Abstract
The synergid cells of the female gametophyte play a role in many steps of the angiosperm fertilization process, including guidance of pollen tube growth to the female gametophyte. However, the mechanisms by which the synergid cells become specified and develop their unique features during female gametophyte development are not understood. We identified MYB98 in a screen for Arabidopsis thaliana genes expressed in the female gametophyte. MYB98 is a member of the R2R3-MYB gene family, the members of which likely encode transcription factors. In the context of the ovule, MYB98 is expressed exclusively in the synergid cells, and mutations in this gene affect the female gametophyte specifically. myb98 female gametophytes are affected in two unique features of the synergid cell, pollen tube guidance and the filiform apparatus, but are otherwise normal. MYB98 also is expressed in trichomes and endosperm. Homozygous myb98 mutants exhibit no sporophytic defects, including trichome and endosperm defects. Together, these data suggest that MYB98 controls the development of specific features within the synergid cell during female gametophyte development.
INTRODUCTION
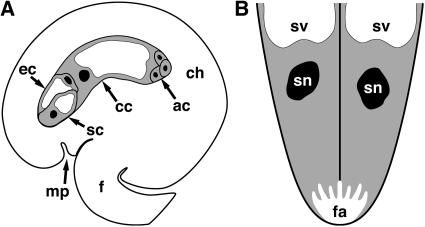
The synergid cells within the female gametophyte (Figure 1A) are essential for the reproductive process in angiosperms. Sexual reproduction is initiated when the male gametophyte is transferred from the anther to the carpel's stigma, whereupon it forms a pollen tube that grows through the carpel's internal tissues to deliver its two sperm cells to the female gametophyte. The pollen tube enters the female gametophyte by growing into one of the two synergid cells. The synergid cell penetrated by the pollen tube typically undergoes cell death before or upon pollen tube arrival. Soon after arrival, the pollen tube ceases growth, ruptures, and releases its two sperm cells into the receptive synergid's cytoplasm, triggering the completion of degeneration. Finally, one sperm cell migrates to the egg cell and the other to the central cell to effect double fertilization of the female gametophyte (Weterings and Russell, 2004).
Figure 1.
Depictions of the Female Gametophyte and Synergid Cells.
(A) Depiction of the Arabidopsis female gametophyte. Modified from Drews et al. (1998).
(B) Depiction of the synergid cells. This view is perpendicular to that in (A).
The cytoplasm is gray, vacuoles are white, and nuclei are black. ac, three antipodal cells; cc, central cell; ch, chalazal region of the ovule; ec, egg cell; f, funiculus; fa, filiform apparatus; mp, micropyle; sc, synergid cell; sn, synergid cell nucleus; sv, synergid cell vacuole.
The synergid cells are required for pollen tube guidance. Using Arabidopsis thaliana mutants, several groups have shown that pollen tubes fail to grow onto ovules containing abnormal female gametophytes, suggesting that the embryo sac is the source of a pollen tube guidance cue (Hulskamp et al., 1995; Ray et al., 1997; Shimizu and Okada, 2000). Using laser ablation in an in vitro pollen germination system in Torenia, Higashiyama and colleagues (2001) showed that the synergid cells, but not other female gametophyte cells, are the source of the pollen tube attractant. Recently, a small maize (Zea mays) protein, EA1, required for pollen tube guidance was reported; however, maize EA1 is not likely to be a universal attractant because no homolog of ea1 is present in Arabidopsis or other dicots (Marton et al., 2005).
The synergid cells also appear to control the late steps of the pollen tube growth pathway. In two Arabidopsis female gametophyte mutants, feronia (Huck et al., 2003) and sirene (Rotman et al., 2003), wild-type pollen tubes enter mutant female gametophytes but fail to cease growth, rupture, and release their contents. Similar pollen tube overgrowths occur in interspecific crosses of Rhododendron (Williams et al., 1986) and in the in vitro Torenia system (Higashiyama et al., 1998). These observations suggest that the female gametophyte contains factors that control the arrest of pollen tube growth and/or the release of pollen tube contents. Because these steps take place within the synergid cell, it is likely that synergid cell–expressed gene products are necessary for these processes (Weterings and Russell, 2004).
The synergid cells have several structural specializations that facilitate the fertilization process. At the micropylar pole, the cell wall is extensively invaginated, forming a structure referred to as the filiform apparatus (Figure 1B). The function of the filiform apparatus is unknown; numerous functions have been proposed, including pollen tube reception and transport of substances into and out of the synergid cells (Willemse and van Went, 1984; Huang and Russell, 1992). At the chalazal pole of the synergid cell, the cell wall is absent or discontinuous, which likely facilitates the fertilization process by providing direct access of the sperm cells to the fertilization targets (Willemse and van Went, 1984; Huang and Russell, 1992). Finally, during cell death, the synergid cytoskeleton reorganizes and forms two actin bands, which are thought to direct the migration of the sperm cells to the fertilization targets (Russell, 1993).
The molecular processes by which the synergid cells acquire their unique structure and function during cell differentiation are not understood. Regulatory molecules controlling these processes have not been identified. Furthermore, only a few genes expressed in synergid cells have been identified, and none of these is expressed exclusively in the synergid cells; these include the Arabidopsis DEMETER (Choi et al., 2002) and AGP18 (Acosta-Garcia and Vielle-Calzada, 2004) genes and the maize ea1 (Marton et al., 2005) and es (Cordts et al., 2001) genes.
Here, we report the identification of a gene, MYB98, that encodes an R2R3-type MYB protein, which likely functions as a transcription factor. In the context of the ovule, MYB98 is expressed exclusively in the synergid cells. MYB98 is required for pollen tube guidance and the formation of the filiform apparatus, both of which are unique features of the synergid cells. Thus, the MYB98 gene appears to encode a regulatory molecule that controls synergid cell differentiation during female gametophyte development.
RESULTS
MYB98 Is Expressed in the Synergid Cells of the Female Gametophyte
We performed a screen to identify genes expressed in the female gametophyte. Our strategy was to identify mRNAs present in wild-type ovules but not in mutant ovules lacking female gametophytes. We used determinant infertile1 (dif1) (Bai et al., 1999; Bhatt et al., 1999; Cai et al., 2003) as the source of mutant ovules. dif1 ovules lack a female gametophyte but are otherwise normal; thus, genes exhibiting reduced expression in dif1 ovules relative to wild-type ovules are likely to be expressed in the female gametophyte but not in the surrounding sporophytic tissue of the ovule.
To identify regulatory genes expressed in the female gametophyte, we harvested pistils from the wild type and dif1, extracted RNA, and used RT-PCR to assay the expression of genes within the R2R3-MYB family (Kranz et al., 2000; Stracke et al., 2001; Jiang et al., 2004). These genes likely encode transcription factors and have been shown to regulate many biological processes (Stracke et al., 2001; Jiang et al., 2004). These assays identified a gene, MYB98, exhibiting reduced expression in dif1 pistils relative to wild-type pistils (Figure 2A).
Figure 2.
RT-PCR Analysis of MYB98 Expression.
(A) Expression of MYB98 in wild-type pistils (WP) and dif1-2/dif1-2 pistils (DP). Expression of the ACTIN2 gene was used as a control.
(B) Real-time RT-PCR analysis of MYB98 expression in pistils (WP), roots (R), floral stems (S), young leaves at <5 mm in length (YL), older leaves at 10 to 20 mm in length (L), flowers at stages 1 to 11 (YF), and siliques at 1 to 2 d after pollination (SL). Each bar represents an average of four to five independent reactions. Error bars indicate sd. The number above each bar represents RNA level relative to pistil (WP).
(C) Expression of MYB98 in wild-type pistils (WP), myb98-1/myb98-1 pistils (98-1), and myb98-2/myb98-2 pistils (98-2). Expression of the ACTIN2 gene was used as a control.
To determine which cells within the female gametophyte express the MYB98 gene, we generated and analyzed transgenic Arabidopsis plants containing ProMYB98:reporter constructs. As shown in Figures 3A and 3B, at the terminal developmental stage (stage FG7), ProMYB98:green fluorescent protein (GFP) and ProMYB98:β-glucuronidase (GUS) were expressed exclusively in the synergid cells. During the fertilization process, one of the synergid cells undergoes cell death and the other synergid cell persists. Expression was detected in the persistent synergid cell for ≥7 d after fertilization (Figure 3D).
Figure 3.
Expression of the ProMYB98:GFP and ProMYB98:GUS Genes.
(A) and (B) Expression of ProMYB98:GFP (A) and ProMYB98:GUS (B) in female gametophytes at the terminal developmental stage (stage FG7). Expression is detected only in the synergid cells. The ovule in (A) is oriented like that in Figure 1A, and the ovule in (B) is oriented like that in Figure 1B. The ovule in (B) was stained in 1 mM 5-bromo-4-chloro-3-indolyl-β-glucuronic acid (X-Gluc) for 30 min.
(C) Expression of ProMYB98:GFP in the synergid cells in a female gametophyte at late stage FG5.
(D) Expression of ProMYB98:GUS in a fertilized embryo sac at 12 h after pollination. Expression is detected only in the persistent synergid cell. This ovule was stained in 1 mM X-Gluc for 4 h.
(E) Expression of ProMYB98:GUS in a fertilized embryo sac at 12 h after pollination. Expression is detected in the persistent synergid cell and endosperm (arrowheads). This ovule was stained in 5 mM X-Gluc for 4 h.
(F) Expression of ProMYB98:GFP in a fertilized embryo sac at 24 h after pollination. Expression is detected in the persistent synergid cell and endosperm (arrowheads).
(G) Expression of ProMYB98:GUS in the trichomes of young (<5 mm) leaves. This leaf was stained in 1 mM X-Gluc for 16 h.
(H) Opened siliques from wild-type (left) and myb98-1/+ (right) plants at 10 d after pollination. Siliques from mutant plants contain desiccated ovules (arrows).
psc, persistent synergid cell; sc, synergid cell.
To determine whether MYB98 is expressed at earlier stages of female gametophyte development, we analyzed expression in flowers at stage 12c, which lack mature (stage FG7) female gametophytes (Christensen et al., 1997). Stage 12c flowers contained ∼33% uncellularized (stages FG4 and early FG5) and 67% cellularized (stages late FG5 and FG6) female gametophytes (n = 159). In flowers at this stage, ProMYB98:GFP expression was detected in ∼60% (n = 200) of the female gametophytes, in contrast with 100% of female gametophytes in mature ovules. All of the female gametophytes expressing ProMYB98:GFP had morphologically identifiable synergid cells (Figure 3C), indicating that cellularization had occurred. Together, these data suggest that MYB98 is expressed in cellularized (late stage FG5 to FG7) but not uncellularized female gametophytes.
To determine whether MYB98 is expressed elsewhere in the plant, we analyzed the ProMYB98:reporter lines and performed real-time RT-PCR with RNA from various organs. During seed development, ProMYB98:GFP and ProMYB98:GUS expression was detected only in the persistent synergid cell (Figure 3D) and the endosperm (Figures 3E and 3F). Expression in the endosperm was weak relative to that in the persistent synergid (cf. the staining conditions summarized in the legends of Figures 3D and 3E). During seed development, expression was highest in very young seeds (≤1 d after pollination) (Figures 3E and 3F) and diminished gradually at progressively older stages; expression was not detected in seeds older than 3 d after pollination.
Within vegetative organs, ProMYB98:GUS expression was detected only in the trichomes of leaves (Figure 3G). The leaf trichome expression was weak relative to the synergid expression (cf. the staining conditions summarized in the legends of Figures 3B and 3G). During leaf development, ProMYB98:GUS expression was highest in very young leaves (<5 mm in length) (Figure 3G) and diminished gradually at progressively older stages; expression was not detected in leaves longer than 8 mm. Within reproductive organs, strong ProMYB98:GUS expression was detected only in the synergid cells (Figure 3B); in addition, very weak expression was detected in the trichomes of sepals (data not shown).
Using real-time RT-PCR (Figure 2B), strong MYB98 expression was detected only in pistils, which contain the synergid cells. In addition, weaker expression was detected in young leaves (<5 mm in length), which correlates with the expression of ProMYB98:GUS in the trichomes of young leaves (Figure 3G); in developing siliques, which correlates with the expression of ProMYB98:GUS in the persistent synergid cell (Figure 3D) and endosperm (Figure 3E); and in young flowers, which correlates with the expression of ProMYB98:GUS in the trichomes of sepals. Expression of MYB98 was not detected in roots, stems, and older leaves. Together, these data suggest that MYB98 is expressed at high levels in the synergid cells and at lower levels in the endosperm of young seeds and the trichomes of young leaves and sepals.
MYB98 Gene Structure
To determine the structure of the MYB98 gene, we isolated a MYB98 cDNA clone and compared its sequence with the genomic sequence. As shown in Figure 4A, MYB98 has three exons and two introns and is predicted to encode a protein of 427 amino acids. As shown in Figure 4B, MYB98 is predicted to contain a Myb domain (Kranz et al., 2000; Stracke et al., 2001) and a putative nuclear localization signal.
Figure 4.
Structure of the MYB98 Protein.
(A) MYB98 gene structure. Black boxes represent coding sequence, and arrowheads represent the T-DNA insertion sites in myb98-1 (40 bp downstream of the ATG) and myb98-2 (195 bp upstream of the ATG). nt, nucleotide.
(B) MYB98 protein structure. MYB98 contains a predicted nuclear localization signal (NLS) between amino acids (aa) 196 and 212 (gray box). The R2 and R3 repeats are represented by black boxes.
(C) Comparison of the R2 and R3 Myb repeats between Arabidopsis MYB98 and Mus musculus c-Myb. Conserved amino acids are shaded in gray. The asterisks denote the conserved Trp residues. The arrow designates the conserved Ala residue in the second helix of R2 that is often a Pro residue in plant R2R3 proteins (Jia et al., 2004). The arrowheads denote the seven amino acids in c-Myb that contact the DNA bases.
The Myb domain is a DNA binding motif found in the vertebrate protooncogene c-myb (Lipsick and Wang, 1999). The Myb domain in c-Myb consists of three imperfect tandem repeats (Myb repeats) of ∼50 amino acids that are referred to as R1, R2, and R3. In c-Myb, R2 and R3 are required for sequence-specific binding and R1 is thought to play a minor role in DNA binding (Ogata et al., 1994). Structural studies of c-Myb have shown that each Myb repeat forms three α-helices, that the second and third α-helices form a helix-turn-helix structure, and that the third helices of R2 and R3 make direct contact with the major groove of the target DNA (Ogata et al., 1992, 1994, 1995).
Most plant MYB proteins, including MYB98, contain only the R2 and R3 Myb repeats (Jin and Martin, 1999; Stracke et al., 2001; Jiang et al., 2004). The R2 and R3 Myb repeats of MYB98 are highly similar to those of c-Myb (66% identical and 86% similar at the amino acid level). The R2 and R3 Myb repeats of c-Myb contain six Trp residues that form the hydrophobic core of the helix-turn-helix structure (Kanei-Ishii et al., 1990) and seven amino acid residues that contact the DNA bases and contribute to DNA binding specificity (Tahirov et al., 2002) (Figure 4C). All of the Trp residues and six of the seven DNA-contacting residues are conserved in MYB98 (Figure 4C). Together, these observations suggest that MYB98 binds DNA and functions as a transcriptional regulator.
We also isolated a cDNA clone from the Arabidopsis Landsberg erecta accession and compared its sequence with that of the Columbia accession. This comparison revealed 16 nucleotide differences that are predicted to result in six amino acid differences (see Supplemental Figure 1 online). None of the amino acid differences are within the R2 and R3 Myb repeats, consistent with the predicted importance of these domains.
The myb98 Mutation Affects the Female Gametophyte Specifically
To determine whether mutations in the MYB98 gene affect the female gametophyte, we obtained lines containing T-DNA insertions in this gene from the Salk Institute Genomic Analysis Laboratory collection (Alonso et al., 2003). We analyzed two independent T-DNA alleles, myb98-1 and myb98-2. The T-DNA in myb98-1 is inserted into the first exon 42 nucleotides downstream of the start codon and 152 nucleotides upstream of the predicted R2 Myb repeat and is associated with a 23-bp deletion. The T-DNA in myb98-2 is inserted into the genomic sequence 195 nucleotides upstream of the start codon and is associated with a 19-bp deletion. With both alleles, we did not detect wild-type MYB98 transcripts by RT-PCR (Figure 2C), suggesting that they are null alleles.
With both alleles, siliques from heterozygous plants contained 50% desiccated ovules (Figure 3H), suggesting that these mutations cause gametophytic lethality. To determine whether the myb98 mutations affect the female gametophyte, we crossed heterozygous mutant plants as female parents with wild-type males and scored the number of F1 progeny that were heterozygous and homozygous wild type. As summarized in Table 1, both alleles exhibited reduced transmission through the female gametophyte, indicating that the myb98-1 and myb98-2 mutations affect the female gametophyte.
Table 1.
Segregation of the myb98-1 and myb98-2 Mutations
| Parental Genotypes
|
Progeny Genotypes
|
|||
|---|---|---|---|---|
| Male | Female | MYB98/MYB98 | myb98/MYB98 | myb98/myb98 |
| myb98-1/MYB98 | myb98-1/MYB98 | 109 | 113 | 5 |
| MYB98/MYB98 | myb98-1/MYB98 | 304 | 3 | – |
| myb98-1/MYB98 | MYB98/MYB98 | 160 | 172 | – |
| myb98-2/MYB98 | myb98-2/MYB98 | 367 | 388 | 13 |
| MYB98/MYB98 | myb98-2/MYB98 | 368 | 5 | – |
| myb98-2/MYB98 | MYB98/MYB98 | 187 | 217 | – |
To determine whether the myb98 mutations also affect the male gametophyte, we crossed heterozygous mutant plants as male parents with wild-type females and scored the number of F1 progeny that were heterozygous and homozygous wild type. With both alleles, heterozygous and homozygous wild-type progeny were present in approximately equal proportions in the F1 generation (Table 1), indicating that the myb98-1 and myb98-2 mutations do not affect the male gametophyte.
As shown in Table 1, plants homozygous for the myb98-1 and myb98-2 mutations were present among the progeny of self-pollinated plants. As expected, homozygous plants had dramatically reduced seed set, as a result of the female gametophyte defect; for example, myb98-1/myb98-1 siliques contained an average of 83% desiccated ovules and 17% green seeds (n = 15 siliques) after self-pollination. Homozygous lines exhibited no sporophytic defects. In particular, homozygous mutants exhibited no overt trichome defects with regard to both density and branching. In addition, mutant siliques lacked abnormal seeds, suggesting that there were no defects in endosperm development. Together, these data indicate that the myb98-1 and myb98-2 mutations affect the female gametophyte specifically.
Molecular Complementation of the myb98-1 Mutation
To determine whether the female gametophyte defect is attributable to disruption of the MYB98 gene, we introduced a wild-type copy of the MYB98 gene into the myb98-1 mutant. We identified plants heterozygous for the myb98-1 allele and hemizygous for the rescue construct and crossed these plants as females with wild-type males. In the F1 generation, myb98-1/MYB98 and MYB98/MYB98 progeny were present in a ratio of ∼1:2. In addition, we identified plants homozygous for the myb98-1 allele and homozygous for the rescue construct; these plants had full seed set. These data indicate that disruption of the MYB98 gene is responsible for the female gametophyte defect in myb98-1 mutants.
Terminal Phenotype of myb98 Female Gametophytes
To determine whether myb98 mutants are affected in female gametophyte development, we analyzed myb98-1 and myb98-2 female gametophytes at the terminal developmental stage (stage FG7) using confocal laser scanning microscopy (CLSM) (Christensen et al., 1997). Wild-type synergid cells are highly polarized, with their nuclei and most of their cytoplasm located at the micropylar pole and their vacuoles at the chalazal poles (Figures 1A, 1B, and 5A). myb98 female gametophytes and myb98 synergid cells in particular (Figure 5B) appeared normal at the level of CLSM, indicating that female gametophyte development and the overall morphology of the synergid cells were not largely affected by the myb98-1 and myb98-2 mutations.
Figure 5.
Microscopic Analysis of myb98 Female Gametophytes.
(A) and (B) CLSM images of female gametophytes at the terminal developmental stage (stage FG7) in the wild type (A) and myb98-1 (B).
(C) and (D) Light micrographs of synergid cells from thick plastic sections of wild-type (C) and myb98-1 (D) ovules.
(E) to (L) Transmission electron microscopy images of wild-type ([E], [G], and [I]), myb98-1 ([F], [H], and [J]), and myb98-2 ([K] and [L]) synergid cells. The asterisks in (D), (F), and (H) denote the abnormal micropylar structure in myb98-1 synergid cells. Within the filiform apparatus of the wild type ([G] and [I]) are electron-translucent (light gray patches) and electron-dense (medium gray patches marked with arrowheads) regions. The dark gray regions are pockets of cytoplasm.
cw, cell wall between the synergid cells; ed, electron-dense region; en, egg cell nucleus; et, electron-translucent region; ev, egg cell vacuole; fa, filiform apparatus; sc, synergid cell; sn, synergid cell nucleus; sv, synergid cell vacuole. Bars = 10 μm in (A) and (B), 5 μm in (C) to (F), 3 μm in (G), (H), and (K), and 1 μm in (I), (J), and (L).
Filiform Apparatus Structure Is Abnormal in myb98 Synergid Cells
To determine whether myb98 female gametophytes have defects at the ultrastructural level, we analyzed mutants using transmission electron microscopy. This analysis showed that the structure of the filiform apparatus is abnormal in myb98 synergid cells.
In the wild type, the filiform apparatus appeared to consist of numerous finger-like projections of the cell wall into the synergid cytoplasm. This was most clearly seen in the chalazal-most region of the filiform apparatus, where individual projections could be observed (Figures 5E and 5G). In the micropylar-most region of the filiform apparatus, the structure was complex and heterogeneous (Figure 5G), suggesting that the finger-like projections are interlaced in this region. Throughout the filiform apparatus, electron-dense and electron-translucent regions were present (Figures 5G and 5I) (Willemse and van Went, 1984; Huang and Russell, 1992).
In contrast with the wild type, the cell wall at the micropylar region of myb98 synergid cells lacked the extensive invaginations characteristic of the filiform apparatus (Figures 5F, 5H, and 5J to 5L). In place of the filiform apparatus was a structure that resembles an extremely thickened cell wall (Figures 5F, 5H, and 5K). In most of the myb98 female gametophytes analyzed (five of five myb98-1 and three of five myb98-2), this micropylar structure lacked cell wall projections completely (Figure 5J) and had uniform electron density (Figure 5H). However, in two of the myb98-2 female gametophytes analyzed, primitive cell wall projections and electron-dense and electron-translucent regions were present (Figures 5K and 5L). In addition to the filiform apparatus defect, the cell wall between the synergid cells was thicker in the mutant than in the wild type (Figures 5E and 5F). Other aspects of synergid cell ultrastructure appeared normal in myb98 synergid cells. Together, these data suggest that MYB98 is required for the formation of the filiform apparatus during synergid cell differentiation.
Pollen Tube Guidance Is Defective in myb98 Synergid Cells
Laser ablation studies have shown that the synergid cells secrete an attractant that guides the pollen tube to the female gametophyte (Higashiyama et al., 2001). To test for defects in pollen tube guidance, we pollinated myb98-1/myb98-1 and myb98-1/MYB98 pistils with wild-type pollen and observed pollen tubes at 12 h after pollination. In the wild type, the pollen tube grew along the ovule's funiculus, through the ovule's micropyle, and into the female gametophyte (Figure 6A). By contrast, wild-type pollen tubes grew abnormally on ovules containing myb98-1 female gametophytes: pollen tubes grew from the placenta to the funiculus but then failed to grow into the micropyle (Figure 6B). Similar results were obtained with myb98-2/MYB98 pistils. Thus, pollen tube guidance is affected in myb98-1 and myb98-2 female gametophytes.
Figure 6.
Pollen Tube Guidance Is Affected in myb98 Female Gametophytes.
(A) Wild-type pollen tube on a wild-type ovule at 12 h after pollination.
(B) Wild-type pollen tube on a myb98-1/myb98-1 ovule at 12 h after pollination.
(C) Wild-type pollen tubes on a myb98-1/myb98-1 ovule at 24 h after pollination. Four pollen tubes (pt1 to pt4) are present on the ovule.
(D) Wild-type pollen tubes on a myb98-1/myb98-1 ovule at 24 h after pollination. Four pollen tubes (pt1 to pt4) are present on the ovule. One of the pollen tubes (pt1) is in the micropyle.
All images are composites composed of CLSM micrographs of the pollen tubes merged with bright-field images of ovules. All images are projections of multiple 1-μm sections. f, funiculus; mp, micropyle; pt, pollen tube; t, tip of pollen tube. Bars = 25 μm.
We also scored pollen tube growth at 24 h after pollination. In the wild type at this time point, 98% (n = 100) of ovules received a single pollen tube. By contrast, multiple (≥3 per ovule) pollen tubes were present on all (n = 200) myb98-1/myb98-1 ovules (Figures 6C and 6D). The majority of these were present at random positions on the ovule. However, in 20% of myb98-1/myb98-1 ovules (n = 100 ovules), one of the pollen tubes had grown into the micropyle (Figure 6D); these most likely become fertilized and give rise to seeds, based on the observed 17% seed set in myb98-1/myb98-1 mutants (discussed above).
The myb98 Mutation Does Not Affect the Fertilization Process
Previous genetic and structural studies suggest that the synergid cells control several steps of the fertilization process, including the arrest of pollen tube growth, the release of pollen tube contents, and the migration of the sperm cells to the egg cell and central cell (Weterings and Russell, 2004). To determine whether myb98 female gametophytes have defects in these steps of the fertilization process, we asked whether mutant female gametophytes could become fertilized and give rise to seeds if penetrated by a pollen tube. Addressing this question was made possible by the partial seed set exhibited by homozygous myb98-1 mutants (discussed above). The partial seed set suggests either that myb98-1 female gametophytes can function normally if penetrated by a pollen tube or that the myb98-1 mutation is partially penetrant. To distinguish between these possibilities, we prepared and analyzed thick plastic sections (Figures 5C and 5D) of 30 myb98-1 female gametophytes. We found that all 30 had defects in the filiform apparatus. Therefore, it is unlikely (P < 0.003) that the 17% seed set is attributable to partial penetrance of the myb98-1 mutation. These data suggest very strongly that female gametophytes affected by the myb98-1 mutation can become fertilized and give rise to seeds. Thus, with the exception of pollen tube guidance, the synergid cells appear to function normally in myb98-1 female gametophytes.
DISCUSSION
MYB98 Is Required for Synergid Cell Differentiation
In the context of the ovule, MYB98 is expressed exclusively in the synergid cells (Figures 3A and 3B). During female gametophyte development, MYB98 is expressed after but not before cellularization (Figure 3C). This specialized expression pattern is consistent with the phenotype of myb98 mutants, which are affected in pollen tube guidance and filiform apparatus structure. MYB98 likely functions as a transcription factor; thus, it is unlikely that this protein plays a direct role in pollen tube guidance and the formation of the filiform apparatus. More likely, MYB98 controls the expression of downstream genes required for these processes.
Other aspects of synergid cell development, including cell specification, appear to be normal in myb98 mutants. This conclusion is based on several observations. First, female gametophyte development appears to be unaffected in myb98 mutants. Second, the overall morphology of the synergid cells is unaffected in myb98 mutants (Figures 5B and 5F). Third, the myb98 mutation does not appear to affect steps of the fertilization process subsequent to pollen tube guidance, including the control of pollen tube growth cessation, pollen tube rupture, and sperm cell migration. Together, these observations indicate that MYB98 controls specific aspects of synergid cell differentiation.
Expression of MYB98 also was detected in trichomes (Figure 3G) and endosperm (Figures 3E and 3F), suggesting a function in the development of these structures. Other MYB genes play a role in trichome (Oppenheimer et al., 1991; Kirik et al., 2005; Matsui et al., 2005) and endosperm (Diaz et al., 2002; Gomez et al., 2002) development. Although we did not detect trichome and endosperm defects in myb98 mutants, MYB98 could function redundantly with other MYB genes.
Function of the Filiform Apparatus
During the fertilization process, the pollen tube enters the female gametophyte by growing through the filiform apparatus and into one of the synergid cells (Weterings and Russell, 2004). This growth pattern suggests that the filiform apparatus plays a role in the entry of the pollen tube into the female gametophyte. For example, it has been proposed that the filiform apparatus protects the female gametophyte from mechanical damage by restricting the pathway of pollen tube growth (Higashiyama, 2002). myb98 embryo sacs have structural defects in the filiform apparatus but can become fertilized. These observations indicate that a fully elaborated filiform apparatus is not required for pollen tube entry into the synergid cell.
The filiform apparatus consists of numerous finger-like projections of the cell wall that extend deeply into the synergid cytoplasm (Figures 5E and 5G). The filiform apparatus greatly increases the surface area of the plasma membrane at the micropylar poles of the synergid cells. Based on these structural observations, it has been proposed that the filiform apparatus facilitates the transport of substances into and out of the synergid cells (Willemse and van Went, 1984; Huang and Russell, 1992; Higashiyama, 2002).
For example, it has been proposed that the filiform apparatus facilitates the export of a pollen tube attractant (Huang and Russell, 1992; Higashiyama, 2002). Consistent with this proposal, myb98 female gametophytes exhibit defects in pollen tube guidance. Thus, the pollen tube guidance defect in myb98 female gametophytes may be a secondary consequence of the filiform apparatus defect. However, at present, we cannot eliminate the possibility that MYB98 is required for the production of a pollen tube attractant in addition to its role in filiform apparatus formation.
It also has been proposed that the filiform apparatus mediates the uptake of metabolites into the embryo sac (Jensen, 1965; Huang and Russell, 1992). If true, myb98 female gametophytes should exhibit reduced viability as a result of nutritional deficiency. However, with the exception of the filiform apparatus defect, myb98 embryo sacs appear structurally normal (Figures 5B and 5F). These observations suggest that the filiform apparatus does not mediate the import of essential metabolites into the female gametophyte.
Models for the Control of Pollen Tube Guidance
Based on an analysis of pollen tube growth patterns in female gametophyte mutants affected at different developmental stages, it has been proposed that the female gametophyte is necessary for pollen tube guidance at two steps: guidance from the placenta to the funiculus (funicular guidance phase), and guidance from the funiculus to the micropyle (micropylar guidance phase) (Shimizu and Okada, 2000). On ovules containing myb98 female gametophytes, wild-type pollen tubes grow from the placenta to the funiculus but then fail to grow into the micropyle (Figure 6B). These data support the two-step hypothesis and indicate that myb98 affects specifically the micropylar guidance phase of pollen tube guidance.
In myb98 female gametophytes, the filiform apparatus is affected dramatically, yet the funicular guidance phase of pollen tube guidance is unaffected. These observations indicate that the funicular guidance phase does not require the filiform apparatus and suggests three possibilities. First, the funicular guidance signal could be produced by the synergid cell but exported by a filiform apparatus–independent pathway. Second, the funicular guidance signal could be produced by a different cell type within the female gametophyte. Third, the female gametophyte could stimulate changes in the surrounding sporophytic tissue that enable pollen tube growth onto the funiculus. Consistent with the third possibility, the sporophytic ovule mutant inner no outer has normal embryo sacs but is impaired in funicular guidance (Baker et al., 1997).
In the wild type, typically only one pollen tube grows onto each ovule (Higashiyama, 2002; Weterings and Russell, 2004). To explain this observation, it has been proposed that the pollen tube guidance cue is eliminated upon fertilization or possibly by the entry of the pollen tube into the micropyle (Higashiyama, 2002; Weterings and Russell, 2004). Ovules containing myb98 female gametophytes generally fail to become fertilized (Figure 6B) and also attract multiple pollen tubes (Figures 6C and 6D). Thus, our observations are consistent with the proposed model.
In ∼20% of myb98-1/myb98-1 ovules, a pollen tube grows into the micropyle and fertilizes the female gametophyte. This occasional fertilization is not attributable to partial penetrance of the myb98-1 mutation because all of 30 embryo sacs analyzed exhibited filiform apparatus defects. The occasional fertilization instead could be the result of random growth of the pollen tubes into the micropyle. The probability of random growth into the micropyle would be increased by the numerous pollen tubes that grow onto myb98-1/myb98-1 ovules. Alternatively, myb98-1 synergid cells could synthesize and/or secrete a reduced level of pollen tube attractant. Distinguishing between these possibilities will likely require identification of the pollen tube attractant(s).
Model for MYB98 Function
The data presented here suggest that MYB98 functions as a transcription factor within the gene regulatory network of the synergid cell and controls the expression of downstream genes required for pollen tube guidance and the formation of the filiform apparatus. Because myb98 synergid cells are largely normal (Figures 5B and 5F), it is unlikely that MYB98 lies at the top of the synergid cell gene regulatory hierarchy. Instead, MYB98 is most likely a midlevel transcription factor controlling a subset of the synergid cell differentiation program; that is, MYB98 appears to control a branch of the synergid cell gene regulatory network involved specifically in pollen tube guidance and the formation of the filiform apparatus.
Identification of factors necessary for the expression of MYB98 in the synergid cell should provide insight into the gene regulatory circuitry specifying synergid cell fate, and identification and analysis of genes downstream of MYB98 should provide insight into the mechanisms that control the generation of the filiform apparatus during synergid cell differentiation and pollen tube guidance by the mature synergid cells. Such studies should ultimately lead to an understanding of the gene regulatory networks controlling the specification and differentiation of this important cell type during female gametophyte development.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana seeds were sterilized in 100% ethanol and germinated on plates containing 0.5× Murashige and Skoog salts (M-9274; Sigma-Aldrich), 0.05% MES, 0.5% sucrose, and 0.8% Phytagar (Life Technologies). Ten-day-old seedlings were transferred to Scott's Redi-Earth and grown under 24-h illumination.
Plant Transformation
T-DNA constructs were introduced into Agrobacterium tumefaciens strain LBA4404 by electroporation. Arabidopsis plants (ecotype Columbia) were transformed using a modified floral dip procedure (Clough and Bent, 1998). Transformed progeny were selected by germinating surface-sterilized T1 seeds on growth medium containing antibiotics. Resistant seedlings were transplanted to soil after 10 d of growth.
Analysis of ProMYB98:GFP and ProMYB98:GUS Expression
The ProMYB98:GFP and ProMYB98:GUS constructs included 1674 bp of sequence upstream of the translational start codon and the first exon of MYB98. This genomic region was obtained by PCR amplification using the primers Myb98BamF (5′-GGGGATCCGCGGCGGAGATAGTGGCTGAGAGGTGGA-3′) and Myb98BamR (5′-TCGGATCCGTGTTCTTGATCACGTGTGAAG-3′). These primers introduced BamHI sites at each end. The resulting PCR product was cloned into pBI101.1 (Clontech) and pBI-GFP(S65T) (Choi et al., 2002) using the BamHI sites, resulting in plasmids pBI101-pMYB98:GUS and pBI-pMYB98:GFP. These constructs were introduced into Arabidopsis plants as described above, and transformed plants were selected by germinating seeds on growth medium containing 40 μg/mL kanamycin. The expression patterns reported in Results are derived from the analysis of five single-insert ProMYB98:GUS lines and seven single-insert ProMYB98:GFP lines.
Tissue from plants containing the pBI101-pMYB98:GUS construct was stained for GUS activity in a solution containing 50 mM sodium phosphate buffer, pH 7.0, 0.2% Triton X-100, 10 mM potassium ferrocyanide, 10 mM potassium ferricyanide, and 1 mM X-Gluc. For staining of ovule tissue, flowers were emasculated at stage 12c (Christensen et al., 1997) and pistils were collected at 24 h after emasculation. Ovules were dissected from the pistil tissue, placed into staining solution on ice, vacuum-infiltrated for 15 min, and stained for 30 min at 37°C. Seeds were processed as above except for the modifications described in the legend of Figure 3. Seedlings (for analysis of expression in leaves and roots) were processed as above except that they were stained for 16 h in a solution containing 2.5 mM potassium ferrocyanide and 2.5 mM potassium ferricyanide. Flower clusters were processed as above except that they were stained for 24 h in a solution containing no potassium ferrocyanide/ferricyanide.
Tissue from plants containing the pBI-pMYB98:GFP construct was initially analyzed using an Olympus SXZ12 compound UV dissecting microscope with epifluorescence. For confocal analysis, we emasculated flowers at stage 12c, waited 24 h, and then removed the flowers from the plants. We then removed sepals, petals, and stamen and dissected off the carpel walls using a 30-gauge syringe needle. The pistils were then mounted on a slide in 10 mM phosphate buffer, pH 7.0. Pistils were analyzed using a Zeiss LSM510 confocal microscope. GFP was excited with an argon laser at a wavelength of 488 nm, and emission was detected between 500 and 530 nm.
RT-PCR
Tissue was harvested from plants and placed immediately into liquid nitrogen. Pistils were harvested from flowers at stage 13 (Smyth et al., 1990). Young flower tissue includes the inflorescence meristem and flowers at stages 1 to 11 (Smyth et al., 1990). Silique tissue includes siliques at 1 to 2 d after pollination. Leaf tissue includes leaves of 10 to 20 mm. Young leaf tissue includes the apical meristem and leaves <5 mm in length. Roots were harvested from seedlings at 10 d after germination. Floral stem tissue includes internodes from 5-week-old plants.
RNA was extracted from these tissues using the Qiagen RNeasy kit according to the manufacturer's instructions (www.qiagen.com). Aliquots of RNA (1 μg) were reverse-transcribed using the RETROscript kit (Ambion) according to the manufacturer's instructions.
For gel-based RT-PCR (Figures 2A and 2C), PCR was performed in a volume of 10 μL on a MJ PTC-200 machine. The PCR mixture consisted of 2 μL of cDNA, 1 μM primers, and 1× master mix. The PCR program consisted of a first step of denaturation and Taq activation (95°C for 4 min) followed by 30 cycles of denaturation (95°C for 5 s), annealing (60°C for 5 s), and extension (72°C for 60 s). We used primers MYB98 0ATGF (5′-CTCCTTTCCAAAACAATGGAGAATTTCGTC-3′) and MYB98 2-3 exR (5′-GTCCTCTTCTTCACTCCATGTTTCTTTCTTA-3′) for amplification of MYB98 RNA, and primers QACT2-F (5′-TCCCTCAGCACATTCCAGCAGAT-3′) and QACT2-R (5′-AACGATTCCTGGACCTGCCTCATC-3′) for amplification of ACTIN2 RNA.
For real-time RT-PCR, PCR was performed using the Roche FastStart DNA Master SYBR Green I master mix (www.roche.com) in a volume of 10 μL on a Roche LightCycler system. The PCR mixture consisted of 0.015 to 0.15 μL of cDNA, 0.5 μM primers, and 1× master mix. In every real-time RT-PCR run, ACTIN2 was used as an internal control to normalize for variation in the amount of cDNA template. The PCR primers used were qMYB98F-IH (5′-AAGACAGGGTACTGATTCAACTCG-3′) and QMYB98-R (5′-AGCGATATGCGACCATTTACGCAA-3′). The PCR program consisted of a first step of denaturation and Taq activation (95°C for 5 min) followed by 45 cycles of denaturation (95°C for 15 s), annealing (60°C for 15 s), and extension (72°C for 10 s). To determine the specificity of the PCR, the amplified products were subjected to melt curve analysis using the machine's standard method. For each gene analyzed, at least four PCR procedures were performed. The reported values are averages of four to five independent trials. We calculated relative expression levels as follows. We first determined the transcript levels of MYB98 relative to a standard using the formula ΔCT = CT(MYB98) − CT(ACTIN2). We next calculated an average ΔCT value for each tissue. We then calculated relative expression levels using the equation 2−(average ΔCT (tissue) – average ΔCT (pistil)).
Cloning the MYB98 cDNA
To amplify a cDNA encompassing the entire open reading frame of MYB98, we used primers MYB98 cDNA F (5′-CAATGGAGAATTTCGTCGACGAGAATGG-3′) and MYB98 cDNA R (5′-CAAATCAGAGTCCATGAACAAAAGCATC-3′). The cDNA was cloned into the pCRII-TOPO vector using the TOPO TA cloning kit (Invitrogen), resulting in plasmid pCRII-cMYB98.
Sequence Analysis
We used pfam 17.0 (http://pfam.wustl.edu/hmmsearch.shtml) to identify protein motifs such as the Myb domains. We used psort (http://psort.nibb.ac.jp/form.html) to identify the putative nuclear localization signal. We used the MegAlign module of the DNAStar Lasergene suite to align sequences; specifically, we used the Lipman-Pearson algorithm for the alignment of protein sequences and the Wilbur-Lipman algorithm for the alignment of nucleotide sequences.
Characterization of the myb98-1 and myb98-2 Alleles
myb98-1 (SALK_020263) and myb98-2 (SALK_147765) were obtained from the Salk Institute Genomic Analysis Laboratory collection (Alonso et al., 2003). The T-DNA insertion in myb98-1 has two left borders. The T-DNA is inserted into the genomic sequence 42 nucleotides downstream of the start codon in MYB98 and is associated with a 23-bp deletion (nucleotides +43 to +65 deleted). The left border–genomic sequence junctions were determined by PCR using the T-DNA–specific primer LBa1 (5′-TGGTTCACGTAGTGGGCCATCG-3′) combined with the genomic sequence primer Myb98-1RP (5′-TCCACTGCTTGGAAATGATGATG-3′) and the T-DNA–specific primer ROK2 6781 RC (5′-ATCGCGCGCGGTGTCATCTA-3′) combined with the genomic sequence primer Myb98-1 LP (5′-TGTTTACCCCAAATTCAACAACACC-3′).
The T-DNA insertion in myb98-2 has two left borders. The T-DNA is inserted into the genomic sequence 195 nucleotides upstream of the start codon of MYB98 and is associated with a 19-bp deletion (nucleotides −196 to −214 deleted). The left border–genomic sequence junctions were determined by PCR using the T-DNA–specific primer LBa1 (see above) combined with the genomic sequence primer Myb98-2RP (5′-GCTTGTAGGTTTTGATTGGTCG-3′) and the T-DNA–specific primer LBa1 (see above) combined with the genomic sequence primer Myb98-2 LP (5′-TTGGTCAGAGACTTTTTGTGAGGG-3′).
Segregation Analysis
Plants segregating the myb98-1 allele were genotyped by PCR using the following primers: LBa1, Myb98-1LP, and Myb98-1RP (see above). Plants segregating the myb98-2 allele were genotyped by PCR using the following primers: LBa1, Myb98-2LP, and Myb98-2RP (see above). Heterozygous plants, identified by PCR, were used in the segregation analysis described below.
For self cross analysis, heterozygous plants were allowed to self-pollinate and progeny seed was collected. The progeny seed was germinated on growth medium containing no antibiotics. The progeny were genotyped and scored with PCR using the primers listed above.
For reciprocal cross analysis, heterozygous plants were crossed with the wild type as outlined in Table 1 and progeny seed was collected. The F1 seed was germinated on growth medium containing 50 μg/mL kanamycin, and the number of resistant and sensitive plants was scored. Resistant and sensitive plants were scored as being genotypes myb98/MYB98 and MYB98/MYB98, respectively.
Molecular Complementation
Molecular complementation was performed using a 3768-bp DNA fragment containing the MYB98 genomic coding sequence (1490 bp) along with 1636 bp of sequence upstream of the translational start codon and 642 bp of sequence downstream of the stop codon. This DNA fragment was amplified by PCR from BAC F28A21 using primers 98ResF (5′-AGGAATTCGAGTAACGGTAACGACGGAG-3′) and 98ResR (5′-CGAAAACCGCAGATGATTTAGAATTCGG-3′). These primers introduced EcoRI sites at each end. The resulting PCR product was cloned into pCAMBIA1300 (CAMBIA) using the EcoRI sites, producing plasmid pCAMBIA1300:MYB98. pCAMBIA1300 contains a marker gene conferring resistance to hygromycin. pCAMBIA1300:MYB98 was introduced into Arabidopsis plants as described above, and transformed plants were selected by germinating seeds on growth medium containing 15 μg/mL hygromycin. Hygromycin-resistant plants also containing the myb98-1 allele were identified by PCR using primers LBa1 (5′-TGGTTCACGTAGTGGGCCATCG-3′) and Myb98-1RP (5′-TCCACTGCTTGGAAATGATGATG-3′).
Plants heterozygous for the myb98-1 allele and hemizygous for the rescue construct were crossed as females with wild-type males. The F1 seed was germinated on growth medium containing 50 μg/mL kanamycin, and the number of resistant and sensitive plants was scored. Resistant and sensitive plants were scored as being genotypes myb98/MYB98 and MYB98/MYB98, respectively. We observed 178 kanamycin-resistant progeny and 302 kanamycin-sensitive progeny. To verify that kanamycin-resistant plants had the rescue construct, we performed PCR using primers pCAMLacZR (5′-CCAGCTGGCGAAAGGGGGAT-3′) and MYB98ATG800R (5′-CCGCATCGTTTATAACAAAGTGTTAACAGTG-3′) to detect the DNA fragment. Of 27 kanamycin-resistant plants analyzed, all contained the rescue construct. Three single-insert lines were analyzed.
To identify plants homozygous for both myb98-1 and the rescue construct, we crossed plants heterozygous for myb98-1 and hemizygous for the rescue construct with myb98-1/myb98-1. In the F2 generation, we identified plants in which 100% of the progeny were resistant to both kanamycin and hygromycin. Two plants of this genotype were identified and analyzed.
Confocal Analysis of Female Gametophyte Structure
Confocal laser scanning microscopy of ovules was performed as described previously (Christensen et al., 1997, 1998, 2002), with the modification that we used a Zeiss LSM510 microscope.
Analysis of Pollen Tube Growth
Flowers at stage 12c (Christensen et al., 1997) were emasculated and then pollinated with wild-type pollen at 24 h after emasculation. At 12 to 24 h after pollination, the flowers were removed from the plant and the sepals, petals, and stamen were removed. The carpel walls were then removed using a 30-gauge syringe needle. The resulting ovules and septum tissue were stained with 0.4% Congo red for 15 min, washed once in 0.5× Murashige and Skoog salts, and then mounted on a glass slide in 0.5× Murashige and Skoog salts. Stained pollen tubes were analyzed using a Zeiss LSM510 confocal microscope. The Congo red dye was excited with a HeNe laser at a wavelength of 543 nm. Emission was detected between 585 and 650 nm.
Plastic Sections and Transmission Electron Microscopy
Flowers from the wild type and myb98 were emasculated at stage 12c (Christensen et al., 1997). At 24 h after emasculation, pistils were collected, cut into 1-mm pieces, and fixed overnight at 4°C in a solution of 4% glutaraldehyde, 1% paraformaldehyde, and 100 mM cacodylate buffer, pH 7.2. After fixation, the tissue was washed three times with 100 mM cacodylate buffer on ice. The tissue segments then were postfixed for 4 h in 1% aqueous osmium tetroxide at room temperature, washed three times in distilled water, postfixed for 1 h in 1% aqueous uranyl acetate at room temperature, and washed three times in distilled water. The tissue segments then were dehydrated in a graded ethanol series (20, 40, 60, 80, 95, 95, 100, 100, and 100% for 20 min each) and then three times in propylene oxide for 10 min. The dehydrated tissues were transferred through intermediate stages of 1:2, 1:1, and 2:1 mixtures of propylene oxide:Spurr's expoxy resin and then embedded in pure Spurr's epoxy resin. The tissue blocks initially were sectioned at 1-μm thickness using an LKB Ultratome III microtome. Sections were stained with 1% toluidine blue and observed under bright-field optics using an Axioplan Zeiss microscope. For ultrastructural analysis, the same blocks were sectioned at 90-nm thickness using a Reichert-Jung Ultracut E microtome, and the sections were transferred to coated slot grids. Ultrastructural analysis was performed using a Hitachi H-7100 electron microscope at 75 kV. Images were quantified using Adobe Photoshop 7 software. Mutant and wild-type ovules were fixed in parallel.
Accession Number
The GenBank accession number for the MYB98 Landsberg erecta ecotype cDNA sequence is DQ198082. The MYB98 gene corresponds to locus At4g18770 from BAC clone F28A21 on chromosome 4.
Supplemental Data
The following material is available in the online version of this article.
Supplemental Figure 1. Comparison of MYB98 cDNA Sequences from Ecotypes Columbia (Col) and Landsberg erecta (Ler).
Supplementary Material
Acknowledgments
We thank Ramin Yadegari and members of the Drews laboratory for critical review of the manuscript. We thank the ABRC for providing the myb98 alleles. We thank Clarissa J. Christensen and Sachi Miyajima for technical assistance. We thank Ed King and the Department of Biology Microscopy Facility for guidance with the microscopic analysis. This work was supported in part by a University of Utah Funding Incentive seed grant to G.N.D. and a USDA Cooperative State Research, Education, and Extension Service fellowship (2004-35304-14931) to M.F.P.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Gary N. Drews (drews@bioscience.utah.edu).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.034603.
References
- Acosta-Garcia, G., and Vielle-Calzada, J.P. (2004). A classical arabinogalactan protein is essential for the initiation of female gametogenesis in Arabidopsis. Plant Cell 16, 2614–2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657. [DOI] [PubMed] [Google Scholar]
- Bai, X., Peirson, B.N., Dong, F., Xue, C., and Makaroff, C.A. (1999). Isolation and characterization of SYN1, a RAD21-like gene essential for meiosis in Arabidopsis. Plant Cell 11, 417–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, S.C., Robinson-Beers, K., Villanueva, J.M., Gaiser, J.C., and Gasser, C.S. (1997). Interactions among genes regulating ovule development in Arabidopsis thaliana. Genetics 145, 1109–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt, A.M., Lister, C., Page, T., Fransz, P., Findlay, K., Jones, G.H., Dickinson, H.G., and Dean, C. (1999). The DIF1 gene of Arabidopsis is required for meiotic chromosome segregation and belongs to the REC8/RAD21 cohesin gene family. Plant J. 19, 463–472. [DOI] [PubMed] [Google Scholar]
- Cai, X., Dong, F., Edelmann, R.E., and Makaroff, C.A. (2003). The Arabidopsis SYN1 cohesin protein is required for sister chromatid arm cohesion and homologous chromosome pairing. J. Cell Sci. 116, 2999–3007. [DOI] [PubMed] [Google Scholar]
- Choi, Y., Gehring, M., Johnson, L., Hannon, M., Harada, J.J., Goldberg, R.B., Jacobsen, S.E., and Fischer, R.L. (2002). DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in Arabidopsis. Cell 110, 33–42. [DOI] [PubMed] [Google Scholar]
- Christensen, C.A., Gorsich, S.W., Brown, R.H., Jones, L.G., Brown, J., Shaw, J.M., and Drews, G.N. (2002). Mitochondrial GFA2 is required for synergid cell death in Arabidopsis. Plant Cell 14, 2215–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen, C.A., King, E.J., Jordan, J.R., and Drews, G.N. (1997). Megagametogenesis in Arabidopsis wild type and the Gf mutant. Sex. Plant Reprod. 10, 49–64. [Google Scholar]
- Christensen, C.A., Subramanian, S., and Drews, G.N. (1998). Identification of gametophytic mutations affecting female gametophyte development in Arabidopsis. Dev. Biol. 202, 136–151. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Cordts, S., Bantin, J., Wittich, P.E., Kranz, E., Lorz, H., and Dresselhaus, T. (2001). ZmES genes encode peptides with structural homology to defensins and are specifically expressed in the female gametophyte of maize. Plant J. 25, 103–114. [DOI] [PubMed] [Google Scholar]
- Diaz, I., Vicente-Carbajosa, J., Abraham, Z., Martinez, M., Isabel-La Moneda, I., and Carbonero, P. (2002). The GAMYB protein from barley interacts with the DOF transcription factor BPBF and activates endosperm-specific genes during seed development. Plant J. 29, 453–464. [DOI] [PubMed] [Google Scholar]
- Drews, G.N., Lee, D., and Christensen, C.A. (1998). Genetic analysis of female gametophyte development and function. Plant Cell 10, 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez, E., Royo, J., Guo, Y., Thompson, R., and Hueros, G. (2002). Establishment of cereal endosperm expression domains: Identification and properties of a maize transfer cell-specific transcription factor, ZmMRP-1. Plant Cell 14, 599–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashiyama, T. (2002). The synergid cell: Attractor and acceptor of the pollen tube for double fertilization. J. Plant Res. 115, 149–160. [DOI] [PubMed] [Google Scholar]
- Higashiyama, T., Kuroiwa, H., Kawano, S., and Kuroiwa, T. (1998). Guidance in vitro of the pollen tube to the naked embryo sac of Torenia fournieri. Plant Cell 10, 2019–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashiyama, T., Yabe, S., Sasaki, N., Nishimura, Y., Miyagishima, S., Kuroiwa, H., and Kuroiwa, T. (2001). Pollen tube attraction by the synergid cell. Science 293, 1480–1483. [DOI] [PubMed] [Google Scholar]
- Huang, B.-Q., and Russell, S.D. (1992). Female germ unit: Organization, isolation, and function. Int. Rev. Cytol. 140, 233–292. [Google Scholar]
- Huck, N., Moore, J.M., Federer, M., and Grossniklaus, U. (2003). The Arabidopsis mutant feronia disrupts the female gametophytic control of pollen tube reception. Development 130, 2149–2159. [DOI] [PubMed] [Google Scholar]
- Hulskamp, M., Schneitz, K., and Pruitt, R.E. (1995). Genetic evidence for a long-range activity that directs pollen tube guidance in Arabidopsis. Plant Cell 7, 57–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, W.A. (1965). The ultrastructure and histochemistry of the synergids of cotton. Am. J. Bot. 52, 238–256. [PubMed] [Google Scholar]
- Jia, L., Clegg, M.T., and Jiang, T. (2004). Evolutionary dynamics of the DNA-binding domains in putative R2R3-MYB genes identified from rice subspecies indica and japonica genomes. Plant Physiol. 134, 575–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, C., Gu, X., and Peterson, T. (2004). Identification of conserved gene structures and carboxy-terminal motifs in the Myb gene family of Arabidopsis and Oryza sativa L. ssp. indica. Genome Biol. 5, R46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, H., and Martin, C. (1999). Multifunctionality and diversity within the plant MYB-gene family. Plant Mol. Biol. 41, 577–585. [DOI] [PubMed] [Google Scholar]
- Kanei-Ishii, C., Sarai, A., Sawazaki, T., Nakagoshi, H., He, D.N., Ogata, K., Nishimura, Y., and Ishii, S. (1990). The tryptophan cluster: A hypothetical structure of the DNA-binding domain of the myb protooncogene product. J. Biol. Chem. 265, 19990–19995. [PubMed] [Google Scholar]
- Kirik, V., Lee, M.M., Wester, K., Herrmann, U., Zheng, Z., Oppenheimer, D., Schiefelbein, J., and Hulskamp, M. (2005). Functional diversification of MYB23 and GL1 genes in trichome morphogenesis and initiation. Development 132, 1477–1485. [DOI] [PubMed] [Google Scholar]
- Kranz, H., Scholz, K., and Weisshaar, B. (2000). c-MYB oncogene-like genes encoding three MYB repeats occur in all major plant lineages. Plant J. 21, 231–235. [DOI] [PubMed] [Google Scholar]
- Lipsick, J.S., and Wang, D.M. (1999). Transformation by v-Myb. Oncogene 18, 3047–3055. [DOI] [PubMed] [Google Scholar]
- Marton, M.L., Cordts, S., Broadhvest, J., and Dresselhaus, T. (2005). Micropylar pollen tube guidance by egg apparatus 1 of maize. Science 307, 573–576. [DOI] [PubMed] [Google Scholar]
- Matsui, K., Hiratsu, K., Koyama, T., Tanaka, H., and Ohme-Takagi, M. (2005). A chimeric AtMYB23 repressor induces hairy roots, elongation of leaves and stems, and inhibition of the deposition of mucilage on seed coats in Arabidopsis. Plant Cell Physiol. 46, 147–155. [DOI] [PubMed] [Google Scholar]
- Ogata, K., Hojo, H., Aimoto, S., Nakai, T., Nakamura, H., Sarai, A., Ishii, S., and Nishimura, Y. (1992). Solution structure of a DNA-binding unit of Myb: A helix-turn-helix-related motif with conserved tryptophans forming a hydrophobic core. Proc. Natl. Acad. Sci. USA 89, 6428–6432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata, K., Morikawa, S., Nakamura, H., Sekikawa, A., Inoue, T., Kanai, H., Sarai, A., Ishii, S., and Nishimura, Y. (1994). Solution structure of a specific DNA complex of the Myb DNA-binding domain with cooperative recognition helices. Cell 79, 639–648. [DOI] [PubMed] [Google Scholar]
- Ogata, K., et al. (1995). Comparison of the free and DNA-complexed forms of the DNA-binding domain from c-Myb. Nat. Struct. Biol. 2, 309–320. [DOI] [PubMed] [Google Scholar]
- Oppenheimer, D.G., Herman, P.L., Sivakumaran, S., Esch, J., and Marks, M.D. (1991). A myb gene required for leaf trichome differentiation in Arabidopsis is expressed in stipules. Cell 67, 483–493. [DOI] [PubMed] [Google Scholar]
- Ray, S., Park, S.-S., and Ray, A. (1997). Pollen tube guidance by the female gametophyte. Development 124, 2489–2498. [DOI] [PubMed] [Google Scholar]
- Rotman, N., Rozier, F., Boavida, L., Dumas, C., Berger, F., and Faure, J.E. (2003). Female control of male gamete delivery during fertilization in Arabidopsis thaliana. Curr. Biol. 13, 432–436. [DOI] [PubMed] [Google Scholar]
- Russell, S.D. (1993). The egg cell: Development and role in fertilization and early embryogenesis. Plant Cell 5, 1349–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu, K.K., and Okada, K. (2000). Attractive and repulsive interactions between female and male gametophytes in Arabidopsis pollen tube guidance. Development 127, 4511–4518. [DOI] [PubMed] [Google Scholar]
- Smyth, D.R., Bowman, J.L., and Meyerowitz, E.M. (1990). Early flower development in Arabidopsis. Plant Cell 2, 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke, R., Werber, M., and Weisshaar, B. (2001). The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 4, 447–456. [DOI] [PubMed] [Google Scholar]
- Tahirov, T.H., et al. (2002). Mechanism of c-Myb-C/EBP beta cooperation from separated sites on a promoter. Cell 108, 57–70. [DOI] [PubMed] [Google Scholar]
- Weterings, K., and Russell, S.D. (2004). Experimental analysis of the fertilization process. Plant Cell 16 (suppl.), S107–S118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemse, M.T.M., and van Went, J.L. (1984). The female gametophyte. In Embryology of Angiosperms, B.M. Johri, ed (Berlin: Springer-Verlag), pp. 159–196.
- Williams, E.G., Kaul, V., Rouse, J.L., and Palser, B.F. (1986). Overgrowth of pollen tubes in embryo sacs of Rhododendron following interspecific pollinations. Aust. J. Bot. 34, 413–423. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.