Abstract
Ribosomal protein L24 (RPL24) is implicated in translation reinitiation of polycistronic genes. A newly isolated Arabidopsis thaliana short valve1 (stv1) mutant, in which one of the RPL24-encoding genes, RPL24B, is deleted, shows specific defects in the apical-basal patterning of the gynoecium, in addition to phenotypes induced by ribosome deficiency. A similar gynoecium phenotype is caused by mutations in the auxin response factor (ARF) genes ETTIN (ETT) and MONOPTEROS (MP), which have upstream open reading frames (uORFs) in their 5′-transcript leader sequences. Gynoecia of a double mutant of stv1 and a weak ett mutant allele are similar to those of a strong ett allele, and transformation with a uORF-eliminated ETT construct partially suppressed the stv1 gynoecium phenotype, implying that STV1 could influence ETT translation through its uORFs. Analyses of 5′-leader-reporter gene fusions showed that the uORFs of ETT and MP negatively regulate the translation of the downstream major ORFs, indicating that translation reinitiation is an important step for the expression of these proteins. Taken together, we propose that perturbation of translation reinitiation of the ARF transcripts causes the defects in gynoecium patterning observed in the stv1 mutant.
INTRODUCTION
The plant hormone auxin plays a crucial role in a wide variety of plant morphogenetic and physiological responses. Proper cellular responses to auxin gradients established by polar transport underlie organ patterning (Berleth et al., 2004). For example, genetic and pharmacological studies have established that apical-basal patterning of the gynoecium, the female reproductive organ of flowering plants, involves both auxin responses and polar auxin transport. The Arabidopsis thaliana gynoecium arises from two congenitally fused carpels and is composed of four distinct parts along the apical-basal axis: stigma (Figures 1A and 1C, orange), style (blue), ovary (magenta), and gynophore (green) (Sessions and Zambryski, 1995; Bowman et al., 1999). The female gametophytes are located in the ovary. Mutations in the PINOID (PID), PIN-FORMED1 (PIN1), ETTIN (ETT), and MONOPTEROS (MP) genes cause the development of abnormal gynoecia, in which the boundary between ovary and gynophore is more distal than in the wild type (cf. Figure 1E with a in Figure 1D, white arrowheads) (Bennett et al., 1995; Sessions and Zambryski, 1995; Przemeck et al., 1996). PID and PIN1 encode a Ser/Thr protein kinase and a membrane-localized protein, respectively, and both genes are regulators of polar auxin transport (Gälweiler et al., 1998; Christensen et al., 2000; Friml et al., 2004). Young flower buds sprayed with a polar auxin transport inhibitor develop abnormal gynoecia much like those of the auxin-related mutants described above (Nemhauser et al., 2000). The ETT and MP genes encode auxin response factors (ARF) 3 and 5, respectively (Sessions et al., 1997; Hardtke and Berleth, 1998). ARF protein is a transcription factor that binds to auxin response elements in the promoters of early auxin response genes and regulates their expression (Ulmasov et al., 1997).
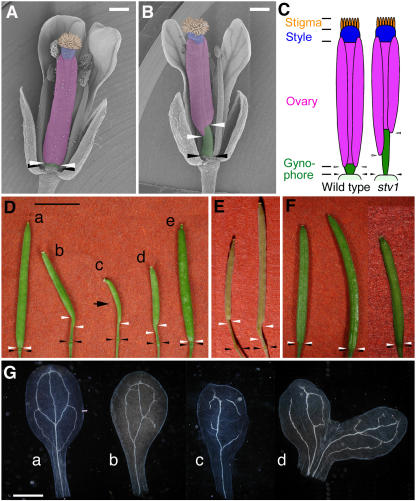
Figure 1.
Apical-Basal Pattern in Gynoecia and Vascular Patterns in Cotyledons of the Wild Type and stv1.
(A) and (B) Scanning electron micrographs of wild-type (A) and stv1-1 (B) gynoecia. The regions of stigma, style, ovary, and gynophore are colored orange, blue, magenta, and green, respectively.
(C) Diagram of gynoecium structures of the wild type and stv1.
(D) Fertilized siliques of wild type (a), stv1-1 (b and c), stv1-2 (d), and stv1-1 transformed with a genomic fragment, including STV1, which is hemizygous for the transgene (e; see Figure 4A). The arrow indicates a complete loss of the ovary region.
(E) Siliques of auxin response (left, ett-2) and polar auxin transport (right, pid-8) mutants.
(F) Siliques of ribosomal protein mutants (left, pfl1/rps18; center, pfl2/rps13; right, aml1/rps5 heterozygote). White and black arrowheads ([A] to [F]) indicate the apical and basal boundaries of the gynophore, respectively.
(G) Vascular patterns of cleared cotyledons from 12-d-old seedlings of the wild type (a) and stv1-1 (b to d).
Bars = 0.5 mm in (A) and (B), 5 mm in (D), and 1 mm in (G).
ARF activity is considered to be partially regulated at the protein level, as it is repressed by interaction with auxin/indole-3-acetic acid proteins (Berleth et al., 2004). Specifically, auxin perception triggers degradation of auxin/indole-3-acetic acid proteins through the ubiquitin-proteasome pathway, which results in ARF activation (Berleth et al., 2004). ARF2 protein, which mediates ethylene and light signaling, is degraded in response to ethylene treatment (Li et al., 2004).
Translational control also contributes to the specific gene regulation associated with developmental programs and environmental responses. For example, several kinds of cis-elements on mRNA are known to affect translation efficiency. One such cis-element is the upstream open reading frame (uORF), which is often found in the 5′-transcript leader sequence (conventionally known as the 5′-untranslated region) of eukaryotic mRNAs. A uORF may regulate the translation of the downstream ORF encoding the major gene product (Geballe and Sachs, 2000; Morris and Geballe, 2000). If uORFs are recognized by a ribosome scanning the mRNA, translation will be terminated at the stop codon of the uORF, and translation of the downstream ORF will require translation reinitiation. For translation reinitiation, the ribosome must form a new initiation complex with eIF2-GTP and Met-tRNA at the start codon of the downstream ORF. Translation efficiency of the downstream ORF can be regulated by several mechanisms, including modulation of the efficiency of reinitiation, stalling of ribosomes due to inhibition of elongation or termination of uORF translation, and exposure of an internal ribosome entry site by ribosome progression along the uORF (Hinnebusch, 1996; Geballe and Sachs, 2000; Morris and Geballe, 2000; Yaman et al., 2003). In some cases, uORFs are involved in mRNA stability (Vilela et al., 1999; Morris and Geballe, 2000; Ruiz-Echevarria and Peltz, 2000).
Cytosolic ribosomal protein L24 (RPL24) belongs to the class of ribosomal proteins present only in archaebacterial and eukaryotic, but not in prokaryotic, ribosomes. An archaebacterial RPL24 homolog, L24e, is located at the surface of the ribosomal large subunit that interacts with the small subunit, close to the main factor binding site (Ban et al., 2000). Yeast mutant analyses showed that RPL24 is involved in the association of large and small ribosome subunits to enhance translation efficiency, although it is not essential for cell viability (Baronas-Lowell and Warner, 1990; Dresios et al., 2000). In plant cells, Arabidopsis RPL24 interacts with transactivator protein (TAV), which is encoded by the cauliflower mosaic virus (CaMV) genome and stimulates translation reinitiation of downstream ORFs of the polycistronic CaMV 35S RNA (Park et al., 2001). Increased expression of RPL24 enhances the TAV-dependent stimulation of translation reinitiation but has no effect on the first translation initiation (Park et al., 2001). These previous studies demonstrated that RPL24 plays a pivotal role in translation reinitiation.
Here, we report the isolation of an Arabidopsis RPL24 mutant, short valve1 (stv1; valve refers to the outer wall of the ovary), which shows defects in apical-basal gynoecium patterning similar to ett and mp mutants. From the results of our genetic and molecular biological analyses, we suggest that ETT and MP expression is regulated by uORFs and requires RPL24-dependent translation reinitiation.
RESULTS
STV1 Is a Novel Factor Involved in Auxin-Mediated Organ Pattern Formation
From a T-DNA–tagged Arabidopsis population, we isolated a recessive mutation, stv1-1, which disturbs the apical-basal development of the gynoecium. Like auxin response and polar transport mutants (Figure 1E) (Bennett et al., 1995; Sessions and Zambryski, 1995; Przemeck et al., 1996), the gynoecium of stv1-1 has a shorter ovary and longer gynophore than the wild type, but the total length of the gynoecium is normal before pollination (Figures 1A to 1C). The extent of the reduction in ovary size varies in each carpel; in some cases, one side of the ovary is missing (c in Figure 1D, arrow), indicating that the apical-basal patterning of the two carpels of the gynoecium develops independently. The stv1-1 phenotype included defects in vascular system and embryo organization, which are also associated with auxin signaling (Friml et al., 2003; Mattsson et al., 2003; Berleth et al., 2004). In the vasculature of wild-type cotyledons, a primary midvein is formed and secondary veins are connected symmetrically (a in Figure 1G). However, most cotyledons of stv1-1 (96%, n = 51) had abnormal vascular patterns that were asymmetric (b in Figure 1G) and/or disconnected (c in Figure 1G). Cotyledons of stv1-1 were occasionally fused or single (3%, n = 98), indicating that stv1-1 is associated with defects in embryo patterning (d in Figure 1G). This series of phenotypic effects suggested that STV1 is involved in auxin-mediated pattern formation.
STV1 Has Pleiotropic Effects on Whole-Plant Development
Morphological defects associated with stv1-1 were also apparent in other plant organs and tissues. After pollination, the gynoecium of stv1-1 failed to elongate, resulting in shorter siliques (a and b in Figure 1D). We found a predominance of arrested seeds in these siliques, resulting in lower fertility (Figure 2A, arrows). Because outcrossing with wild-type pollen resulted in the production of arrested seeds similar to those of self-pollinated plants (data not shown), we predicted that there would be defects in stv1-1 ovules. In fact, as observed in cleared gynoecia, the stv1-1 ovules had shorter integuments than wild-type ones, and in some extreme cases, the gametophyte was protruding from the integuments (Figure 2B). This abnormal development of ovules in stv1-1 mutants may explain the reduction in female fertility.
Figure 2.
Pleiotropic Effects of the stv1 Mutation during Plant Development.
(A) Seeds in elongated siliques of the wild type (left) and stv1-1 (right). Arrested seeds are shown in stv1-1 (arrows).
(B) Nomarski micrographs of ovules from cleared wild-type (left) and stv1-1 (right) siliques. Inner and outer integuments are shortened, and the female gametophyte is exposed in the stv1-1 ovule (arrow).
(C) Aerial parts of 44-d-old wild-type (left) and stv1-1 (right) plants.
(D) Elongation of primary root of the wild type (circle), stv1-1 (square), and stv1-2 (triangle). Error bars indicate sd (n = 39 for the wild type, n = 18 for stv1-1, and n = 30 for stv1-2).
(E) Epidermal cell length of wild-type, stv1-1, and stv1-2 roots. Root hair-forming cells 5 to 7 mm from the root tip of 10-d-old seedlings were measured. Each bar represents the mean for 10 to 14 individual plants. Error bars indicate sd (n = 51 for the wild type, n = 37 for stv1-1, and n = 47 for stv1-2).
(F) Aerial parts of 23-d-old wild-type (left) and stv1-1 (right) plants.
Bars = 0.5 mm in (A), 0.1 mm in (B), 2 cm in (C), and 5 mm in (F).
Growth was retarded in both aerial and underground parts of stv1-1 mutants, resulting in plants that were smaller overall than the wild type (Figure 2C). In stv1-1 roots, growth rates were reduced (Figure 2D). However, mature root epidermal cells had the same size as those in the wild type (Figure 2E), indicating that cell division but not cell elongation were affected in the mutant root tips. stv1-1 leaves were smaller than wild-type ones and pointed (Figure 2F). These phenotypic observations suggested that STV1 had pleiotropic effects on whole-plant development in addition to specific effects on organs and tissues affected by known auxin signaling mutations.
Genetic Interactions between STV1 and ETT
To investigate the genetic interactions between STV1 and a gene involved in auxin signaling, we examined the phenotype of an stv1-1 ett-2 double mutant. ett-2 is a weak allele mutant of ett, whose gynoecium resembles that of stv1-1, as described above (Figure 1E) (Sessions and Zambryski, 1995). The ett-2 allele has a point mutation at a splice junction that may reduce the amount of ETT mRNA (Sessions et al., 1997). The gynoecia of stv1-1 ett-2 double mutants showed severe reduction of the ovary and morphological abnormalities in the apical region that were similar to ett strong allele mutant phenotypes (Figure 3A) (Sessions and Zambryski, 1995). This result indicated that the stv1-1 mutation enhanced the gynoecium phenotype of the ett-2 mutation. Occasionally, inflorescences of the double mutant failed to produce any normal flower buds, resulting in pin-like structures (Figure 3C) that were not found in either stv1 or ett single mutants (Figures 2C and 3B). This observation indicated that both STV1 and ETT function in flower bud formation. Pin-like inflorescences are formed in mp, pid, and pin1 mutants (Okada et al., 1991; Bennett et al., 1995; Przemeck et al., 1996) and in wild-type plants treated with polar auxin transport inhibitors (Okada et al., 1991). These results supported our proposition that STV1 is associated with auxin signaling.
Figure 3.
Genetic Interaction of STV1 with ETT.
(A) Gynoecia of stv1-1 ett-2 double mutants (left) and ett-1 (right), a strong allele of ett. Bar = 1 mm.
(B) and (C) Shoot apices of ett-2 (B) and stv1-1 ett-2 double mutant (C).
Molecular Cloning of STV1
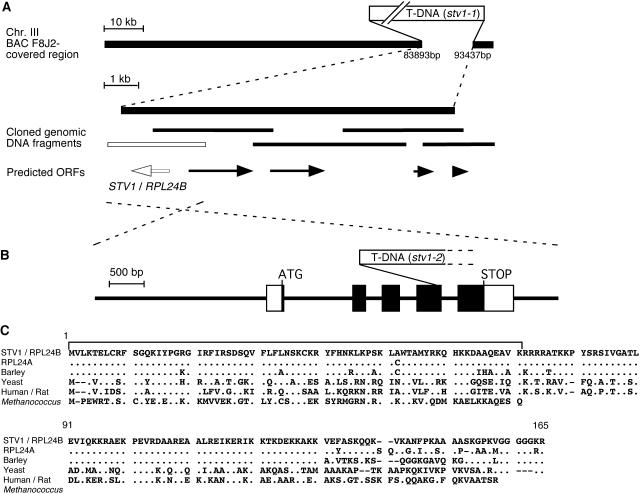
We isolated genomic DNA fragments flanking the T-DNA insert by screening a genomic library made from stv1-1 genomic DNA. We pinpointed the STV1 locus within the region covered with an annotated BAC clone, F8J2, on chromosome III, and found that the T-DNA insertion resulted in the deletion of an entire 10-kb region containing five predicted genes (Figure 4A). We then transformed stv1-1 mutants with wild-type genomic DNA fragments, each containing one of the five predicted genes of the replaced region, and identified a locus, RPL24B (At3g53020), that restored the phenotype of stv1-1 to wild type (e in Figure 1D). Another T-DNA–tagged line of this gene was isolated from a T-DNA insertion collection and was found to have the same phenotype as stv1-1 (d in Figure 1D, and Figures 2D and 2E). We concluded that STV1 is the RPL24B gene and named the new line stv1-2. RPL24B encodes an Arabidopsis homolog of cytosolic RPL24, which is found in archea and higher eukaryotes (Figure 4C). There is another homolog encoded by the RPL24A gene (At2g36620) in Arabidopsis. The STV1 protein and its homolog have highly conserved amino acid sequences in their N-terminal halves (Figure 4C). stv1-1 plants transformed with a truncated STV1 gene encoding only the N-terminal half of the protein (amino acids 1 to 71), which differs from that of the homolog by only one amino acid, were indistinguishable from wild-type plants (data not shown), which suggested that the STV1 protein and its homolog possess equivalent functions.
Figure 4.
Molecular Cloning of STV1.
(A) Genomic structure of the BAC F8J2-covered region. Open boxes represent inserted T-DNAs. Predicted ORFs and cloned genomic DNA fragments for phenotype-rescue experiments are shown below the map as arrows and bars, respectively. Transformation with the DNA fragment represented by the white bar conferred a wild-type phenotype on stv1 plants (see e in Figure 1D). White arrow represents the STV1/RPL24B gene.
(B) Schematic structure of STV1. The solid line corresponds to the cloned STV1 genomic fragment in (A). Closed and open rectangles indicate the protein coding regions and untranslated regions, respectively. A T-DNA inserted in the fourth exon in stv1-2 is shown.
(C) Alignment of RPL24 amino acid sequences from Arabidopsis (STV1/RPL24B and RPL24A), barley, yeast, human/rat, and Methanococcus. Dots represent residues identical to those of STV1. The bracket indicates the amino acid sequences in the C terminus–truncated protein sufficient for STV1 functions (see text).
Expression Pattern of STV1
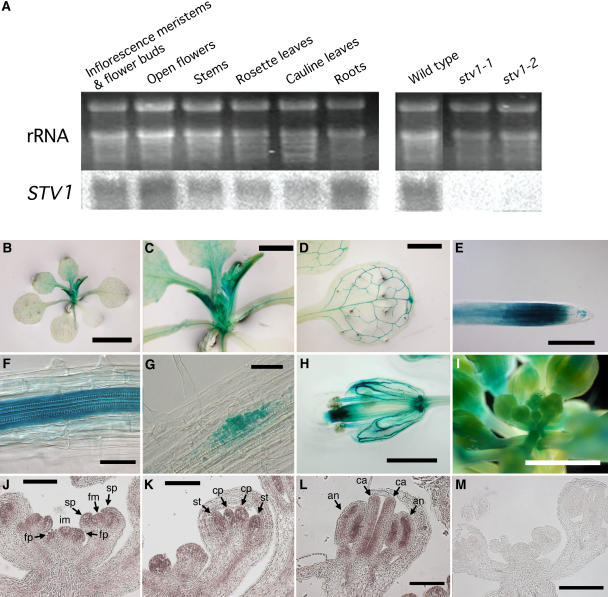
RNA gel blot hybridization using the 3′-half sequence as a probe revealed that STV1 was transcribed in all tested organs (Figure 5A, left panel). STV1 transcripts were not detected in stv1-1 and stv1-2 by the same probe, indicating that this probe did not recognize the transcripts of the RPL24A encoding the homolog (Figure 5A, right panel). However, it remained unknown whether truncated STV1 transcripts were expressed in stv1-2 or not. To determine the temporal and spatial pattern of STV1 expression, we histochemically analyzed transgenic plants carrying the β-glucuronidase (GUS) gene under the control of a 1.1-kb STV1 promoter. Strong GUS staining was observed in shoot apices (Figures 5B and 5C), young developing leaves (Figures 5B and 5C), the vasculature of leaves (Figure 5B and 5D), the cell division region of root tips (Figure 5E), the vasculature of roots (Figure 5F), lateral root primordia (Figure 5G), pollen (Figure 5H), the vasculature of floral organs (Figure 5H), inflorescence apices (Figure 5I), and young flower buds (Figure 5I). Many parts of the stained tissues are known to have high cell proliferation activity. STV1 expression in the early stages of flower development was examined more closely by mRNA in situ hybridization. The STV1 transcript was detected throughout the shoot apices and in young developing flowers (cf. Figures 5J to 5L with 5M), although the signal was strongest in regions with vigorously dividing cells, such as inflorescence and flower meristems (Figure 5J), primordia of floral organs (Figures 5J and 5K), and regions expected to develop male and female gametophytes within anthers and carpels, respectively (Figures 5L). The results showed that STV1 was transcribed in the flower at the early stages of carpel development, including the tissues in which ETT and MP are transcribed (Sessions et al., 1997; Hardtke and Berleth, 1998). STV1 expression in meristematic cells and vasculature is consistent with the typical expression pattern of ribosomal protein genes (Van Lijsebettens et al., 1994; Williams and Sussex, 1995; Ito et al., 2000; Weijers et al., 2001).
Figure 5.
Expression Pattern of STV1.
(A) RNA gel blot analysis of STV1 expression in various organs of the wild type (left panel) and in inflorescence meristems and young flower buds of the wild type, stv1-1, and stv1-2 (right panel). The complementary strand of the 3′-half of the STV1 cDNA was used as a specific probe. Ethidium bromide staining of rRNA serves as a loading control.
(B) to (I) Histochemical GUS staining patterns of transgenic plants carrying the ∼1.1-kb STV1 promoter:GUS fusion construct. T2 plants derived from at least four independent T1 lines were observed.
(B) Aerial parts of a 15-d-old plant.
(C) High-magnification image of (B).
(D) Rosette leaf of a 13-d-old plant.
(E) Root tip.
(F) Root vasculature.
(G) Lateral root primordium.
(H) Open flower.
(I) Inflorescence with young flower buds.
(J) to (M) In situ hybridization analysis of STV1 mRNA abundance in shoot apices and young flowers. im, inflorescence meristem; fm, floral meristem; fp, flower primordium; sp, sepal primordium; cp, carpel primordium; st, stamen primordium; ca, carpel; an, anther.
(J) A longitudinal section of an inflorescence meristem and young flower buds with sepal primordia.
(K) A longitudinal section of a young flower bud with carpel and stamen primordia.
(L) A longitudinal section of a flower initiating the formation of gametophytes.
(M) Sense probe was hybridized as a control.
Bars = 3 mm in (B), 1 mm in (C), (D), (H), and (I), 0.2 mm in (E), 50 μm in (F) and (G), and 0.1 mm in (J) to (M).
Phenotypic Comparison of stv1 with Other Ribosomal Protein Gene Mutants
Previous reports showed that three Arabidopsis ribosomal protein gene mutants, pfl1/rps18, pfl2/rps13, and aml1/rps5, showed growth retardation, reduced fertility, narrow leaves, and defects in vascular patterning resembling that observed in the stv1 mutant (Van Lijsebettens et al., 1994; Ito et al., 2000; Weijers et al., 2001). The overlapping phenotypes of these mutants are to be expected because the genes involved encode components of the same cellular machinery, the ribosome. However, none of these known mutants has any defects in gynoecium patterning (Figure 1F), indicating that STV1 has a unique function in gynoecium development that distinguishes it from other ribosomal protein genes.
ETT and MP Have uORFs
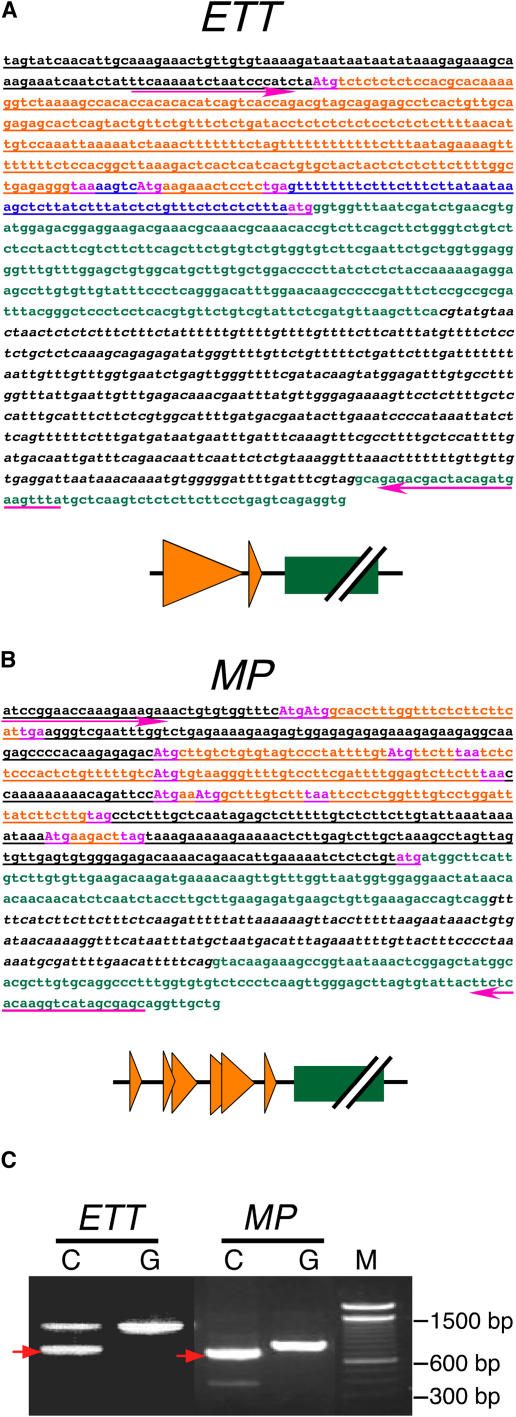
Park et al. (2001) reported that Arabidopsis RPL24, encoded by RPL24A, is associated with translation reinitiation of viral polycistronic RNA in plant cells. This suggested that the stv1 mutation might cause defects in the expression of polycistronic genes or genes with a uORF. Because some uORFs are predicted in the 5′-genomic regions of ETT and MP (Figures 6A and 6B, orange characters), we investigated whether the regions containing these putative uORFs are actually transcribed. Using the primers (Figures 6A and 6B, magenta arrows) designed to cover the region containing the putative uORFs and the first intron (Figures 6A and 6B, italic characters), the 5′-transcript leader sequences were amplified from cDNA derived from young flower buds (Figure 6C). Subsequent sequence analyses for these PCR fragments verified that they were amplified from cDNAs without introns. These studies of the 5′-transcript leader sequences revealed that ETT has at least two and MP at least six uORFs in their 5′-transcript leader sequences (Figures 6A and 6B).
Figure 6.
ETT and MP uORFs.
(A) and (B) Sequences of the 5′-genomic region, including the transcript leader (top) and summary of gene structure (bottom) of ETT (A) and MP (B). Underlined sequences were subcloned into a reporter construct for the transient expression assay and in vitro translation (see Figures 8 and 9). Magenta arrows represent primers used for RT-PCR in (C). Italics represent the first intron. Orange, magenta, and green characters represent uORF sequences, start or stop codons, and main ORF coding regions, respectively. Uppercase characters represent nucleotides mutated in the uORF-eliminated constructs (see Figures 7 to 9). In the summary diagrams, orange triangles represent uORFs, and green boxes represent coding regions.
(C) Amplification of the 5′-transcript leader sequences of ETT and MP by RT-PCR. Total RNA for the template was isolated from wild-type inflorescence meristems and young flower buds (Columbia ecotype). C and G represent cDNA and genomic DNA, respectively. M represents the molecular marker, a 100-bp ladder. Bands indicated by red arrows were excised and sequenced. The resulting sequences were consistent with their predicted 5′-transcript leader sequences.
Partial Suppression of the stv1 Phenotype by uORF-Eliminated ETT Expression
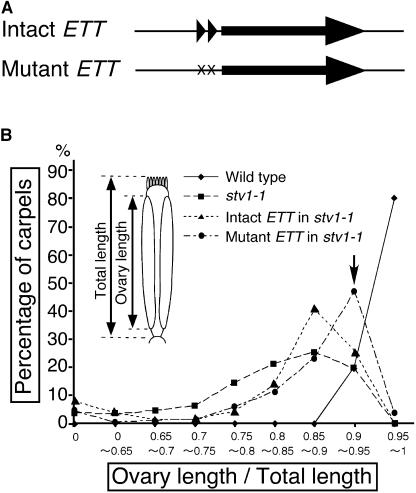
Based on the findings that ETT and MP have uORFs, we hypothesized that full expression of ETT and MP involves translation reinitiation and would thus require STV1. To test this hypothesis, we expressed an ETT version lacking the uORFs in the stv1 background. If the gynoecium defect in stv1 is caused by a perturbation in the efficiency of translation reinitiation of ETT, then making ETT expression independent of translation reinitiation should affect the stv1 gynoecium phenotype. First, we constructed a modified ETT genomic clone in which the start codons of the two uORFs were changed from ATG to TTG, to prevent uORF translation initiation (Figures 6A and 6B, uppercase characters, and Figure 7A), and then we transformed either the modified clone (mutant ETT) or the unmodified clone (intact ETT) into an ett-2 mutant to confirm that the genomic regions introduced are sufficient for ETT function. The T1 lines expressing both constructs showed complementation of the gynoecium defect (data not shown). There were apparent morphological differences between plants carrying intact or mutant ETT genes. We then crossed two independent T1 lines for each construct with stv1-1 and examined the gynoecium structures of the established F3 and F4 generations in the stv1-1 background. The most frequent ratio of ovary length to total gynoecium length observed in stv1-1 lines carrying the mutant ETT construct was intermediate between those of stv1-1 and the wild type (Figure 7B), suggesting that expression of the form of ETT lacking the uORFs partially suppressed the stv1 gynoecium phenotype. Plants expressing the intact ETT showed a slight suppression of the stv1 gynoecium phenotype, but the effect was weaker than in plants expressing the mutant ETT (Figure 7B). Additional copies of the ETT gene might affect the gynoecium patterning in stv1 by increasing ETT expression levels.
Figure 7.
Transformation with ETT Lacking the uORFs Partially Suppresses the stv1 Gynoecium Phenotype.
(A) Overview of the genomic subclone constructs used for the transformation experiments in (B). Constructs include an ∼7.2-kb genomic fragment from the ETT locus, with (intact ETT) or without (mutant ETT) uORFs. The construct without uORFs has point mutations in the upstream ATGs (see Figure 6A). Triangles and arrows represent the uORFs and ETT ORFs, respectively.
(B) Frequency distribution of the ratio of ovary length to total carpel length of the wild type (diamond), stv1-1 (square), and stv1-1 transformed with intact ETT (intact ETT in stv1-1; triangle) or with mutant ETT (mutant ETT in stv1-1; circle). The abscissa is the ratio of ovary length to total length (diagram shown in the chart), and the ordinate is the percentage of carpels in each ratio category. Ovary and total lengths were from fully elongated siliques. The arrow indicates the most frequent ratio observed in stv1-1 transformed with mutant ETT.
As mentioned above, one side of the ovary of stv1-1 gynoecia was completely lost in extreme cases (4%). The frequency at which this severe malformation occurred was not decreased in the transgenic plants (7.8% in plants expressing intact ETT; 4.5% in plants expressing mutant ETT), indicating that the complete loss of ovaries occasionally observed in stv1 may be caused by different mechanisms than the defect of apical-basal patterning. These findings were consistent with our hypothesis that STV1 is involved in uORF-mediated expression of functional ETT.
The uORFs of ETT and MP Repress Translation of Downstream ORFs
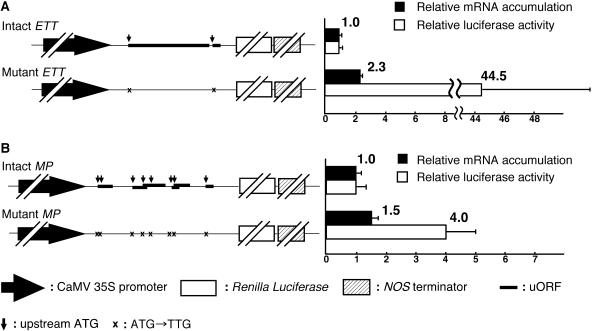
It is widely thought that in some cases uORFs can affect the translation of a downstream major ORF specifying a gene product. Therefore, we examined whether the uORFs associated with ETT or MP regulate the expression of the major downstream ORF. We inserted the ETT and MP 5′-upstream sequences including the 5′-transcript leader sequences (Figures 6A and 6B, underlined characters), with or without mutations in the translation start codons of the uORFs (Figures 6A and 6B, uppercase characters), between a constitutive promoter and the luciferase coding sequence (Figures 8A and 8B, left panels). We then introduced the constructs into Arabidopsis mesophyll protoplasts. Right panels in Figures 8A and 8B show relative luciferase activities (white bars) and mRNA accumulations (black bars) measured after overnight incubation. The luciferase activity produced from the ETT construct without uORFs was 44.5-fold higher than that from the intact construct. An increase of the luciferase activity was also observed in the case of MP, where the construct lacking the uORF gave fourfold higher activity than the intact one. On the other hand, the amounts of mRNA derived from the constructs in which uORFs had been eliminated were 2.3- and 1.5-fold higher than those from the intact ETT and MP, respectively. Thus, the major cause for the enhancement of luciferase activity in the absence of uORFs was the efficiency of translation and not the accumulation of mRNA.
Figure 8.
Effects of ETT and MP uORFs on the Expression of Downstream ORFs in Mesophyll Protoplasts.
Constitutive CaMV 35S promoter–driven Renilla luciferase genes that had been fused to intact (with uORFs) or mutant forms (without uORFs) of the ETT (A) and MP (B) 5′-transcript leader sequences were transiently expressed in Arabidopsis mesophyll protoplasts, and luciferase activity and mRNA accumulation were analyzed after overnight incubation (see Methods). Symbols used in the description of constructs on the left are defined below the figure. Open and closed bars in the chart on the right indicate luciferase activity and mRNA accumulation, respectively. Results for the Renilla luciferase activity and mRNA accumulation were normalized with respect to the activity and mRNA accumulation, respectively, of the cointroduced firefly luciferase gene. Then, the luciferase activity and mRNA accumulation of the mutant fusion constructs relative to those of the intact fusion constructs were calculated. The data are given as means of three independent assays, and error bars indicate sd.
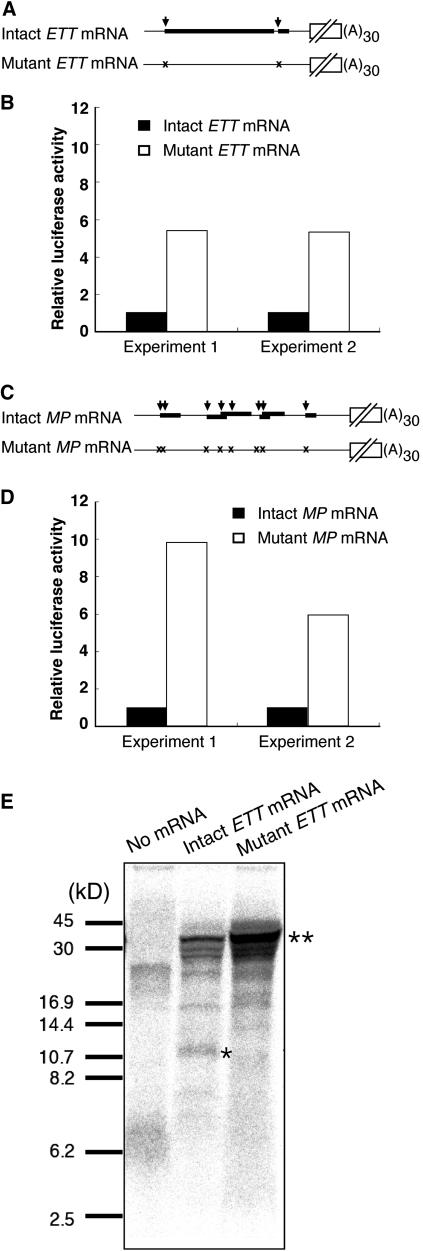
We tested the hypothesis that the ETT and MP uORFs decrease the translation levels of the downstream ORFs using wheat germ extract as an in vitro translation system. We synthesized luciferase mRNA fused to ETT or MP 5′-upstream sequences with or without uORFs (Figures 6A and 6B, underlined characters) in vitro and added a 5′-cap structure and a poly(A) tail (Figures 9A and 9C). Duplicate assays using independently synthesized mRNA showed increased translation efficiency of ETT when the uORF had been eliminated (Figure 9B). Similar results were obtained for MP (Figure 9D). These in vivo and in vitro analyses suggested that the ETT and MP uORFs control the expression of the downstream main ORFs by repression of their translation.
Figure 9.
ETT and MP uORFs Affect the Translation of Downstream ORFs in an in Vitro System.
(A) to (D) Equivalent amounts of Renilla luciferase mRNAs fused to intact (with uORFs) or mutant forms (without uORFs) of the ETT ([A] and [B]) or MP ([C] and [D]) 5′-transcript leader sequences were subjected to in vitro translation using wheat germ extract, and luciferase activity was measured. Structures of the mRNAs used for the assay ([A] and [C]) and relative luciferase activities ([B] and [D]) are shown. Symbols in (A) and (C) are the same as in Figure 8. Open and closed bars in (B) and (D) indicate relative luciferase activity of intact and mutant form mRNA, respectively. Renilla luciferase activity was normalized against the activity of cotranslated firefly luciferase mRNA. The luciferase activity relative to that of the intact form mRNA was then calculated. Assays were performed twice for independently synthesized mRNAs.
(E) 35S-Met–labeled products of in vitro translation were analyzed by SDS-PAGE on a 16.5% Tris-Tricine SDS polyacrylamide gel. Molecular masses of protein standards are shown on the left. The predicted size of the ETT first uORF is 11 kD (single asterisk) and that of the Renilla luciferase is 36 kD (double asterisk).
The First ETT uORF Is Translated in Vitro
To investigate whether uORFs are translated, in vitro translation was performed in the presence of 35S-Met. A peptide of the size predicted from the first uORF sequence of the ETT gene (11 kD) was detected only when the uORF was actually present, indicating that this uORF sequence was translated (Figure 9E, single asterisk). However, we could not find a peptide corresponding to the second uORF, possibly because the size of the predicted peptide, 0.5 kD, was too small for detection. The amount of peptide corresponding to luciferase (36 kD) synthesized from the ETT fusion mRNA that lacked the uORFs was larger than that from the intact mRNA (Figure 9E, double asterisk). This result was consistent with the difference of luciferase activity. In the case of MP, the amount of luciferase increased similarly, but no small peptides that might have been encoded by the uORFs (predicted sizes were below 2.9 kD) could be detected in our system (data not shown).
DISCUSSION
Specific Role of RPL24 in Gynoecium Pattern Formation
Some ribosomal proteins have specific functions in addition to those required for general protein synthesis (Wool, 1996). Arabidopsis RPL24 plays a pivotal role in translation reinitiation of polycistronic CaMV 35S RNA in plant cells (Park et al., 2001). In this paper, phenotypic analyses of stv1, a mutant of RPL24, revealed that in addition to its ribosome-related role in whole-plant growth and the development of several organs, RPL24 functions specifically in auxin-mediated apical-basal patterning of the gynoecium. This additional function has not been shown for other ribosomal proteins so far and could be explained if RPL24 affected the translation reinitiation of genes responsible for gynoecium development.
We suggest that ETT and MP, mutants of which show similar gynoecium phenotypes as stv1, are candidate genes whose translation could be affected by RPL24. This hypothesis is supported by three observations in this study: (1) ETT and MP possess uORFs that affect the expression of the downstream ORF, (2) the gynoecium phenotype of a weak ett allele was enhanced by a stv1 mutation, and (3) the gynoecium phenotype of stv1 was partially complemented by transformation with the ETT gene lacking its uORFs. The enhancement of the weak ett allele by stv1 indicated that a loss of RPL24 activity might cause a decrease of ETT translation, while the partial complementation of stv1 by the expression of uORF-less ETT suggested that the removal of the uORFs abolished the dependence of ETT expression on RPL24 and increased the level of ETT protein in stv1. The quantification of ETT and MP protein levels in wild-type and stv1 plants will clarify the role of RPL24 in ETT and MP translation. We do not, however, discard the possibility that other genes are also involved in the RPL24-dependent regulation of gynoecium development.
The N-terminal half of RPL24, the amino acid sequence of which is highly conserved in archea and eukaryotes, is sufficient to bind to the CaMV TAV protein that controls translation reinitiation (Park et al., 2001). Our study demonstrated that the N-terminal half is sufficient for all STV1 functions in Arabidopsis. These findings suggest that the specific role of RPL24 in translation reinitiation might also be conserved in other organisms. Indeed, mammal and fungal cells contain a subset of genes whose expression is regulated by uORFs (Geballe and Sachs, 2000; Morris and Geballe, 2000). Recently, it was reported that a mouse mutant lacking RPL24 shows slow progression of the cell cycle and tissue-specific defects during development (Oliver et al., 2004). It would be interesting to test whether some aspects of the mutant phenotype are owing to altered expression of uORF-regulated genes.
uORFs of the ETT and MP Genes
There are several uORFs in the ETT and MP 5′-transcript leader sequences. We demonstrated that a peptide encoded by the first uORF of ETT was translated in vitro, but peptides from the second ETT uORF and the MP uORFs were not detected. Investigation of the sequence context around the start codons (uATGs) of the ETT and MP uORFs showed that there are purine residues at position −3 (with respect to the A of the ATG codon) in the second uATG of ETT and in five of the eight uATGs of MP. This purine residue at position −3 is the most important nucleotide for the initiation of translation by scanning ribosomes (Kozak, 1986). We assume that the uORFs carrying a purine at position −3 probably were translated and that the peptides could not be detected because of their small sizes (the predicted molecular mass of the largest peptide was only 2.9 kD).
uORFs are involved not only in the regulation of translation but also in the control of mRNA stability (Vilela et al., 1999; Ruiz-Echevarria and Peltz, 2000). The elimination of the uORFs of ETT and MP resulted in 1.5- to 2.3-fold accumulation of mRNAs in protoplasts transiently expressing the 5′-leader-reporter gene fusions, indicating that the ETT and MP uORFs might affect mRNA stability. On the other hand, the reporter activities of translation products were increased by point mutations of the uATGs to a greater extent than the accumulation of mRNA. In fact, mRNAs that lacked the uORFs were translated more effectively by an in vitro translation system than the complete mRNAs. These findings suggested that the ETT and MP uORFs repress the expression of the downstream ORF at the translational level.
After termination of uORF translation, the efficiency of the reinitiation of the translation of the downstream ORF may depend mainly on the length of the uORF and the intercistronic distance between the uORF stop codon and the start codon of the main ORF (Hinnebusch, 1996; Geballe and Sachs, 2000; Morris and Geballe, 2000). Previous observations that increases in the length of uORFs or in the size of the intercistronic spacer resulted in the inhibition or enhancement, respectively, of downstream ORF translation gave rise to the hypothesis that recharging of the ribosome with initiation factors is required for translation reinitiation. Ribosomes translating longer uORFs may take longer time for recharging because they shed the factors, and longer intercistronic spacers may permit additional time for recharging. The second ETT and all MP uORFs consist of <75 nucleotides, short enough to reinitiate translation at the downstream ORF (Rajkowitsch et al., 2004). However, the first uORF of ETT is 279 nucleotides long. In the case of CaMV 35S RNA, ORF VII, a 291-nucleotide ORF, is upstream of ORF I, which is translated effectively with TAV (Bonneville et al., 1989), and RPL24 is involved in its translation reinitiation (Park et al., 2001). Therefore, RPL24 might play a role in the translation regulation by long uORFs in ETT as in 35S RNA. The lengths of intercistronic spaces in the ETT 5′-leader and in MP are 61 and 91 nucleotides, respectively. They appear long enough for efficient reinitiation, compared with the intercistronic length of other uORF-regulated genes, such as ORF I of CaMV 35S RNA (60 nucleotides; Bonneville et al., 1989).
There are two types of uORF-mediated translational control, one dependent on and the other independent of the amino acid sequences of the peptides encoded by the uORFs (Geballe and Sachs, 2000; Morris and Geballe, 2000). The peptide sequence encoded by the ATB2/(At)bZIP11 uORF is conserved in the 5′-transcript leader sequences of some other basic domain/leucine zipper (bZIP) genes in Arabidopsis and other plants, suggesting that these amino acid sequences could be crucial for the uORF-mediated translational control of ATB2/(At)bZIP11 (Wiese et al., 2004). On the other hand, the translational control of the maize Lc gene does not depend on the amino acid sequences of the uORF peptides (Wang and Wessler, 1998). In the case of the ETT and MP uORFs, there is no conservation of the number, length, or amino acid sequence of the encoded peptides, even though the uORFs of both genes affect the translation of the downstream major ORF. These differences raise the possibility that translational control by ETT and MP uORFs might be independent of the amino acid sequences. Alternatively, translation of ETT and MP could be regulated by different mechanisms, which might be related to the finding that ETT and MP have distinct effects, namely activation and repression of the transcription of auxin response genes (Ulmasov et al., 1999; Tiwari et al., 2003).
uORF-Mediated Translational Control and the Auxin Response
uORF-mediated translational control is responsible for polyamine homeostasis and sucrose sensing in plants (Hanfrey et al., 2002; Wiese et al., 2004). In these cases, polyamine and sucrose induce the uORF-mediated translation repression of the S-adenosylmethionine decarboxylase and ATB2/(At)bZIP11 genes, respectively. Similarly, the uORF-mediated translational control of ETT and/or MP expression could be regulated by extracellular or intracellular signals. Specifically, these signals might be associated with the auxin response because ETT and MP encode ARF proteins, which can regulate the expression of auxin response genes (Sessions et al., 1997; Hardtke and Berleth, 1998).
The Arabidopsis Information Resource database (http://www.arabidopsis.org/home.html) thus far includes nine ARF genes with apparent uORFs (see Supplemental Figure 1 online). The other ARF genes could also have uORFs, but their 5′-transcript leader sequences are not known. It is plausible, therefore, that uORF-dependent translational regulation may be a common aspect of the auxin response. An Arabidopsis mutant with a lesion in a subunit of the translation initiation factor eIF3h, which is responsible for uORF-mediated translation, shows altered transcription of early auxin response genes (Kim et al., 2004), a finding that supports our hypothesis. Another possibility is that other environmental and hormone signals could regulate the uORF-controlled translation of ARF proteins because there is crosstalk between the response pathways for auxin and these other signals (Swarup et al., 2002; Halliday, 2004; Li et al., 2004). Further analyses would clarify the significance of uORF-mediated translational control in auxin signaling.
METHODS
Plant Material and Growth Conditions
stv1-1 was isolated by screening a CaMV 35S promoter:CPC transgenic plant population, in which the absence of trichomes is a dominant visible marker for the transgene (Wada et al., 1997). stv1-2 is a line, CS11180, identified by PCR screening of K. Feldmann's T-DNA insertion collection, provided by the ABRC (Feldmann, 1991; McKinney et al., 1995). ett-2 and pfl1 were also provided by the ABRC. pid-8 was kindly provided by D. Smyth, pfl2 by T. Ito and K. Shinozaki, and aml1 by D. Weijers and R. Offringa. The ecotypes of the wild type, stv1-1, stv1-2, ett-2, and pid-8 are Wassilewskija, pfl1 and aml1 are C24, and pfl2 is Nossen. The genotypes of each mutation in the double mutant were verified by PCR. Primers used for the stv1-1 mutation were 5′-TGCGGATAAGGTAATTGCC-3′, 5′-CCAATATTCATGGTGTTCCT-3′, and 5′-AAGAAGAGAGCTGAGAAGCC-3′, and for the ett-2 mutation, 5′-GGTCTAATACTCCTCACATG-3′ and 5′-CTTAAGGCTTCAGGAGATTC-3′. Amplified DNA fragments were digested with AflII for ett-2 analysis. The genotype of the aml1 heterozygote was verified by PCR using the following primers: 5′-GCTTCGTCAATCATTTACCATATT-3′, 5′-AAATAGTATCATTGGAACAAGGAA-3′, and 5′-CACAGTTTTCGCGATCCAGACTG-3′.
Seeds were sown on the surface of vermiculite in small pots and incubated for 3 d at 4°C. Plants were grown under continuous white fluorescent light at 22°C. Growth conditions for observation of underground plant parts were described previously (Nishimura et al., 2003).
Phenotypic Analyses of stv1
Siliques, leaves, and roots were studied with an M420 microscope (Leica). Microscopy images were taken with a Coolpix 990 digital camera (Nikon). Scanning electron microscopy was performed as described previously (Matsumoto and Okada, 2001). For the observation of vasculature, 12-d-old seedlings were fixed overnight in acetic acid:ethanol (9:1, v:v). After replacement with 70, 50, and 30% ethanol, samples were cleared in chloral hydrate:glycerol:water (8:1:2, w:v:v).
Molecular Cloning of STV1
A genomic region containing the STV1 locus was isolated by screening an stv1-1 genomic library constructed in the λFIXII vector (Stratagene) using the T-DNA region as a probe. Genomic DNA fragments covering the deleted region in stv1-1, corresponding to regions 83663 to 86400, 84881 to 88202, 87679 to 91911, 90270 to 93560, and 92558 to 94619 of the F8J2 BAC vector, were amplified by PCR from F8J2 using the Expand High-Fidelity PCR system (Roche Diagnostics) and subcloned into the pPZP221 binary vector (Hajdukiewicz et al., 1994). Subcloned DNA fragments were transformed into stv1-1 by vacuum infiltration using Agrobacterium tumefaciens strain ASE for phenotype rescue. cDNA of STV1 was amplified by RT-PCR from total RNA isolated from inflorescence meristem and young flower buds of Landsberg erecta ecotype with the Superscript II first-strand synthesis system (Invitrogen), cloned into the pT7 blue vector (Novagen), and sequenced. The construct for expression of the N-terminal half of the STV1 protein was made by self-ligation of a DNA fragment amplified from the STV1 genomic clone using KOD plus polymerase (Toyobo) and the inversely directed primers 5′-CTTCTTGGTGGCACGTCTCCTTCT-3′ and 5′-TGAAGAGCTTAAAGCCATCTTTTC-3′.
Expression Analyses of STV1
For RNA gel blot analyses, total RNA was isolated with the RNeasy plant mini kit (Qiagen). Total RNA (10 μg) was separated on a 1.5% agarose/formaldehyde gel, transferred to a Hybond N+ membrane (Amersham Biosciences), and probed with the 3′-sequences of STV1 cDNA. The sequences for the probe were amplified from the cDNA clone by PCR using the primers 5′-AGGACGAGAAGAAGGCAAAGAAGG-3′ and 5′-CTCAAGTCTGATATTATAAGAGTAGCAAAA-3′.
A promoter-GUS construct for STV1 was made by insertion of the promoter region of STV1 (∼1.1 kb), amplified by PCR, in front of the GUS gene of the pBI101 binary vector (Clontech). The primers were 5′-AGCTGTTTATGCAAGTAGTC-3′ and 5′-CATGGTAGTAGCTGCTCCTC-3′. The construct was transformed into the Columbia ecotype by vacuum infiltration as described above. Histochemical GUS staining was performed by the methods described previously by Donnelly et al. (1999) for aerial parts and by Malamy and Benfey (1997) for roots.
In situ mRNA hybridization was performed as described previously (Matsumoto and Okada, 2001). The probe for the 3′-sequence was amplified from the cDNA clone using the primers 5′-GGCAAACGCTGAAGAGCTTAAA-3′ and 5′-AGGACGAGAAGAAGGCAAAGAAGG-3′.
Sequencing of the 5′-Transcript Leaders of ETT and MP
The 5′-transcript leader sequences of ETT and MP were amplified from first-strand cDNAs derived from inflorescence meristems and young flower buds of the Columbia ecotype using the primers shown in Figure 6 (magenta arrows), subcloned into the pT7 Blue vector, and sequenced.
Transient Expression Assay
The 5′-upstream regions of ETT and MP (Figures 6A and 6B, underlined characters) were amplified from the BAC clones T1B8 and F6K9, respectively, using the KODplus DNA polymerase and inserted between the constitutive CaMV 35S promoter and the Renilla luciferase gene of the 221-hRL vector. The 221-hRL vector, which contains the CaMV 35S promoter, the Renilla luciferase gene, and the nopaline synthase gene (NOS) terminator, was made from pBI221 (Clontech) by replacing GUS with the Renilla luciferase gene derived from the hRL-null vector (Promega). The subcloned regions are shown in Figure 6. Site-directed mutagenesis was achieved by PCR. The mutagenized DNA fragments were amplified from the subclones using the KODplus DNA polymerase and inversely directed primers carrying point mutations and self-ligated. The mutagenesis was repeated in the case of multipoint mutations. The 221-luc+ vector harboring a firefly luciferase gene driven by the 35S promoter (Matsuo et al., 2001) was used as an internal control.
Transient transformation of the reporter fusion genes into Arabidopsis thaliana mesophyll protoplasts was based on the method for suspension cells described previously (Ueda et al., 2001). More than 20 rosette leaves of plants (Wassilewskija ecotype) growing under continuous white light were cut into narrow strips with a razor blade and were incubated in 25 mL of enzyme solution (0.4 M mannitol, 5 mM EGTA, 1% [w/v] cellulase Y-C [Kyowa Chemical Products], and 0.05% [w/v] pectolyase Y-23 [Kyowa Chemical Products]) for 30 min at room temperature under vacuum and then for 1 h at 30°C with gentle agitation. The enzyme solution was passed through nylon mesh (120-μm pore size) and centrifuged at 100g for 5 min to harvest mesophyll protoplasts. The protoplasts were washed twice with 25 mL of solution A (0.4 M mannitol, 70 mM CaCl2, and 5 mM MES, pH 5.7) and resuspended in MS-mannitol medium (Murashige and Skoog cell culture medium [Murashige and Skoog, 1962] supplemented with 0.4 M mannitol and 5 mM MES, pH 5.7). Five micrograms of the Renilla luciferase-fusion construct, 3 μg of 221-luc+, and 2 μg of salmon sperm DNA were added to 0.2 mL of the MS-mannitol solution containing 106 to 107 protoplasts, together with 0.2 mL of polyethylene glycol solution [25% PEG6000, 100 mM Ca(NO3)2, and 450 mM mannitol, pH 9]. The mixture was incubated at room temperature for 15 min and then diluted with 5 mL of Ca(NO3)2. The protoplasts were collected by centrifugation at 100g for 5 min, resuspended in 8 mL of the MS-mannitol solution, and incubated at 22°C for 16 h in the dark. After washing with 5 mL of 0.4 M mannitol, reporter activity was measured by the Dual-Reporter assay system (Promega) with a Lumat LB9507 luminometer (Berthold Technology).
mRNA accumulation was quantified by real-time RT-PCR. Total RNA was isolated from protoplasts with the RNeasy plant mini kit. On-column DNase digestion (Qiagen) was performed during the RNA purification. After an additional DNase I treatment (Invitrogen), the first-strand cDNA was synthesized as described above. All reactions were performed also without the reverse transcriptase as a control. Real-time PCR was subjected by the Taqman sequence detection system with an ABI-Prism 7500 sequence detector (Applied Biosystems). The Renilla luciferase gene was amplified with the primers 5′-GCGACGATCTGCCTAAGATGT-3′ and 5′-CGACAATAGCGTTGGAAAAGAA-3′ and detected with the FAM-labeled minor groove binder probe 5′-TCGAGTCCGACCCTGG-3′. The firefly luciferase gene was amplified with the primers 5′-TGCACATATCGAGGTGGACATC-3′ and 5′-GCCAACCGAACGGACATTT-3′ and detected with the VIC-labeled minor groove binder probe 5′-CTTACGCTGAGTACTTC-3′. Amounts of mRNA were calculated by subtraction of the value of the reaction without reverse transcriptase from that with reverse transcriptase, normalized with respect to the cointroduced firefly luciferase gene. cDNA from protoplasts transformed with the 221-hRL vector and the 221-luc+ vector was used as a standard control for each independent experiment.
Construction and Observation of Transgenic stv1 Plants Expressing ETT with or without uORFs
The ETT genomic region corresponding to 79,439 to 86,552 of the T1B8 BAC clone was amplified from T1B8 by PCR with the KODplus DNA polymerase and subcloned into the pPZP221NP binary vector. The pPZP221NP binary vector was made from pPZP221 by replacing the CaMV 35S promoter with the NOS promoter derived from pBI101 (Nishimura et al., 2003). The mutant ETT genomic clone was constructed by complete replacement of the 5′-upstream region of the ETT genomic clone with that of the mutant 5′-upstream-luciferase fusion construct described above. These ETT genomic clones were transformed into ett-2 plants by vacuum infiltration to verify the recovery of the wild-type phenotype. For each construct, two independent transgenic ett-2 plants were crossed with the stv1-1 mutant to generate stv1-1 homozygotes with the transgene. From F3 or F4 transgenic stv1-1 homozygotes, we dissected ∼10 fully elongated siliques per individual from the 6th to 15th flowers on the primary or secondary shoots. The total number of dissected carpels was 400 for the wild type, 396 for stv1-1, 238 and 280 for the two independent transgenic lines of intact ETT, and 720 and 380 for the two independent transgenic lines of mutant ETT. Lengths of the whole gynoecium and ovary region were determined from micrographs with the scientific image analysis program Image-Pro Plus (Media Cybernetics).
In Vitro Transcription and Translation
In vitro transcription and translation were performed as described previously (Chiba et al., 2003), with a minor modification. Constructs for in vitro transcription are made by inserting the 5′-upstream-Renilla luciferase fusion gene described above for the transient assay between the XbaI and SacI sites of the pSP64 poly(A) vector (Promega) containing the SP6 promoter and poly(A)30. The plasmid DNAs were linearized with EcoRI and transcribed using the AmpliCap SP6 high yield message maker kit (Epicentre Technologies). After treatment with RNase-free DNase I, the transcripts were purified using the RNeasy plant mini kit and the QuickPrep Micro mRNA purification kit (Amersham Biosciences). In vitro translation was performed in a reaction mixture containing 12.5 μL of wheat germ extract (Promega), 2 μL of 1 mM amino acid mixture lacking Met (Promega), 2.5 μL of 1 mM l-Met, 25 units of RNasin (Promega), 200 fmol of Renilla luciferase fusion mRNA, and 400 fmol of the Luciferase control RNA (Promega) as a control, for total volume of 25 μL. Reactions were performed at 25°C for 25 min. Luciferase activities were measured as described above.
For electrophoresis of translated products, the reaction was performed in a reaction mixture containing 12.5 μL of wheat germ extract, 2 μL of 1 mM amino acid mixture lacking Met, 25 units of RNasin, 25 μCi of 35S-Met (Amersham Bioscience), Complete Mini EDTA-free protease inhibitor cocktail (Roche Diagnostics), and 1 pmol of Renilla luciferase fusion mRNA in a total volume of 25 μL. Reactions were performed at 25°C for 40 min, and samples were separated on a 16.5% Tris-Tricine SDS polyacrylamide gel (Schägger and von Jagow, 1987). The gel was treated with a fluorographic enhancer, ENLIGHTNING (Perkin-Elmer Life and Analytical Sciences), and examined with a BAS1500 Bio-Imaging analyzer (Fuji Photo Film).
Accession Numbers
Sequence data from this article can be found in the GenBank data library under accession numbers AB199790 (STV1 cDNA), AB199791 (the 5′-region of ETT cDNA), and AB199792 (the 5′-region of MP cDNA). Protein IDs of the amino acid sequences used for the alignment are AAD20138 (RPL24A), P50888 (barley), P04449 (yeast), P83731 (human), P83732 (rat), and P54064 (Methanococcus). Accession numbers of the BAC clone sequences are AL132969 (F8J2), U78721 (T1B8), and AC007797 (F6K9).
Supplemental Data
The following material is available in the online version of this article.
Supplemental Figure 1. uORFs of Arabidopsis ARF Genes.
Supplementary Material
Acknowledgments
The authors thank David R. Smyth, Takuya Ito, Kazuo Shinozaki, Dolf Weijers, Remko Offringa, Kazuyuki Hiratsuka, Hidetaka Kaya, Masaaki Umeda, Sayaka Inada, Ryoko Sakurai, Hitoshi Onouchi, Satoshi Naito, Tatsuya Sakai, Keisuke Matsui, Sumie Ishiguro, Seiji Takeda, Yuhei Tsuchida, Noritaka Matsumoto, Ryuji Tsugeki, and all members of the Okada laboratory for materials, helpful discussions, and technical advice. We also thank the ABRC at Ohio State University and Kazusa DNA Research Institute for providing seeds and DNA. This work was supported by grants from the Japanese Ministry of Education, Science, Culture, and Sports (a Grant-in-Aid for Science Research on Priority Area 14036220 and the Grant for Biodiversity Research of the 21st Century Centers of Excellence [A14]). T.N. was a Junior Research Associate recipient of RIKEN and a research fellow of the Japan Society for the Promotion of Science.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Kiyotaka Okada (kiyo@ok-lab.bot.kyoto-u.ac.jp).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.036533.
References
- Ban, N., Nissen, P., Hansen, J., Moore, P.B., and Steitz, T.A. (2000). The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289, 905–920. [DOI] [PubMed] [Google Scholar]
- Baronas-Lowell, D.M., and Warner, J.R. (1990). Ribosomal protein L30 is dispensable in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 10, 5235–5243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett, S.R.M., Alvarez, J., Bossinger, G., and Smyth, D.R. (1995). Morphogenesis in pinoid mutants of Arabidopsis thaliana. Plant J. 8, 505–520. [Google Scholar]
- Berleth, T., Krogan, N.T., and Scarpella, E. (2004). Auxin signals—Turning genes on and turning cells around. Curr. Opin. Plant Biol. 7, 553–563. [DOI] [PubMed] [Google Scholar]
- Bonneville, J.M., Sanfaçon, H., Fütterer, J., and Hohn, T. (1989). Postrranscriptional trans-activation in cauliflower mosaic virus. Cell 59, 1135–1143. [DOI] [PubMed] [Google Scholar]
- Bowman, J.L., Baum, S.F., Eshed, Y., Putterill, J., and Alvarez, J. (1999). Molecular genetics of gynoecium development in Arabidopsis. Curr. Top. Dev. Biol. 45, 155–205. [DOI] [PubMed] [Google Scholar]
- Chiba, Y., Sakurai, R., Yoshino, M., Ominato, K., Ishikawa, M., Onouchi, H., and Naito, S. (2003). S-adenosyl-L-methionine is an effector in the posttranscriptional autoregulation of the cystathionine γ-synthase gene in Arabidopsis. Proc. Natl. Acad. Sci. USA 100, 10225–10230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen, S.K., Dagenais, N., Chory, J., and Weigel, D. (2000). Regulation of auxin response by the protein kinase PINOID. Cell 100, 469–478. [DOI] [PubMed] [Google Scholar]
- Donnelly, P.M., Bonetta, D., Tsukaya, H., Dengler, R.E., and Dengler, N.G. (1999). Cell cycling and cell enlargement in developing leaves of Arabidopsis. Dev. Biol. 215, 407–419. [DOI] [PubMed] [Google Scholar]
- Dresios, J., Derkatch, I.L., Liebman, S.W., and Synetos, D. (2000). Yeast ribosomal protein L24 affects the kinetics of protein synthesis and ribosomal protein L39 improves translational accuracy, while mutants lacking both remain viable. Biochemistry 39, 7236–7244. [DOI] [PubMed] [Google Scholar]
- Feldmann, K.A. (1991). T-DNA insertion mutagenesis in Arabidopsis: Mutational spectrum. Plant J. 1, 71–82. [Google Scholar]
- Friml, J., Vieten, A., Sauer, M., Weijers, D., Schwarz, H., Hamann, T., Offringa, R., and Jürgens, G. (2003). Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426, 147–153. [DOI] [PubMed] [Google Scholar]
- Friml, J., et al. (2004). A PINOID-dependent binary switch in apical-basal PIN polar targeting directs auxin efflux. Science 306, 862–865. [DOI] [PubMed] [Google Scholar]
- Gälweiler, L., Guan, C., Müller, A., Wisman, E., Mendgen, K., Yephremov, A., and Palme, K. (1998). Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282, 2226–2230. [DOI] [PubMed] [Google Scholar]
- Geballe, A.P., and Sachs, M.S. (2000). Translational control by upstream open reading frames. In Translational Control of Gene Expression, N. Sonenberg, J.W.B. Hershey, and M. Mathews, eds (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp. 595–614.
- Hajdukiewicz, P., Svab, Z., and Maliga, P. (1994). The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25, 989–994. [DOI] [PubMed] [Google Scholar]
- Halliday, K.J. (2004). Plant hormones: The interplay of brassinosteroids and auxin. Curr. Biol. 14, R1008–R1010. [DOI] [PubMed] [Google Scholar]
- Hanfrey, C., Franceschetti, M., Mayer, M.J., Illingworth, C., and Michael, A.J. (2002). Abrogation of upstream open reading frame-mediated translational control of a plant S-adenosylmethionine decarboxylase results in polyamine disruption and growth perturbations. J. Biol. Chem. 277, 44131–44139. [DOI] [PubMed] [Google Scholar]
- Hardtke, C.S., and Berleth, T. (1998). The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J. 17, 1405–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch, A.G. (1996). Translational control of GCN4: Gene-specific regulation by phosphorylation of eIF2. In Translational Control, J.W.B. Hershey, M. Mathews, and N. Sonenberg, eds (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp. 199–244.
- Ito, T., Kim, G.-T., and Shinozaki, K. (2000). Disruption of an Arabidopsis cytoplasmic ribosomal protein S13-homologous gene by transposon-mediated mutagenesis causes aberrant growth and development. Plant J. 22, 257–264. [DOI] [PubMed] [Google Scholar]
- Kim, T.-H., Kim, B.-H., Yahalom, A., Chamovitz, D.A., and von Arnim, A.G. (2004). Translational regulation via 5′ mRNA leader sequences revealed by mutational analysis of the Arabidopsis translation initiation factor subunit eIF3h. Plant Cell 16, 3341–3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak, M. (1986). Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell 44, 283–292. [DOI] [PubMed] [Google Scholar]
- Li, H., Johnson, P., Stepanova, A., Alonso, J.M., and Ecker, J.R. (2004). Convergence of signaling pathways in the control of differential cell growth in Arabidopsis. Dev. Cell 7, 193–204. [DOI] [PubMed] [Google Scholar]
- Malamy, J.E., and Benfey, P.N. (1997). Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124, 33–44. [DOI] [PubMed] [Google Scholar]
- Matsumoto, N., and Okada, K. (2001). A homeobox gene, PRESSED FLOWER, regulates lateral axis-dependent development of Arabidopsis flowers. Genes Dev. 15, 3355–3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo, N., Minami, M., Maeda, T., and Hiratsuka, K. (2001). Dual luciferase assay for monitoring gene expression in higher plants. Plant Biotechnol. 18, 71–75. [Google Scholar]
- Mattsson, J., Ckurshumova, W., and Berleth, T. (2003). Auxin signaling in Arabidopsis leaf vascular development. Plant Physiol. 131, 1327–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney, E.C., Ali, N., Traut, A., Feldmann, K.A., Belostotsky, D.A., McDowell, J.M., and Meagher, R.B. (1995). Sequence-based identification of T-DNA insertion mutations in Arabidopsis: Actin mutants act2-1 and act4-1. Plant J. 8, 613–622. [DOI] [PubMed] [Google Scholar]
- Morris, D.R., and Geballe, A.P. (2000). Upstream open reading frames as regulators of mRNA translation. Mol. Cell. Biol. 20, 8635–8642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassay with tobacco tissue cultures. Physiol. Plant 15, 473–497. [Google Scholar]
- Nemhauser, J.L., Feldman, L.J., and Zambryski, P.C. (2000). Auxin and ETTIN in Arabidopsis gynoecium morphogenesis. Development 127, 3877–3888. [DOI] [PubMed] [Google Scholar]
- Nishimura, T., Yokota, E., Wada, T., Shimmen, T., and Okada, K. (2003). An Arabidopsis ACT2 dominant-negative mutation, which disturbs F-actin polymerization, reveals its distinctive function in root development. Plant Cell Physiol. 44, 1131–1140. [DOI] [PubMed] [Google Scholar]
- Okada, K., Ueda, J., Komaki, M.K., Bell, C.J., and Shimura, Y. (1991). Requirement of the auxin polar transport system in early stages of Arabidopsis floral bud formation. Plant Cell 3, 677–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver, E.R., Saunders, T.L., Tarlé, S.A., and Glaser, T. (2004). Ribosomal protein L24 defect in Belly spot and tail (Bst), a mouse Minute. Development 131, 3907–3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, H.-S., Himmelbach, A., Browning, K.S., Hohn, T., and Ryabova, L.A. (2001). A plant viral “reinitiation” factor interacts with the host translational machinery. Cell 106, 723–733. [DOI] [PubMed] [Google Scholar]
- Przemeck, G.K.H., Mattsson, J., Hardtke, C.S., Sung, Z.R., and Berleth, T. (1996). Studies on the role of the Arabidopsis gene MONOPTEROS in vascular development and plant cell axialization. Planta 200, 229–237. [DOI] [PubMed] [Google Scholar]
- Rajkowitsch, L., Vilela, C., Berthelot, K., Ramirez, C.V., and McCarthy, J.E.G. (2004). Reinitiation and recycling are distinct processes occurring downstream of translation termination in yeast. J. Mol. Biol. 335, 71–85. [DOI] [PubMed] [Google Scholar]
- Ruiz-Echevarria, M.J., and Peltz, W. (2000). The RNA binding protein Pub1 modulates the stability of transcripts containing upstream open reading frames. Cell 101, 741–751. [DOI] [PubMed] [Google Scholar]
- Schägger, H., and von Jagow, G. (1987). Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166, 368–379. [DOI] [PubMed] [Google Scholar]
- Sessions, R.A., Nemhauser, J.L., McCall, A., Roe, J.L., Feldmann, K.A., and Zambryski, P.C. (1997). ETTIN patterns the Arabidopsis floral meristem and reproductive organs. Development 124, 4481–4491. [DOI] [PubMed] [Google Scholar]
- Sessions, R.A., and Zambryski, P.C. (1995). Arabidopsis gynoecium structure in the wild type and in ettin mutants. Development 121, 1519–1532. [DOI] [PubMed] [Google Scholar]
- Swarup, R., Parry, G., Graham, N., Allen, T., and Bennett, M. (2002). Auxin cross-talk: Integration of signaling pathways to control plant development. Plant Mol. Biol. 49, 411–426. [DOI] [PubMed] [Google Scholar]
- Tiwari, S.B., Hagen, G., and Guilfoyle, T.J. (2003). The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 15, 533–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda, T., Yamaguchi, M., Uchimiya, H., and Nakano, A. (2001). Ara6, a plant-unique novel type Rab GTPase, functions in the endocytic pathway of Arabidopsis thaliana. EMBO J. 20, 4730–4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov, T., Hagen, G., and Guilfoyle, T.J. (1997). ARF1, a transcription factor that binds to auxin response elements. Science 276, 1865–1868. [DOI] [PubMed] [Google Scholar]
- Ulmasov, T., Hagen, G., and Guilfoyle, T.J. (1999). Activation and repression of transcription by auxin-response factors. Proc. Natl. Acad. Sci. USA 96, 5844–5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Lijsebettens, M., Vanderhaeghen, R., De Block, M., Bauw, G., Villarroel, R., and Van Montagu, M. (1994). An S18 ribosomal protein gene copy at the Arabidopsis PFL locus affects plant development by its specific expression in meristems. EMBO J. 13, 3378–3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilela, C., Ramirez, C.V., Linz, B., Rodrigues-Pousada, C., and McCarthy, J.E.G. (1999). Post-termination ribosome interactions with the 5′UTR modulate yeast mRNA stability. EMBO J. 18, 3139–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada, T., Tachibana, T., Shimura, Y., and Okada, K. (1997). Epidermal cell differentiation in Arabidopsis determined by a Myb homolog, CPC. Science 277, 1113–1116. [DOI] [PubMed] [Google Scholar]
- Wang, L., and Wessler, S.R. (1998). Inefficient reinitiation is responsible for upstream open reading frame-mediated translational repression of the maize R gene. Plant Cell 10, 1733–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers, D., Dijk, M.F.-v., Vencken, R.-J., Quint, A., Hooykaas, P., and Offringa, R. (2001). An Arabidopsis Minute-like phenotype caused by a semi-dominant mutation in a RIBOSOMAL PROTEIN S5 gene. Development 128, 4289–4299. [DOI] [PubMed] [Google Scholar]
- Wiese, A., Elzinga, N., Wobbes, B., and Smeekens, S. (2004). A conserved upstream open reading frame mediates sucrose-induced repression of translation. Plant Cell 16, 1717–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, M.E., and Sussex, I.M. (1995). Developmental regulation of ribosomal protein L16 genes in Arabidopsis thaliana. Plant J. 8, 65–76. [DOI] [PubMed] [Google Scholar]
- Wool, I.G. (1996). Extraribosomal functions of ribosomal proteins. Trends Biochem. Sci. 21, 164–165. [PubMed] [Google Scholar]
- Yaman, I., Fernandez, J., Liu, H., Caprara, M., Komar, A.A., Koromilas, A.E., Zhou, L., Snider, M.D., Scheuner, D., Kaufman, R.J., and Hatzoglou, M. (2003). The zipper model of translational control: A small upstream ORF is the switch that controls structural remodeling of an mRNA leader. Cell 113, 519–531. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.