curli subunit protein (CsgA) and a nucleator protein (CsgB). A transcriptional regulator of curli production (CsgD), an outer membrane lipoprotein (CsgG), and two putative curli assembly factors (CsgE and CsgF) are encoded on the csgDEFG operon.
Recently, we reported that, during in vitro growth, curli fibers were infrequently expressed in strains of enterohemorrhagic E. coli serotype O157:H7 (23). However, E. coli O157:H7 strains ATCC 43894 and ATCC 43895 (American Type Culture Collection, Rockville, Md.) both produced noncurliated and temperature-independent, curliated phenotypes in a phase-variant manner as detected by binding to Congo red dye (23). Red, curliated variants of both strains remained stable on Congo red indicator (CRI) plates at 28°C but switched to a mixed population of red and white variants following passage in Luria-Bertani (LB) (Difco Laboratories, Detroit, Mich.) broth at 37°C. The red-to-white phenotype switch was accompanied by specific base pair changes in the csgD promoter; an A-to-T transversion at base −7 from the putative csgD transcriptional start for strain ATCC 43894 and a T-to-G transversion at position −9 for strain ATCC 43895 (23). The csgD promoter changes in both were associated with additional functional differences as suggested by differing biochemical substrate utilization patterns (23).
In this study, we investigated in vitro and in vivo functional differences between the red and white variants of the E. coli O157:H7 strains ATCC 43894 and ATCC 43895.
Bacterial strains, cell lines, and culture conditions.
E. coli variants 43895OR, 43895OW, 43894OR1, and 43894OW1 have been described previously and were maintained on CRI plates at 28°C (8, 23). These strains were derived from E. coli O157:H7 strains ATCC 43894 (CDC EDL 932) and ATCC 43895 (CDC EDL 933). Strains 43895ORs and 43895OWs were made streptomycin resistant by passage on brain heart infusion (Difco Laboratories) agar at 30°C containing increasing streptomycin concentrations (10, 20, 50, and 100 μg/ml). For cell invasion assays, control bacteria were propagated on brain heart infusion agar. Salmonella enterica serovar Typhimurium strain ATCC 14028 and enteroinvasive E. coli (EIEC) strain ATCC 43893 (CDC EDL 1284 [929-78] O124:NM) were used as positive control (invasive) strains, and E. coli DH5α was used as a negative (noninvasive) control strain. Bacterial strains tested in invasion assays, microscopic studies, Shiga toxin assays, and mouse challenge studies were harvested from YESCA plates incubated for 48 h at 28°C and resuspended in LB broth (8). The aggregation of bacteria was eliminated by vigorous vortexing and verified by direct visualization using light microscopy. Bacterial plate counts were performed on LB agar plates. HEp-2 cells and Vero cells were obtained from the American Type Culture Collection. HEp-2 cells were maintained in minimal essential medium α medium (Gibco BRL, Grand Island, N.Y.) supplemented with 10% fetal bovine serum (FBS) (HyClone, Logan, Utah) at 37°C in a 5% CO2 atmosphere. Vero cells were grown in Medium 199 (Gibco BRL) containing 5% FBS at 37°C in a 5% CO2 atmosphere.
PFGE of red and white variants.
Red and white variants of E. coli O157:H7 strains ATCC 43894 and ATCC 43895 were analyzed by pulsed-field gel electrophoresis (PFGE) to compare strain relatedness and to detect band differences between the red and white phenotypic variants within strains. PFGE was performed as described previously by using the restriction enzyme XbaI (10). Comparison of the PFGE banding patterns showed zero or two fragment differences between the red and white variants within individual strains, demonstrating a common origin of the variants (results not shown) (22). Comparison of the red and white variants between strains ATCC 43894 and ATCC 43895 also suggested that these strains are indistinguishable or very closely related.
HEp-2 cell gentamicin invasion assay and microscopic studies.
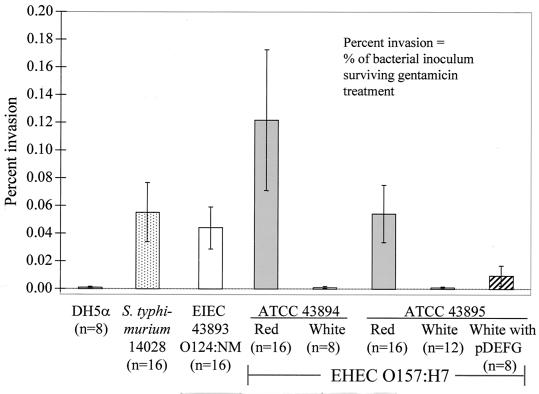
E. coli O157:H7 strains are considered noninvasive when cultured with HEp-2 cells (4, 19). We compared the relative invasiveness of the red and white variants of ATCC 43894 and ATCC 43895 for HEp-2 cells (with EIEC ATCC 43893 and S. enterica serovar Typhimurium ATCC 14028 as positive invasion controls and E. coli DH5α as a negative control), by using a previously described procedure with modifications (9). We also measured the invasiveness of an ATCC 43895 white variant transformed to a Congo red-binding phenotype with the cloned csgDEFG operon (plasmid pDEFG) (23). HEp-2 cells were seeded onto 24-well plates at a density of 5 × 104 cells/well, infected with 20 bacteria/cell, and incubated for 3 h at 37°C in 5% CO2 in minimal essential medium with 10% FBS. Cells were washed to remove nonadherent bacteria and then incubated for an additional hour in culture medium containing 100 μg of gentamicin (Gibco BRL)/ml to kill all extracellular bacteria. After washing, cells were lysed with 500 μl of 0.25% (vol/vol) Triton X-100 (Boehringer Mannheim Corporation, Indianapolis, Ind.) to release any internalized bacteria, which were enumerated by serial dilution and plate counts. The mean percentages of invasion (the percentage of bacterial inoculum that survived gentamicin treatment) with 95% confidence intervals were calculated by using public domain statistical software (Pepi, version 2; USD, Inc., Stone Mountain, Ga.) and plotted. The Games-Howell multiple comparison procedure was used to make pairwise comparisons of each percent invasion group mean by using Pepi (5). As shown in Fig. 1, the red variants of ATCC 43894 and ATCC 43895 were more invasive than their respective white variants: red ATCC 43894 was 227 times more invasive than its white noncurliated variant, and red ATCC 43895 was 94 times more invasive than its white counterpart. The red variants of ATCC 43894 and ATCC 43895, S. enterica serovar Typhimurium ATCC 14028, and EIEC 929-78 were all significantly more invasive than DH5α, white ATCC 43894, and white ATCC 43895 in all pairwise comparisons (P was <0.01 in all cases). Furthermore, the red variants of ATCC 43894 and ATCC 43895, S. enterica serovar Typhimurium ATCC 14028, and EIEC 929-78 did not significantly differ from each other in percent invasion in all pairwise comparisons among these strains (P > 0.5). Thus, the white variants of ATCC 43894 and ATCC 43895 were no more invasive than the noninvasive control strain E. coli DH5α (Fig. 1). However, the red variants of both strains had significantly greater percentages of invasion than the white variants and were statistically as invasive as both positive control strains under the conditions tested. Similar results were obtained using cultured HeLa cells (results not shown).
FIG. 1.
Comparative percentage of HEp-2 cell invasion (with 95% confidence intervals [error bars]) by various bacterial strains. The number in parentheses below each strain is the number of experimental replicates. EHEC, enterohemorrhagic E. coli.
In gentamicin protection assays, the percentage of the initial bacterial inoculum recovered for a typical invasive organism ranges from approximately 0.5 to 25% (17). Thus, the HEp-2 cell percentages of invasion that we observed for the positive controls EIEC and Salmonella and for the red variant E. coli O157:H7 were relatively low (2). However, invasion assay conditions have a great effect on percent invasion for a given bacterial strain and host cell line (21). All strains in this study were grown under conditions designed to inhibit phase variation of curli expression (i.e., on fixed media at 28°C to the stationary growth phase), and infected HEp-2 cells were incubated with gentamicin for only 1 h (instead of the usually employed 3 h) to minimize intercellular replication of internalized bacteria. These conditions have been shown to substantially reduce bacterial internalization by HEp-2 cells (2, 21).
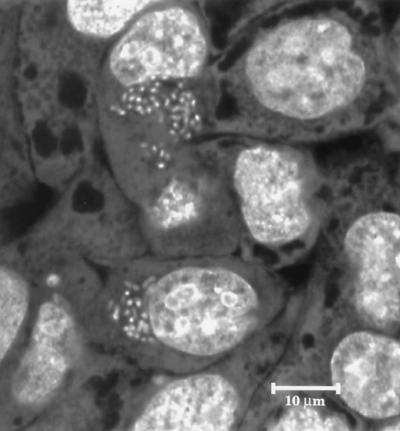
We used confocal microscopy on HEp-2 cells grown in glass bottom microwell dishes (MatTek Corp., Ashland, Mass.) to confirm bacterial internalization following the gentamicin protection assay. Intracellular bacteria were labeled with the LIVE/DEAD BacLight bacterial viability kit (Molecular Probes, Inc., Eugene, Oreg.) and visualized on a Leica TCS SP confocal microscope (Leica Microsystems, Heidelberg, Germany). Although relatively few HEp-2 cells contained internalized bacteria, the number of red variant E. coli O157:H7 bacteria per infected cell was high (Fig. 2). Approximately 99% of the internalized bacteria were viable based on viability staining. No internalization of white variants was detected in parallel samples.
FIG. 2.
Confocal microscopic image of HEp-2 cells infected with the ATCC 43895 Congo red-binding variant. The optical section includes an intracellular plane that contains numerous bacterial cells adjacent to nuclei.
These findings suggest that the Congo red-binding variants of strains ATCC 43894 and ATCC 43895 may express factors that enhance invasion of certain cell types. However, an ATCC 43895 white variant transformed to the Congo red-binding phenotype with plasmid pDEFG was no more invasive than the white variant (P > 0.5), indicating that the mechanism of red variant invasion is more complex than a simple overexpression of the csgDEFG operon.
The ability of the red or white variants of ATCC 43895 to induce localized cytoskeletal rearrangements and condensation of filamentous actin at the sites of bacterial attachment was determined as described previously (9) by using HEp-2 cells grown in microwell dishes. Filamentous actin was stained with Alexa Fluor 488 phalloidin (Molecular Probes, Inc.) and visualized on a Leica TCS SP confocal microscope. Although the level of adhesion of the red variants to the cultured cells appeared to be greater than that of the white variants, quantitation was difficult because both strains showed inconsistent cell binding and readily adhered to exposed glass surfaces. With both variants, only a small number of dense actin plaques (the fluorescent actin staining phenotype) were observed and correlation of fluorescent actin staining to individual attached bacterial cells was often difficult (results not shown). Under the conditions studied, neither of the variants appeared to induce remarkable actin rearrangements in HEp-2 cells.
The red variant of ATCC 43895 has increased virulence in a mouse model.
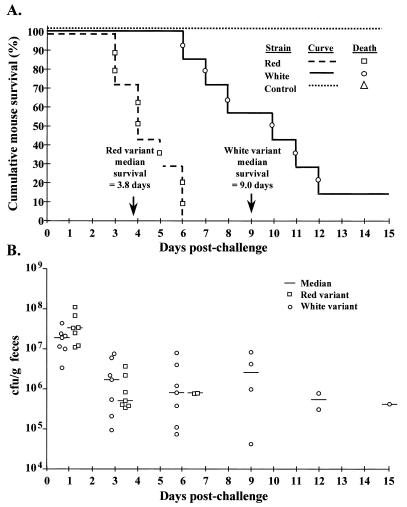
Virulence differences between the red and white variants were compared by oral inoculation of streptomycin-treated mice with streptomycin-resistant strains of ATCC 43895 using the model described by Wadolkowski et al. (25). Three groups of seven individually housed female CD-1 mice were challenged by oral gavage with 0.5 ml of either the red or the white variant suspended in LB broth at a concentration of 109 CFU/ml or with 0.5 ml of sterile LB broth only. Mice were monitored daily for morbidity and mortality. Affected mice exhibited lethargy and hind-limb paralysis. Moribund mice were euthanatized. Kaplan-Meier survival curves were generated for each mouse challenge group, where the outcome event of interest was the number of days postchallenge until individual mouse death. Median survival times were estimated by interpolation (Pepi), and the log rank test was used to make pairwise comparisons between challenge group survival curves (Pepi). In addition, the duration of intestinal colonization was estimated by measuring fecal shedding (reported in colony-forming units per gram of feces) of the inoculated strains on days 1, 3, 6, 9, 12, and 15 postchallenge. All fecal samples collected on day 3 postchallenge were also examined for Congo red dye-binding E. coli O157:H7 growth on CRI plates.
All seven mice challenged with the red variant ATCC 43895 died within 6 days (median survival = 3.8 days), whereas six of seven mice challenged with the white variant died within 12 days postchallenge (median survival = 9.0 days) (Fig. 3A). None of the control mice died during the 15 days of follow-up. Log rank test comparisons of survival curves demonstrated that the red variant-challenged mice had significantly shorter survival times than did those challenged with the white variant (P = 0.003) or the control group (P = 0.001), and the white variant-challenged mice also had shorter survival times than did the control mice (P = 0.005). These results indicate an increased virulence associated with the Congo red-binding variants. Fecal cultures indicated that both red and white variants colonized the gastrointestinal tract at similar levels, which was compared to no growth in the control group (Fig. 3B). Isolated colonies from all seven mice in the white variant-challenged group failed to bind Congo red. In the red variant-challenged group, fecal samples from mouse no. 1 had white colonies only, samples from mouse no. 3 had both red and white colonies, and samples from the other mice in the group had red colonies only. This suggests that the red variants are capable of phenotypic switching in vivo.
FIG. 3.
Survival curves of (A) and fecal shedding by (B) CD-1 mice orally challenged with red or white variants of streptomycin-resistant ATCC 43895 E. coli O157:H7 (concentration, 109 CFU/ml) compared to those of control mice (LB broth). Control mice did not shed E. coli O157:H7; each data point represents an individual mouse. There were seven mice per group.
Assay of Shiga toxin production.
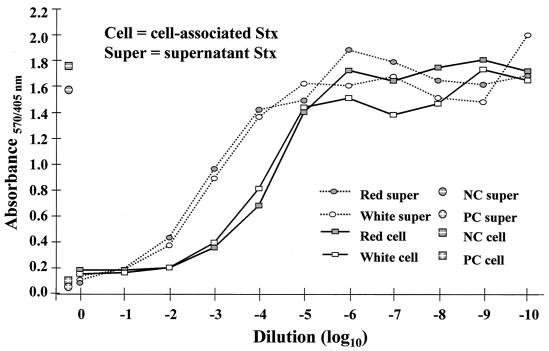
Previous studies have shown that the lethality of enterohemorrhagic E. coli strains tested in the described mouse model was dependent on type II Shiga toxin (Stx) production (11). To determine if the virulence differences (i.e., mouse survival and HEp-2 cell invasion) between the red and white variants resulted from relative differences in Stx production, we compared Stx production in strains 43895OR, 43895OW, 43894OR1, and 43894OW1. Bacteria suspended in LB broth were quantitated by plate counts and incubated for 4 h at 37°C. Culture supernatants and cell lysates from 10 ml of each culture were collected as described previously (13). The cytotoxicities for both the culture supernatants and the bacterial lysates of Vero cells were determined in 96-well microtiter plates by using a previously described method with modification (6, 18). Following incubation with the toxin source and washing, the attached cells were fixed in 2% formalin in phosphate-buffered saline and stained with 100 μl of 0.13% crystal violet in 5% ethanol-2% formalin-phosphate-buffered saline for 2 min. After stain removal and water rinsing, the stain was eluted by the addition of 200 μl of 50% ethanol for 20 min and the absorbance of the wells was determined at dual wavelengths of 570 and 405 nm. The dilution required to kill 50% of the cells in a given well (50% cytotoxic dose [CD50]) was determined in comparison with the negative control (i.e., wells containing Vero cells not exposed to toxin). The CD50, for both secreted and cell-associated Stx, determined for the red variants of each strain were not different from the CD50 of the counterpart white variants of each strain (Fig. 4). Assays performed on the streptomycin-resistant red and white variants of ATCC 43895 showed similar results, confirming that induction of streptomycin resistance in the strains did not alter their Stx-producing properties (results not shown). These results indicate that the in vitro invasion and in vivo mouse mortality differences between the red and white variants were not due to differential Stx production.
FIG. 4.
Comparative Vero cell Stx assay for red (curliated) and white (noncurliated) variants of ATCC 43895 E. coli O157:H7. NC, negative control; PC, positive control.
The results of this study indicate that the Congo red dye-binding variants of E. coli O157:H7 strains ATCC 43894 and ATCC 43895, which contain promoter alterations allowing for greater expression from csgD, display functional in vitro and in vivo differences that could reflect increased virulence of the red variants, compared to that of the white variants, during natural infections. However, whether the observed differences in cell invasion and mouse virulence in E. coli O157:H7 ATCC 43894 and ATCC 43895 are related to increased expression of curli fibers or to another coregulated factor remains to be determined.
Acknowledgments
We thank Carol Chitko-McKown for assistance with cultured cell assays; Ron Mlejnek, Tammy Sorensen, and Tod Stewart for technical assistance; and Joan Rosch for manuscript preparation. We also thank Peter Cooke for assistance with confocal microscopic studies.
Editor: B. B. Finlay
REFERENCES
- 1.Bian, Z., A. Brauner, Y. Li, and S. Normark. 2000. Expression of and cytokine activation by Escherichia coli curli fibers in human sepsis. J. Infect. Dis. 181: 602–612. [DOI] [PubMed] [Google Scholar]
- 2.Boudeau, J., A. L. Glasser, E. Masseret, B. Joly, and A. Darfeuille-Michaud. 1999. Invasive ability of an Escherichia coli strain isolated from the ileal mucosa of a patient with Crohn’s disease. Infect. Immun. 67: 4499–4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collinson, S. K., L. Emödy, K. H. Muller, T. J. Trust, and W. W. Kay. 1991. Purification and characterization of thin, aggregative fimbriae from Salmonella enteritidis. J. Bacteriol. 173: 4773–4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Donnenberg, M. S., A. Donohue-Rolfe, and G. T. Keusch. 1989. Epithelial cell invasion: an overlooked property of enteropathogenic Escherichia coli (EPEC) associated with the EPEC adherence factor. J. Infect. Dis. 160: 452–459. [DOI] [PubMed] [Google Scholar]
- 5.Games, P. A., and J. F. Howell. 1976. Pairwise multiple comparison procedures with unequal n’s and/or variances. J. Ed. Stat. 1: 113–125. [Google Scholar]
- 6.Gentry, M. K., and J. M. Dalrymple. 1980. Quantitative microtiter cytotoxicity assay for Shigella toxin. J. Clin. Microbiol. 12: 361–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hammar, M., A. Arnqvist, Z. Bian, A. Olsén, and S. Normark. 1995. Expression of two csg operons is required for production of fibronectin- and Congo red-binding curli polymers in Escherichia coli K-12. Mol. Microbiol. 18: 661–670. [DOI] [PubMed] [Google Scholar]
- 8.Hammar, M., Z. Bian, and S. Normark. 1996. Nucleator-dependent intercellular assembly of adhesive curli organelles in Escherichia coli. Proc. Natl. Acad. Sci. USA 93: 6562–6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kresse, A. U., M. Rohde, and C. A. Guzman. 1999. The EspD protein of enterohemorrhagic Escherichia coli is required for the formation of bacterial surface appendages and is incorporated in the cytoplasmic membranes of target cells. Infect. Immun. 67: 4834–4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laegreid, W. W., R. O. Elder, and J. E. Keen. 1999. Prevalence of Escherichia coli O157:H7 in range beef calves at weaning. Epidemiol. Infect. 123: 291–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lindgren, S. W., A. R. Melton, and A. D. O’Brien. 1993. Virulence of enterohemorrhagic Escherichia coli O91:H21 clinical isolates in an orally infected mouse model. Infect. Immun. 61: 3832–3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nasr, A. B., A. Olsén, U. Sjöbring, W. Müller-Esterl, and L. Björck. 1996. Assembly of human contact phase proteins and release of bradykinin at the surface of curli-expressing Escherichia coli. Mol. Microbiol. 20: 927–935. [DOI] [PubMed] [Google Scholar]
- 13.O’Brien, A. D., and G. D. Laveck. 1982. Immunochemical and cytotoxic activities of Shigella dysenteriae 1 (Shiga) and Shiga-like toxins. Infect. Immun. 35: 1151–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olsén, A., A. Arnqvist, M. Hammar, S. Sukupolvi, and S. Normark. 1993. The RpoS sigma factor relieves H-NS-mediated transcriptional repression of csgA, the subunit gene of fibronectin-binding curli in Escherichia coli. Mol. Microbiol. 7: 523–536. [DOI] [PubMed] [Google Scholar]
- 15.Olsén, A., A. Jonsson, and S. Normark. 1989. Fibronectin binding mediated by a novel class of surface organelles on Escherichia coli. Nature 338: 652–655. [DOI] [PubMed] [Google Scholar]
- 16.Olsén, A., M. J. Wick, M. Mörgelin, and L. Björck. 1998. Curli, fibrous surface proteins of Escherichia coli, interact with major histocompatibility complex class I molecules. Infect. Immun. 66: 944–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robins-Browne, R. M., and V. Bennett-Wood. 1992. Quantitative assessment of the ability of Escherichia coli to invade cultured animal cells. Microb. Pathog. 12: 159–164. [DOI] [PubMed] [Google Scholar]
- 18.Schmitt, C. K., M. L. McKee, and A. D. O’Brien. 1991. Two copies of Shiga-like toxin II-related genes common in enterohemorrhagic Escherichia coli strains are responsible for the antigenic heterogeneity of the O157:H− strain E32511. Infect. Immun. 59: 1065–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sherman, P., R. Soni, M. Petric, and M. Karmali. 1987. Surface properties of the Vero cytotoxin-producing Escherichia coli O157:H7. Infect. Immun. 55: 1824–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sjöbring, U., G. Pohl, and A. Olsén. 1994. Plasminogen, absorbed by Escherichia coli expressing curli or by Salmonella enteritidis expressing thin aggregative fimbriae, can be activated by simultaneously captured tissue-type plasminogen activator (t-PA). Mol. Microbiol. 14: 443–452. [DOI] [PubMed] [Google Scholar]
- 21.Small, P. L. C., R. R. Isberg, and S. Falkow. 1987. Comparison of the ability of enteroinvasive Escherichia coli, Salmonella typhimurium, Yersinia pseudotuberculosis, and Yersinia enterocolitica to enter and replicate within HEp-2 cells. Infect. Immun. 55: 1674–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulse-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33: 2233–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uhlich, G. A., J. E. Keen, and R. O. Elder. 2001. Mutations in the csgD promoter associated with variations in curli expression in certain strains of Escherichia coli O157:H7. Appl. Environ. Microbiol. 67: 2367–2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vidal, O., R. Longin, C. Prigent-Combaret, C. Dorel, M. Hooreman, and P. Lejeune. 1998. Isolation of an Escherichia coli K-12 mutant strain able to form biofilms on inert surfaces: involvement of a new ompR allele that increases curli expression. J. Bacteriol. 180: 2442–2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wadolkowski, E. A., J. A. Burris, and A. D. O’Brien. 1990. Mouse model for colonization and disease caused by enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 58: 2438–2445. [DOI] [PMC free article] [PubMed] [Google Scholar]