Abstract
In plants, secondary wall thickenings play important roles in various biological processes, although the factors regulating these processes remain to be characterized. We show that expression of chimeric repressors derived from NAC SECONDARY WALL THICKENING PROMOTING FACTOR1 (NST1) and NST2 in Arabidopsis thaliana resulted in an anther dehiscence defect due to loss of secondary wall thickening in anther endothecium. Plants with double, but not single, T-DNA–tagged lines for NST1 and NST2 had the same anther-indehiscent phenotype as transgenic plants that expressed the individual chimeric repressors, indicating that NST1 and NST2 are redundant in regulating secondary wall thickening in anther walls. The activity of the NST2 promoter was particularly strong in anther tissue, while that of the NST1 promoter was detected in various tissues in which lignified secondary walls develop. Ectopic expression of NST1 or NST2 induced ectopic thickening of secondary walls in various aboveground tissues. Epidermal cells with ectopic thickening of secondary walls had structural features similar to those of tracheary elements. However, among genes involved in the differentiation of tracheary elements, only those related to secondary wall synthesis were clearly upregulated. None of the genes involved in programmed cell death were similarly affected. Our results suggest NAC transcription factors as possible regulators of secondary wall thickening in various tissues.
INTRODUCTION
In contrast with primary cell walls, which are synthesized in basically all plant cells, lignified secondary walls develop only in cells that have ceased to expand (Turner et al., 2001). After the cessation of expansion and division, a secondary wall is synthesized within the bounds of the primary wall. The successive addition of secondary xylem with heavily thickened secondary walls forms wood, which accounts for a major part of terrestrial biomass and is widely used as a renewable material and source of energy for humans. In plants, secondary wall thickenings play important roles in various biological processes, such as the dehiscence of anthers, the shattering of silique pods, and the formation of tracheary elements and fibers. The lignification in the endodermal layer of the valve margin of silique pods is necessary for their dehiscence, generating tension via desiccation and leading to pod shattering (Spence et al., 1996; Liljegren et al., 2000, 2004). Secondary wall thickenings in the xylem, including the vessels, parenchyma, and the interfascicular region of inflorescence stems, help vascular plants to withstand the negative pressure in vessels generated through transpiration and provide mechanical strength to stems. Xylem vessels are composed of tracheary elements whose secondary wall thickenings have elaborate striated patterns (Fukuda, 1997; Ye, 2002), which are formed by microtubules (Ye, 2002; Oda et al., 2005). Unlike other cells with secondary wall thickening, the formation of tracheary elements is immediately followed by programmed cell death during the differentiation process (Fukuda, 1997; Ye, 2002), and several proteases and nucleases involved in programmed cell death have been identified (Perez-Amador et al., 2000; Funk et al., 2002; Ito and Fukuda, 2002).
The secondary walls of anther endothecium have striated patterns similar to those in tracheary elements. These secondary wall thickenings are necessary for anther dehiscence, and they generate the tensile force necessary for the rupture of the stomium (Keijzer, 1987). A loss-of-function mutation in the Arabidopsis thaliana MYB26 gene has been shown to induce a defect in the secondary wall thickening of anther walls with resultant indehiscent anthers (Steiner-Lange et al., 2003). Defects in DEFECTIVE IN ANTHER DEHISCENCE1 (DAD1), CORONATINE INSENSITIVE1 (COI1), DELAYED DEHISCENCE1, or FATTY ACID DESATURATION3/7/8 also affect anther dehiscence. The products of these genes are involved in the jasmonic acid signaling that had been suggested to control dehydration during the maturation of anthers but not the thickening of secondary walls (McConn and Browse, 1996; Xie et al., 1998; Sanders et al., 2000; Ishiguro et al., 2001).
In spite of substantial structural similarities among the secondary wall thickenings of various tissues, no common mechanism or factor that regulates secondary wall thickening has been fully characterized. In this report, we provide evidence that two NAC transcription factors, NAC SECONDARY WALL THICKENING PROMOTING FACTOR1 (NST1) and NST2, act redundantly to regulate the secondary cell wall thickenings in anther walls that are necessary for anther dehiscence. We also show that NAC transcription factors are possible regulators of secondary wall thickening not only in the anther wall but also in various tissues, including tracheary elements.
RESULTS
Expression of Chimeric NST1 and NST2 Repressors Induces Defects in Anther Dehiscence
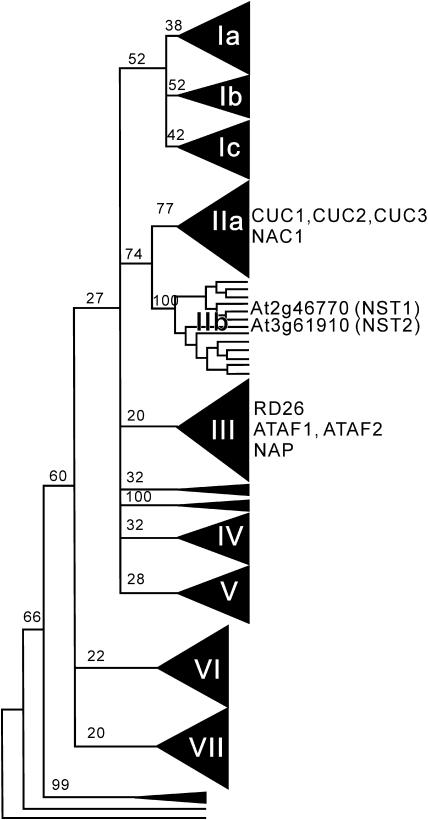
In the Arabidopsis genome, we found 110 genes that encode NAC transcription factors by computational analysis. A phylogenetic tree of the NAC family of transcription factors revealed seven different groups (Figure 1). CUP-SHAPED COTYLEDON1 (CUC1), CUC2, and CUC3, which regulate organ separation and meristem formation (Aida et al., 1997; Takada et al., 2001; Vroemen et al., 2003), and NAC1, which regulates lateral root development (Xie et al., 2000), are classified into subgroup IIa. In addition, RD26 and NAC-LIKE ACTIVATED BY AP3/PI (NAP), which are involved in abscisic acid signaling and in stamen elongation, respectively (Sablowski and Meyerowitz, 1998; Fujita et al., 2004), belong to group III. However, most members of the NAC family have not yet been characterized.
Figure 1.
Schematic Diagram of an Unrooted Phylogenetic Tree of NAC Transcription Factors from Arabidopsis.
Some bootstrap values from 100 trials are shown on the tree. The branches with <20% support have been collapsed to give polytomies. NAC transcription factors whose functions have been characterized, namely, CUC1, CUC2, CUC3, and NAC1, which belong to subgroup IIa, and RD26, ATAF1, ATAF2, and NAP, which belong to subgroup III, are shown. NST1 and NST2, which were characterized in this study, are also shown. Triangles with Roman numerals represent subgroups.
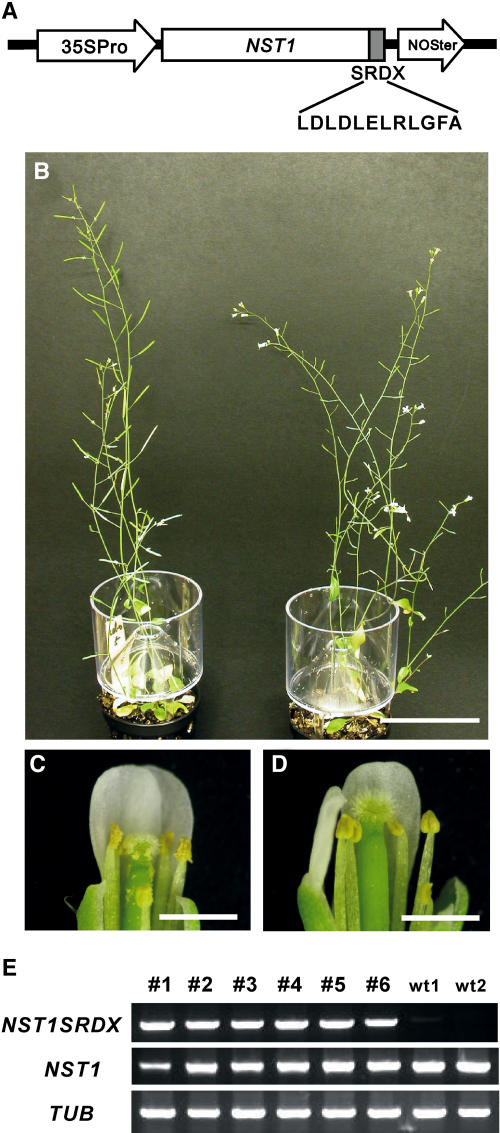
For functional analysis of NAC transcription factors, we applied our chimeric repressor silencing technology and converted individual NAC transcription factors into dominant repressors by fusion with the EAR-motif repression domain (SRDX; Hiratsu et al., 2003, 2004). We found that a NAC transcription factor in subgroup IIb (At2g46770) induced anther indehiscence when it was converted to a chimeric repressor and expressed under control of the cauliflower mosaic virus (CaMV) 35S promoter (35S:NST1SRDX) in Arabidopsis (Figure 2). We designated the At2g46770 NAC transcription factor NST1. Transgenic plants expressing the chimeric NST1 repressor were morphologically indistinguishable from the wild type except for the indehiscent anthers (Figures 2B to 2D), even though NST1 belongs to a neighboring cluster of CUC genes and the expression of the chimeric CUC1 repressor induces organ fusion and a defect in meristems, as in cuc1 cuc2 double mutants (Aida et al., 1997; Hiratsu et al., 2003). We confirmed that both the NST1SRDX transgene and the endogenous NST1 gene were expressed in the inflorescences of 35S:NST1SRDX plants (Figure 2E), and this observation suggested that the indehiscent anther phenotype of 35S:NST1SRDX plants was probably not due to cosuppression effects.
Figure 2.
Expression of the Chimeric NST1 Repressor Induces Indehiscent Anthers in Arabidopsis.
(A) Schematic diagram of the 35S:NST1SRDX transgene. 35SPro, NOSter, and SRDX represent the CaMV 35S promoter, the terminator sequence of the NOS gene, and the repression domain of 12 amino acids, respectively.
(B) Six-week-old wild-type (left) and 35S:NST1SRDX (right) plants are shown. The 35S:NST1SRDX transgenic plants rarely set elongated siliques.
(C) Top half of a wild-type flower. Some of the petals and sepals have been removed. Spilt pollen can be seen on the anthers and pistil of the wild type, while no such pollen grains are observed in the transgenic plant (D).
(D) The top half of a transgenic flower. Some of the petals and sepals have been removed.
(E) The expression of the NST1SRDX transgene and the endogenous NST1 gene in 35S:NST1SRDX plants was examined by RT-PCR analysis. Numbers above lanes (1 to 6) indicate individual T1 transgenic plants with indehiscent anthers. wt1 and wt2 indicate wild-type plants. TUB indicates the gene for β-tubulin, which was used as an internal control.
Bars = 5 cm in (B) and 1 mm in (C) and (D).
To analyze the function of NST2 (At3g61910), a homolog of NST1 with 63% identity at the amino acid level, we also expressed the chimeric NST2 repressor in Arabidopsis. The expression of the chimeric NST2 repressor induced no obviously unusual phenotype when the gene was driven by the CaMV 35S promoter (35S:NST2SRDX). To investigate the biological function of the NST transcription factors in more detail, we fused an upstream region of ∼3 kb of NST1 and NST2 to NST1SRDX and NST2SRDX, respectively, to express the chimeric NST1 and NST2 repressors under the control of their own promoters (ProNST1:NST1SRDX and ProNST2:NST2SRDX). Both lines of transgenic plants had indehiscent anthers, resembling 35S:NST1SRDX plants. The frequency of indehiscent anthers in these transgenic plants was higher than in 35S:NST1SRDX plants, suggesting that the promoters of these genes are appropriately active in anthers.
NST1 and NST2 Regulate Secondary Wall Thickening in the Anther Endothecium
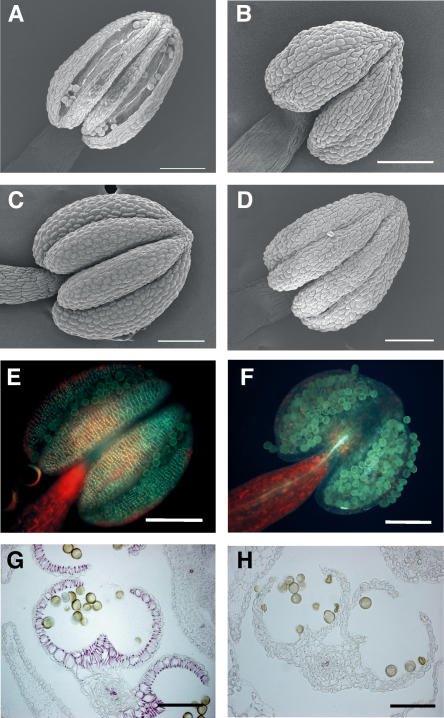
Among mutants with indehiscent anthers, the dad1 mutant was shown to have a defect in the dehydration process that is necessary for the maturation of anthers (Ishiguro et al., 2001), while the Arabidopsis MYB26 mutant, like 35S:MYB26SRDX plants, has a defect in the thickening of the secondary wall of anther walls (Steiner-Lange et al., 2003; our unpublished data). To identify the way in which NST1SRDX and NST2SRDX induce indehiscent anthers, we investigated the morphology of anthers of indehiscent mutants and transgenic plants. The anthers of the dad1 mutant, as well as those of the coi1 mutant (data not shown), were turgid and failed to mature, even when the flower was fully open, as a result of a defect in dehydration (Figure 3C). By contrast, the anthers of 35S:MYB26SRDX plants were desiccated and mature, as in the wild type, when the flower was fully open, but anthers were indehiscent because of the absence of secondary wall thickening (Figures 3A and 3D). When we compared the indehiscent anthers of these two different types of mutant with the anthers of 35S:NST1SRDX plants, we found that 35S:MYB26SRDX had similar morphological features to those of 35S:NST1SRDX plants (Figures 3B and 3D). To determine whether the indehiscent anthers of 35S:NST1SRDX plants were due to the loss of secondary wall thickening, we investigated the lignified material that is a major component of secondary walls by cytological analysis. In the anthers of wild-type plants, the distinct net-like autofluorescence of lignin was clearly visible under UV illumination, and the deep red staining of lignified material by phloroglucinol was clearly recognized in the endothecium layer (Figures 3E and 3G). However, little or no net-like structure and staining were observed in the anthers of 35S:NST1SRDX plants (Figures 3F and 3H). The same results were recorded for ProNST1:NST1SRDX and ProNST2:NST2SRDX plants (data not shown). These observations indicated that the indehiscent anthers that were induced by the chimeric NST1 and NST2 repressors were due to a defect in secondary wall thickening in the anther endothecium and that NST1 and NST2 are positive regulators of secondary wall thickening in anther walls. Since the xylem vessels in the stamens of 35S:NST1SRDX plants were stained by phloroglucinol (Figure 3H), it appeared that the chimeric NST1 repressor had not suppressed the secondary wall thickening anywhere other than in the anther walls in stamens.
Figure 3.
Microscopy Observations of Dehiscent and Indehiscent Anthers.
(A) Scanning electron microscopy image of wild-type anther showing longitudinal cleavage in the center of the locule and the retracted anther wall.
(B) 35S:NST1SRDX anther with an indehiscent but desiccated and mature appearance.
(C) dad1 mutant anther with an indehiscent and turgid appearance.
(D) 35S:MYB26SRDX anther with an indehiscent but desiccated and mature appearance.
(E) Wild-type anther showing the net-like autofluorescence of lignin under UV illumination.
(F) 35S:NST1SRDX anther under UV illumination with no autofluorescence.
(G) Cross section of wild-type anther stained with phloroglucinol. Lignified material is stained deep red.
(H) Cross section of a 35S:NST1SRDX anther stained with phloroglucinol. No deep red staining is evident.
Bars = 100 μm.
NST1SRDX and NST2SRDX Act as Repressors in Plants
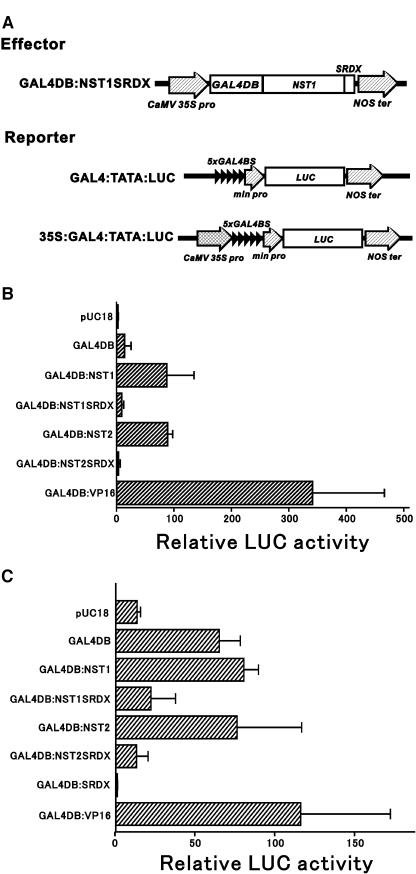
To confirm that the indehiscent anther phenotype of the transgenic plants had been induced by the repressive activity of each transgene, we examined the repressive activity of NST1SRDX and NST2SRDX by transient reporter analysis. As shown in Figure 4, the coding region of NST1 or NST2 was fused to the yeast GAL4 DNA binding domain in both of the effector plasmids. The luciferase reporter gene that was driven by a minimal promoter that contained the GAL4 binding site was then activated when Arabidopsis leaves were cobombarded with each pair of plasmids. Moreover, while neither the effector plasmid for NST1SRDX nor that for NST2SRDX activated the GAL4:TATA:LUC reporter gene (Figure 4B), both plasmids significantly suppressed the expression of the reporter gene construct that contained the CaMV 35S promoter with GAL4 binding sites (Figure 4C). These data indicate that NST1 and NST2 have activation activity and that both gene products with the SRDX repression domain act as repressors in plant cells. These observations suggest that the indehiscent phenotype observed in this study was induced by the suppression of the target genes of NST1 and NST2.
Figure 4.
Functional Analysis of NST1 and NST2 and the Corresponding Chimeric Repressors by Transient Expression Assays.
(A) Schematic representation of the constructs for expression of the effector and reporter genes. Only one example of an effector construct is shown. The reporter contains repeated GAL4 binding sites and a minimal CaMV 35S promoter (−46) upstream of the luciferase reporter with or without the enhancer region (−800 to −46) of the CaMV 35S promoter. GAL4DB, 5xGAL4BS, VP16, and 35S represent the DNA binding domain of yeast GAL4 transcription factor, five repeated GAL4 binding sites, the transcriptional activation domain of Herpes simplex virus, and the enhancer region of the CaMV 35S promoter, respectively.
(B) Relative luciferase activities after cobombardment of Arabidopsis leaves with GAL4DB fusion effectors and the GAL4:TATA:LUC reporter gene.
(C) Relative luciferase activities after cobombardment of Arabidopsis leaves with GAL4DB fusion effectors and the 35S:GAL4:TATA:LUC reporter gene. All luciferase activities are expressed relative to the value obtained with the reporter construct alone. Error bars indicate sd (n = 3).
Promoter Activities of the NST1 and NST2 Genes
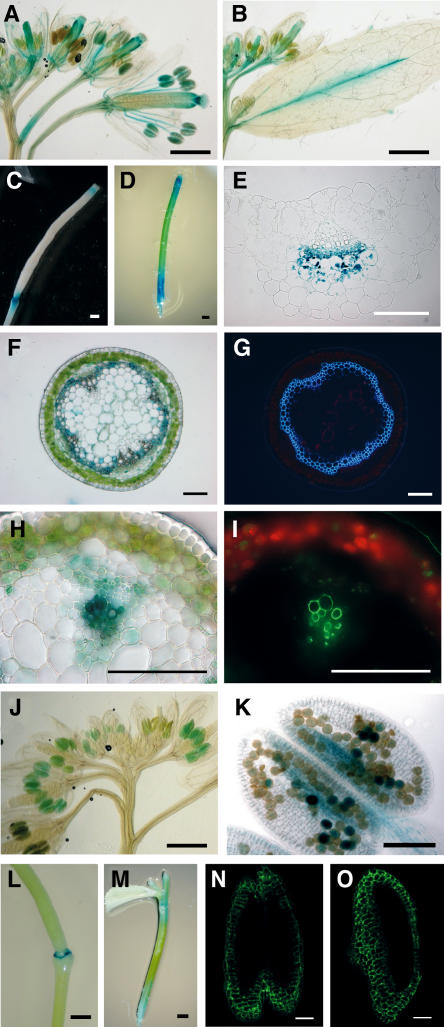
We examined the promoter activities of the NST1 and NST2 genes in promoter-reporter experiments in an effort to define the biological functions of these genes in more detail. As in the ProNST1:NST1SRDX and ProNST2:NST2SRDX constructs, we fused an upstream region of ∼3 kb from the codon for initiation of translation of the NST1 and, separately, of the NST2 gene to a reporter gene for the β-glucuronidase (GUS) reporter gene and expressed the constructs in Arabidopsis. As shown in Figures 5A to 5D, the GUS activity that was induced by the promoter of the NST1 gene was found in various aboveground tissues, including anthers, filaments of stamens, the base of carpels, styles, the boundaries between siliques and pedicels, the midrib of leaf veins, and inflorescence stems. Sections of leaf petioles of ProNST1:GUS plants revealed GUS activity on the phloem side of vascular bundles in the leaf midrib (Figure 5E). In the basal part of inflorescence stems, GUS activity was detected in a circular pattern on the adaxial side of the cambium (Figure 5F), while in the upper part of these stems immediately below the flower buds, activity was detected in the vascular bundles (Figure 5H). Illumination by UV light revealed that the region in which the GUS activity was detected in the inflorescence stem corresponded clearly to the region that was undergoing thickening of the lignified secondary wall, suggesting that NST1 is expressed in tissues in which secondary walls are developing (Figures 5F to 5I). By contrast, strong activity of the NST2 promoter was detected in anther walls and pollen grains (Figures 5J and 5K). Occasionally, we detected the activity of the NST2 promoter in the basal part of siliques and in inflorescence stems (Figures 5L and 5M).
Figure 5.
Promoter Activities of the NST1 and NST2 Genes.
(A) Promoter activity of NST1 in inflorescences. GUS activity is evident in anthers, filaments of stamens, and carpels of ProNST1:GUS plants.
(B) Promoter activity of NST1 in cauline leaves. GUS activity is evident in the midrib of leaf veins of ProNST1:GUS plants.
(C) Promoter activity of NST1 in a silique. GUS activity was evident in the upper region and at boundaries between siliques and pedicels of ProNST1:GUS plants.
(D) Promoter activity of NST1 in an inflorescence stem.
(E) Cross section of a petiole of a cauline leaf of a ProNST1:GUS plant. GUS activity is evident on the phloem side of vascular bundles.
(F) Cross section of the basal region of an inflorescence stem of a ProNST1:GUS plant. GUS activity is evident in a circular pattern on the adaxial side of the cambium.
(G) Cross section of the inflorescence stem in (F) under UV illumination. The circular region undergoing lignified secondary wall thickening emitted fluorescence that corresponded to the region in which GUS was expressed in (F).
(H) Cross section of the uppermost part of an inflorescence stem of a ProNST1:GUS plant. GUS activity is evident in the vascular bundle.
(I) Cross section of the inflorescence stem in (H) under UV illumination. The lignified materials in vessels of the vascular bundle emit fluorescence that corresponds to the region in which GUS was expressed in (H).
(J) Promoter activity of the NST2 gene in inflorescences. Strong GUS activity is evident in the anthers of a ProNST2:GUS plant.
(K) Promoter activity of NST2 in an anther. GUS activity is evident in the anther wall and pollen grains
(L) Promoter activity of NST2 in the basal region of a silique.
(M) Promoter activity of NST2 in an inflorescence stem.
(N) Confocal image of GFP fluorescence in the anther wall of a ProNST1:GFP plant.
(O) Confocal image of GFP fluorescence in the anther wall of a ProNST2:GFP plant.
Bars = 100 μm in (E) to (I), (K), (N), and (O) and 1 mm in (A) to (D), (J), (L), and (M).
We examined the expression of NST1 and NST2 in the anther walls using, as a reporter, the gene for green fluorescent protein (GFP). Obvious fluorescence in anther walls was observed in both ProNST1:GFP and ProNST2:GFP transgenic plants. While obvious fluorescence due to GFP was observed in the anthers of most of the ProNST2:GFP lines, GFP fluorescence driven by the NST1 promoter was weaker than that in ProNST2:GFP plants and was detected in only a limited number of ProNST1:GFP transgenic lines (Figures 5N and 5O). These results indicate that the promoter activity of NST2 in anther walls was much higher than that of NST1. However, GFP fluorescence in pollen grains was rarely detectable in ProNST2:GFP plants (Figure 5O), even though the GUS activity in pollen grains was detected in some ProNST2:GUS transgenic lines (Figure 5K). This discrepancy might have been due to differences in stability between GUS and GFP.
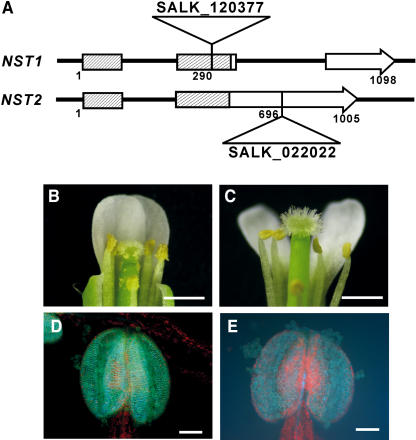
Double T-DNA–Tagged Lines for the NST1 and NST2 Genes Yield the Same Indehiscent Phenotype as NST1SRDX and NST2SRDX Transgenic Plants
To examine whether the phenotype induced by the chimeric NST1 and NST2 repressors corresponded to that of NST1 and NST2 knockout lines, we prepared stable T-DNA–tagged lines for the NST1 and NST2 genes, in which we confirmed the insertion of the T-DNA tag and the absence of expression of the correct transcript of NST1 and NST2, respectively (Figure 6A). Neither the homozygous NST1 single T-DNA–tagged line (SALK_120377) nor the NST2 single T-DNA–tagged line (SALK_022022) had a visibly defective phenotype (data not shown). To obtain the double T-DNA–tagged lines homozygous both for NST1 and NST2, we crossed the single T-DNA–tagged lines and self-pollinated the resultant F1 progeny. The resultant NST1 and NST2 double T-DNA–tagged plants had exclusively indehiscent anthers (Figure 6C), and light microscopy revealed that the indehiscent anthers were due to the loss of secondary wall thickening in the anther endocethuim (Figure 6E), as had been the case in the transgenic plants that expressed the chimeric NST1 or NST2 repressor (Figure 3F). No other recognizable defects in other tissues were found. These results demonstrated that the loss-of-function phenotype induced by each chimeric repressor corresponded to that of the double T-DNA–tagged line and that NST1 and NST2 are functionally redundant at least in the anther.
Figure 6.
NST1 and NST2 Double T-DNA–Tagged Lines Have Indehiscent Anthers.
(A) Schematic diagram of the structure of the NST1 and NST2 genes and the sites of insertion of T-DNA (SALK_120377 and SALK_022022) in the corresponding genes. Boxes and arrows represent coding regions, and thin lines represent noncoding regions. Hatched boxes represent conserved NAC domains.
(B) Wild-type flower.
(C) An NST1 and NST2 double T-DNA–tagged flower showing indehiscent anthers.
(D) Wild-type anther under UV illumination. Net-like autofluorescence is visible in the anther wall, an indication of the presence of lignified secondary walls.
(E) A double knockout anther under UV illumination.
Bars = 1 mm in (B) and (C) and 100 μm in (D) and (E).
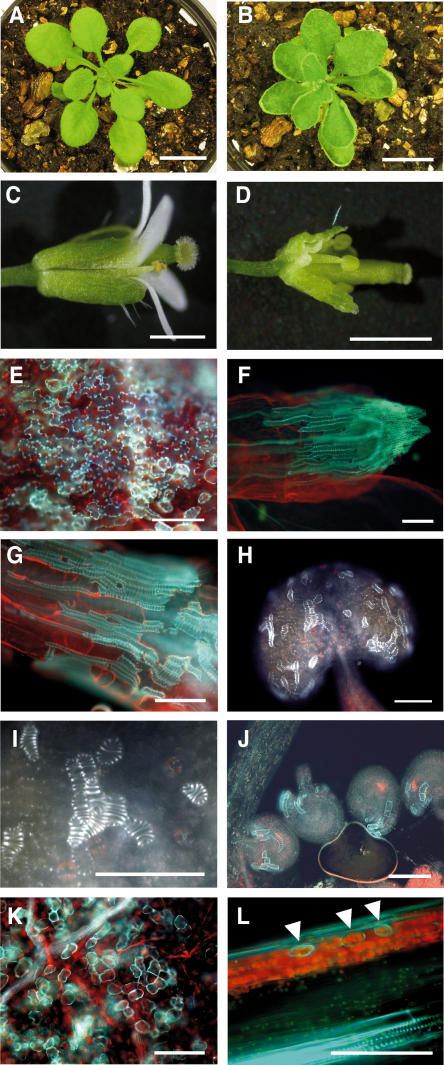
Ectopic Expression of NSTs Induces Ectopic Thickening of Secondary Walls in Various Tissues
Since our loss-of-function analysis showed that NST1 and NST2 are possible regulators of secondary wall thickening, we expressed these genes ectopically under the control of the CaMV 35S promoter (35S:NST1 and 35S:NST2) to investigate the molecular function of NST1 and NST2 by gain-of-function analysis. Ectopic expression of NST1 or NST2 was found to induce ectopic lignified secondary wall thickening in various tissues, including anthers, stamens, ovules, stems and leaves, and, less frequently, root tissues. The proportion of cells with secondary wall thickening was more frequent in 35S:NST1 plants than in 35S:NST2 plants, suggesting that the activity of NST1 might be higher than that of NST2. Moreover, most epidermal cells with ectopic secondary wall thickening had a striated appearance very similar to that of tracheary elements (Figures 7E to 7J). However, no similar striations were observed in the ectopic secondary walls in the mesophyll cells of rosette leaves and the cortical region of inflorescence stems (Figures 7K and 7L).
Figure 7.
Ectopic Expression of NST1 Induces Ectopic Secondary Wall Thickening in Various Tissues.
(A) Wild-type plant.
(B) 35S:NST1 plant showing upwardly curled rosette leaves.
(C) Wild-type flower.
(D) A flower from a 35S:NST1 plant. Sepals and petals were abnormally bent.
(E) Epidermal cells in a rosette leaf of a 35S:NST1 plant under UV illumination. Ectopic lignified secondary wall thickening is apparent as autofluorescence in the epidermal cells.
(F) A sepal of a 35S:NST1 plant under UV illumination. Ectopically and heavily lignified secondary walls with a striated pattern similar to that of tracheary elements are evident on the epidermis.
(G) Epidermis of a pedicel of a 35S:NST1 plant under UV illumination. An ectopic tracheary element-like structure is evident on the epidermis of the pedicel.
(H) An anther of a 35S:NST1 plant under UV illumination. A lignified ectopic tracheary element-like structure is evident on the epidermis of the anther.
(I) Magnified view of (H). The striated pattern of the ectopic secondary wall is evident.
(J) Ovules of a 35S:NST1 plant under UV illumination.
(K) Mesophyll cells of a rosette leaf of a 35S:NST1 plant under UV illumination.
(L) Longitudinal section of an inflorescence stem of a 35S:NST1 plant. Arrowheads indicate ectopic lignified secondary walls in the cortical region that do not exhibit a striated pattern.
Bars = 1 cm in (A) and (B), 1 mm in (C) and (D), and 100 μm in (E) to (L).
The 35S:NST1 and 35S:NST2 plants were usually smaller and grew more slowly than wild-type plants. The rosette leaves of the transgenic plants often curled upwards, and their marginal regions were extensively lignified (Figures 7B and 7E). Their floral organs were also smaller than those of wild-type plants, and their sepals and petals were often bent (Figure 7D). In these floral tissues, ectopically and heavily lignified secondary walls developed with striated patterns similar to those of tracheary elements (Figure 7F). These observations indicate that the abnormal appearance of leaves and floral organs of 35S:NST1 and 35S:NST2 plants was due to the ectopic accumulation of lignified materials, reflecting a previous report of a correlation between the extent of lignification and cell expansion (Cano-Delgado et al., 2000).
Enhanced Gene Expression in 35S:NST1 Plants
To characterize genes that are regulated by NST1, we performed microarray and comprehensive gene group analyses. Among 20,084 genes that we examined, the expression of 701 genes was enhanced twofold or more with Q-values under 0.01 in 35S:NST1 plants, as compared with the wild type (see Supplemental Table 1 online). Comparative analysis revealed that upregulated genes in 35S:NST1 plants significantly overlapped several groups of genes known to be related to secondary wall thickening (Table 1; see Supplemental Table 2 online). These genes include those that are specifically upregulated in xylem (Zhao et al., 2005; xylem biased gene set) and in weight-treated or intermediate stems (Ko et al., 2004) as well as genes involved in the biosynthesis and modification of xyloglucan and the cinnamate monolignol pathway. Thus, the entire transcriptome of 35S:NST1 plants is similar, to some extent, to that of xylem and to those of weight-treated or intermediate stems. It appears, therefore, that NST1 can activate genes related to secondary wall thickening. In addition, groups of genes that are upregulated by various abiotic stresses, such as drought, wounding, osmotic stress, and cold stress, significantly overlap the genes that are upregulated in 35S:NST1 plants. The genes related to abiotic stress responses also overlapped, to a significant extent, with those genes that are specifically upregulated in xylem and in weight-treated or intermediate stems (data not shown). These observations indicate that secondary wall thickening might be associated with stress responses or that genes related to responses to abiotic stress might be involved in secondary wall synthesis.
Table 1.
List of Groups of Genes That Overlap Significantly the Genes Whose Expression Is Enhanced in 35S:NST1 Plants
| Name of Gene Group | Number of Genes | Number of Genes Overlapped | Q-Value | Odds Ratio | Resource or Reference |
|---|---|---|---|---|---|
| Response to abscisic acid stimulus (GO:0009737) | 50 | 13 | 1.36E-08 | 9.69 | http://www.arabidopsis.org/ |
| Response to dessication (GO:0009269) | 14 | 6 | 3.86E-06 | 20.50 | http://www.arabidopsis.org/ |
| Response to wounding (GO:0009611) | 41 | 9 | 8.65E-06 | 7.71 | http://www.arabidopsis.org/ |
| Response to salt stress (GO:0009651) | 17 | 6 | 1.37E-05 | 14.91 | http://www.arabidopsis.org/ |
| Response to cold (GO:0009409) | 39 | 8 | 4.18E-05 | 7.07 | http://www.arabidopsis.org/ |
| Response to oxidative stress (GO:0006979) | 115 | 14 | 4.29E-05 | 3.82 | http://www.arabidopsis.org/ |
| Secondary wall biosynthesis (GO:0009834) | 4 | 3 | 0.000122 | 81.47 | http://www.arabidopsis.org/ |
| Lignin biosynthesis (GO:0009809) | 26 | 4 | 0.00640 | 4.95 | http://www.arabidopsis.org/ |
| Xylem biased genes | 283 | 57 | <2.96E-16 | 7.36 | Zhao et al. (2005) |
| Upregulated genes in weight-treated or intermediate stem | 587 | 80 | <2.96E-16 | 4.70 | Ko et al. (2004) |
| Genes involving xyloglucan biosynthesis and modification | 113 | 16 | 2.13E-06 | 4.55 | Tokimatsu et al. (2005) |
| Genes involving cinnamate monolignol pathway | 72 | 12 | 6.60E-06 | 5.50 | Tokimatsu et al. (2005) |
| Glycoside hydrolase | 345 | 30 | 1.00E-05 | 2.65 | Tokimatsu et al. (2005) |
| Peroxidase class III | 64 | 11 | 1.12E-05 | 5.70 | Tokimatsu et al. (2005) |
Selected groups of genes that significantly overlap the genes that are upregulated (twofold or more with Q-value <0.01) in 35S:NST1 plants are listed. The number of genes in each group that was examined by microarray analysis in this study is given in the second column, and of these, the number of genes that were the same as genes upregulated in 35S:NST1 plant is listed in the third column. Q-values (see Methods) and odds ratios (= number of genes that actually overlapped/number of genes expected by chance) from Fisher's exact test are listed in the fourth and fifth columns, respectively. The data resource or reference is listed in the sixth column. GO and <2.96E-016 represent gene ontology defined by the consortium and values below 2.96E-016, respectively.
Since the ectopic expression of NST1 or NST2 induced tracheary element-like structures in epidermal cells, we compared the profiles of expression of genes related to the differentiation of tracheary elements in two microarray experiments, namely, 35S:NST1 versus the wild type in rosette leaves (this study) and xylem versus nonvascular tissues in root hypocotyls (Zhao et al., 2005). Table 2 shows that, among the genes related to the differentiation of tracheary elements, those involved in secondary wall thickening, such as genes for cellulose synthase, glycosyl transferase, and laccase, were significantly upregulated in both experiments. By contrast, genes involved in programmed cell death, such as genes for endopeptidases and nucleases, and those for early markers of differentiation of tracheary elements were significantly upregulated in xylem, while not one of them was upregulated in 35S:NST1 plants.
Table 2.
List of the Extent of Change in the Level of Expression of Genes That Might Be Involved in the Differentiation of Tracheary Elements in 35S:NST1 Plants and in the Xylem of Root Hypocotyl
| Locus | Common Name | Description | Fold Change in 35S:NST1 | Fold Change in Xylem | Reference |
|---|---|---|---|---|---|
| At4g18780 | IRX1 | Cellulose synthase, catalytic subunit (IRX1) | 8.31* | 13.83* | Taylor et al. (2000) |
| At5g17420 | IRX3 | Cellulose synthase, catalytic subunit (IRX3) | 6.21* | 16.98* | Taylor et al. (1999) |
| At5g44030 | IRX5 | Cellulose synthase, catalytic subunit (IRX5) | 3.12* | 17.24* | Taylor et al. (2003) |
| At5g54690 | IRX8 | Glycosyl transferase family 8 protein | 5.80* | 19.28* | Persson et al. (2005) |
| At5g03170 | IRX13 | Fasciclin-like arabinogalactan-protein (FLA11) | 2.05* | 21.14* | Persson et al. (2005) |
| At1g15950 | IRX4 | Cinnamoyl-CoA reductase, putative | 1.65* | 0.92 | Jones et al. (2001) |
| At2g38080 | IRX12 | Laccase, putative/diphenol oxidase, putative | 11.48* | 35.97* | Brown et al. (2005); Sawa et al. (2005) |
| At4g35350 | XCP1 | Cys endopeptidase, papain-type (XCP1) | 0.44* | 75.16* | Funk et al. (2002) |
| At1g20850 | XCP2 | Cys endopeptidase, papain-type (XCP2) | 0.42* | 92.12* | Funk et al. (2002) |
| At4g00230 | XSP1 | Subtilisin-like Ser endopeptidase (XSP1) | 0.82 | 3.28* | Zhao et al. (2000) |
| At1g11190 | BFN1 | Bifunctional nuclease (BFN1), putative ZEN1 ortholog | 1.06 | 3.65 | Perez-Amador et al. (2000); Ito and Fukuda (2002) |
| At4g08160 | AtXyn3 | Glycosyl hydrolase family 10 protein | 0.66 | 18.69* | Sawa et al. (2005) |
| At3g62160 | Transferase family protein | 0.62 | 2.11 | Sawa et al. (2005) | |
| At4g32880 | AtHB-8 | Homeobox-Leu zipper transcription factor (HB-8) | 0.83 | 21.66* | Baima et al. (2001) |
| At1g19850 | MP | Transcription factor MONOPTEROS (MP) | 0.73 | 3.24 | Hardtke and Berleth (1998) |
Changes in 35S:NST1 plants relative to the level in wild-type plants and in xylem relative to the nonvascular tissue of the root hypocotyl (Zhao et al., 2005) are listed. These values in 35S:NST1 plant are ratios of means. The description of each gene is taken, basically, from The Arabidopsis Information Resource (TAIR) website (http://www.arabidopsis.org/) with slight modification. A reference for the evidence that the listed gene might be involved in the differentiation of tracheary elements is given in the sixth column. The asterisks indicate statistically significantly changed (Q-value < 0.01).
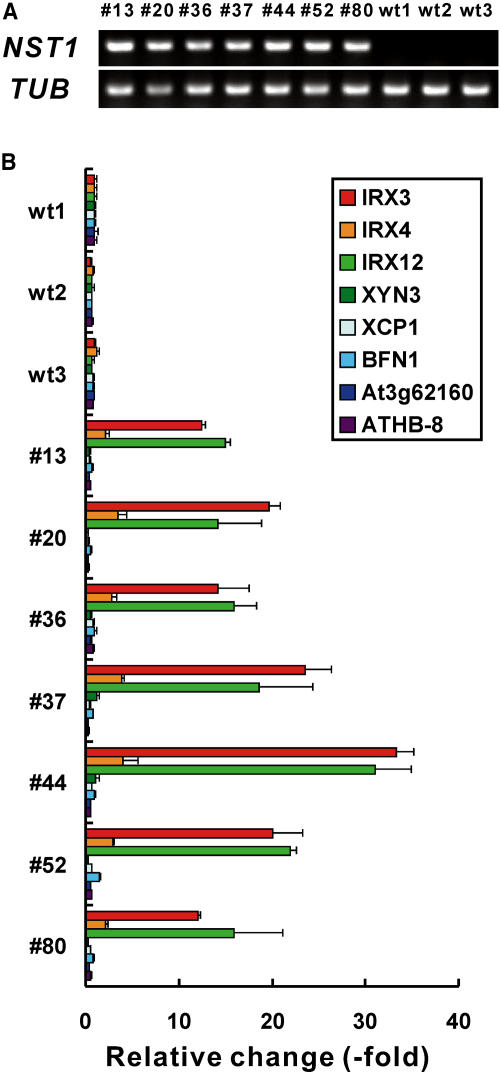
We next examined the expression of IRREGULAR XYLEM3 (IRX3), IRX4, IRX12, XYLANASE3 (XYL3), XYLEM-SPECIFIC PAPAIN-LIKE CYSTEIN PEPTIDASE1 (XCP1), BIFUNCTIONAL NUCLEASE1 (BFN1), At3g62160, and HOMEOBOX GENE8 (ATHB-8), which are involved in the differentiation of tracheary elements, by quantitative RT-PCR analysis. The IRX3, IRX4, and IRX12 genes encode a cellulose synthase that is specific for secondary walls (Taylor et al., 1999), cinnamoyl-CoA reductase, which is involved in lignin biosynthesis (Jones et al., 2001), and a putative laccase that is involved in lignin biosynthesis (Brown et al., 2005), respectively. A mutation in IRX3, IRX4, or IRX12 was shown to affect the morphology of tracheary elements and resulted in collapsed xylem (Taylor et al., 1999; Jones et al., 2001; Brown et al., 2005). XYN3 encodes a xylanase and is expressed specifically during the programmed cell death that is associated with the differentiation of tracheary elements (Sawa et al., 2005). XCP1 encodes a peptidase specific for tracheary elements (Funk et al., 2002), and BFN1 encodes a homolog of ZINNIA ENDONUCLEASE1 (ZEN1), a nuclease in Zinnia elegans (Perez-Amador et al., 2000; Ito and Fukuda, 2002). Both these enzymes are involved in the programmed cell death of tracheary elements. At3g62160 encodes a putative acetyltransferase and is upregulated during the transdifferentiation of tracheary elements (Sawa et al., 2005), and ATHB-8 is a marker of procambial cells for cambium during vascular regeneration (Baima et al., 1995, 2001).
As shown in Figure 8 and consistent with the results of the microarray analysis, the expression of the IRX3, IRX4, and IRX12 genes, all of which are involved in secondary wall thickening, was enhanced 2- to 30-fold in all seven of the independent 35S:NST1 transgenic lines examined, as compared with the wild type. By contrast, neither the genes involved in programmed cell death nor genes for early markers of differentiation of tracheary elements, namely, XYN3, XCP1, BFN1, At3g62160, and ATHB-8, were upregulated in 35S:NST1 plants. Our results indicate that NST1 and NST2 can be positive regulators of secondary wall thickening in tracheary elements but that the entire process of differentiation of tracheary elements is unlikely to be regulated by NST1 and NST2.
Figure 8.
Expression of Genes Related to the Differentiation of Tracheary Elements.
(A) Expression of NST1 in rosette leaves of 35S:NST1 transgenic and wild-type plants, as revealed by RT-PCR. Overexpression of NST1 is evident in all the 35S:NST1 transgenic lines. Numbers above the lanes indicate individual 35S:NST1 transgenic lines. wt1, wt2, and wt3 indicate three independent wild-type plants. TUB indicates the gene for β-tubulin, which was used as an internal control.
(B) Expression of genes related to the differentiation of tracheary elements, as revealed by quantitative RT-PCR. Each bar represents the amount of the transcript of a gene relative to that of the internal control. The relative level of expression of each gene in wt1 was set at 1. Numbers below the vertical axis correspond to the numbers shown in (A). Error bars represent ±sd (n = 3).
DISCUSSION
NST1 and NST2 Act Redundantly to Regulate Secondary Wall Thickening in the Anther Endothecium
We showed here that the expression of chimeric NST1 and NST2 repressors suppressed secondary wall thickening in anther walls and resulted in indehiscent anthers, while ectopic expression of NST1 or NST2 induced ectopic secondary wall thickening in various tissues via upregulation of genes related to secondary wall synthesis. The indehiscent anther phenotype was observed only in the NST1 and NST2 double T-DNA-tagged lines but not in the single T-DNA–tagged lines. These results demonstrate that NST1 and NST2 act redundantly to regulate anther dehiscence by promoting secondary wall thickening in the anther wall and, moreover, that the chimeric NST1 and NST2 repressors dominantly suppressed the expression of genes related to secondary wall thickening in the presence of endogenous and redundant factors, as demonstrated previously (Hiratsu et al., 2003; Matsui et al., 2004, 2005). NAC transcription factors have been shown to be involved in various aspects of plant growth and development and in stress responses (Olsen et al., 2005), but, to our knowledge, a role for NAC transcription factors in the regulation of secondary wall thickening has not been reported previously. To date, Arabidopsis MYB26 has been shown to be necessary for secondary wall thickening in the anther wall, but it has not been determined whether it can induce ectopic secondary wall thickening, like NST1, when overexpressed (Steiner-Lange et al., 2003). Because the expression of MYB26 is enhanced in 35S:NST1 plants (data not shown), it is possible that NST1 might regulate the expression of MYB26.
Although NST1 and NST2 have similar amino acid sequences and both regulate secondary wall thickening, the two genes are transcribed differently and the transcription factors encoded by them seem to have different activities. Our promoter-reporter experiment has suggested that the promoter activity of NST2 is somewhat anther specific and is more prominent than that of the NST1 gene in anther walls. By contrast, because indehiscent anthers were induced in 35S:NST1SRDX plants and not in 35S:NST2SRDX plants, and ectopic secondary wall thickening was much more frequent in 35S:NST1 plants than in 35S:NST2 plants, NST2 may be a much less potent transcription factor than NST1 in the regulation of genes involved in secondary wall thickening, possibly due to lower DNA binding activity and/or protein stability. Thus, 35S:NST2SRDX may have failed to suppress expression of the target genes and to dominate the activation activity of endogenous NST1 and NST2 probably due to the intrinsically lower potency of NST2SRDX. The activity of the NST2 promoter in anther walls appears to be significantly stronger than that of the CaMV 35S promoter because indehiscent anthers were indeed induced in ProNST2:NST2SRDX plants.
Thus, anther dehiscence in Arabidopsis is probably regulated by two closely related but complicatedly redundant NAC transcription factors, NST1, which is expressed only modestly in anther walls, and NST2, which is strongly expressed in anthers but has limited activity.
NSTs Induce Secondary Wall Thickening but Not the Entire Process of Differentiation of Tracheary Elements
We demonstrated that NST1 and NST2 are positive regulators of secondary wall thickening. In addition, they induce striated tracheary element-like structures in epidermal cells when expressed ectopically. The epidermal cells with ectopic striated structures closely resembled tracheary elements induced from suspension cultured cells by hormonal treatment (Oda et al., 2005). This observation indicates that similar regulatory mechanisms might control secondary wall thickening in anther endothecium and in tracheary elements. This notion is supported by reports of morphological observations in the literature that demonstrate that anther endothecium develops striated secondary wall thickening, with the patterns depending on the genus or species and resembling those of tracheary elements (Manning, 1996). Furthermore, profiles of the transcriptome of 35S:NST1 plants revealed that a large number of the genes that were upregulated in 35S:NST1 plants were also upregulated in wild-type xylem. It is likely that cells that develop striated secondary wall thickening, such as cells in anther walls and trarcheary elements, share similar molecular events in the process of secondary wall formation.
However, there was a striking difference in terms of the expression of genes specific for the differentiation of tracheary elements between 35S:NST1 plants and wild-type xylem. Genes involved in secondary wall synthesis, namely, genes for the synthesis of cellulose and lignin, were clearly upregulated in 35S:NST1 plants and in wild-type xylem. By contrast, genes related to programmed cell death, such as genes for xylanase, peptidases and nucleases, and genes for early markers of differentiation of tracheary elements, were clearly upregulated in wild-type xylem but none of them were upregulated in 35S:NST1 plants. This pattern of expression is remarkably different from that in the ate mutant, in which ectopic tracheary elements differentiated in the epidermis of cotyledons and hypocotyls (Sawa et al., 2005). By contrast with those in 35S:NST1 plants, genes related to secondary wall synthesis and to programmed cell death in the differentiation of tracheary elements were upregulated in the ate mutant (Sawa et al., 2005).
Our observations indicate that the tracheary element-like epidermal cells in 35S:NST1 plants are not likely to be true tracheary elements because they do not undergo programmed cell death, the maturation step in the differentiation of tracheary elements, although they have a very similar appearance to that of tracheary elements. NST1 and NST2 appear to function in the formation of tracheary elements by promoting secondary wall thickening but are unlikely to have the ability to regulate the entire process of differentiation of tracheary elements. This notion is consistent with the fact that the promoter activities of NST1 and NST2 are evident in the tissues that do not undergo programmed cell death immediately, such as interfascicular fibers. Secondary wall thickening and programmed cell death during differentiation of tracheary elements might operate independently or might involve separable mechanisms as suggested previously (Turner and Hall, 2000).
Do NST and/or Related Molecules Play Pivotal Roles in Secondary Wall Formation in Tissues Other Than Anthers?
In this article, we showed that NST1 is a possible regulator of secondary wall thickening in xylem, including tracheary elements, because analysis of transcriptomes revealed that similar sets of genes related to secondary wall thickening were upregulated both in wild-type xylem and in 35S:NST1 plants and, moreover, that strong activity of the NST1 promoter was also associated with xylem, in particular, with interfascicular fibers and differentiating tracheary elements. In fact, when we carefully examined transverse sections of inflorescence stems, we occasionally observed the abnormal development of secondary walls in the inflorescence stems in ProNST1:NST1SRDX plants, but no such abnormal phenotype was evident in 35S:NST1SRDX plants, in ProNST2:NST2SRDX plants, or in the NST1 and NST2 double T-DNA–tagged lines (data not shown). Since expression of NST1SRDX only partially suppressed secondary wall thickening in inflorescence stems and since double T-DNA–tagged plants had no abnormalities in their stems, it is possible that another NST factor might exist that plays a major role in the secondary wall thickening in inflorescence stems.
According to the available results of expression profiling with microarrays (Schmid et al., 2005), expression of one NAC transcription factor in subgroup IIb, which we refer to as NST3, is strongly enhanced in stems but not in stamens. This observation suggests that NST3 might be involved in secondary wall thickening in stem organs. In addition, among the members of subgroup IIb, the profiling data indicate that only NST1 and NST2 are strongly upregulated in stamens at stage 12, as would be expected from the results obtained in this study.
Plants have several types of secondary walls. Tracheary elements have striated secondary walls and their differentiation is associated with immediate programmed cell death. Like tracheary elements, anther endothecia also have a striated pattern in their secondary walls. On the other hand, interfascicular fibers neither have a striated pattern in their secondary walls nor are they associated with immediate programmed cell death (Turner and Hall, 2000). Because ectopic expression of NST1 induces ectopic secondary walls in various tissues but does not always induce the striation of secondary walls or programmed cell death, it is reasonable to consider that NSTs can regulate secondary wall thickening in every tissue. NAC transcription factors that belong to the same group as NST1 might be master regulators of secondary wall thickening in plants.
METHODS
Computer Analysis
Arabidopsis thaliana genes that encode NAC transcription factors were collected from the whole genome set of amino acids sequences retrieved from TAIR ftp site (ATH1_pep_cm_20040228, available at ftp://ftp.arabidopsis.org/home/tair/sequences/blast_datasets/) using the NAC domain as the query by FASTA search (detection E-value set to 0.001; Pearson and Lipman, 1988). The conserved domains of each NAC domain transcription factor were extracted, and a phylogenetic tree of NAC transcription factors was constructed with the ClustalW program. Information regarding gene duplication within the Arabidopsis genome was obtained from The Institute for Genomic Research ftp site (segmentally_duplicated_genes.Arab_v5.txt; available at ftp://ftp.tigr.org/pub/data/a_thaliana/ath1/DATA_RELEASE_SUPPLEMENT).
Plasmids
The protein coding regions of NST1 and NST2 were amplified from a flower cDNA library using appropriate primers. Each amplified fragment was cloned into the SmaI site of the p35SSRDXG vector to produce p35S:NST1SRDX and p35S:NST2SRDX or into the p35SSG vector to produce p35S:NST1 and p35S:NST2, and the region corresponding to each transgene was transferred into the pBCKH plant expression vector using the Gateway system (Invitrogen). To drive each transgene by its own promoter, we amplified the 5′ upstream region of 2837 bp from the site of translational initiation of the NST1 gene and of 2710 bp of the NST2 gene and replaced the CaMV 35S promoter of p35S:NST1SRDX and p35S:NST2SRDX, respectively. These promoter regions were also fused, separately, to reporter genes for GUS and GFP, respectively.
For transient effector-reporter analysis, we inserted the coding sequences of NST1, NST2, NST1SRDX, and NST2SRDX, separately, into the SmaI and SalI sites of the p35S-GAL4DB plasmid (Ohta et al., 2000) to generate fusion proteins with GAL4DB. The reporter gene constructs GAL4:TATA:LUC and 35S:GAL4:TATA:LUC were described previously (Ohta et al., 2000; Hiratsu et al., 2002). All the synthetic primers used in this study are listed in Supplemental Table 3 online.
Plant Growth Conditions and Transformation
Arabidopsis plants were grown in soil at 22°C with 16 h of light daily. For plant transformation, a T-DNA vector carrying the appropriate construct was introduced into Agrobacterium tumefaciens strain GV3101 by electroporation, and the resultant Agrobacterium was infiltrated into Arabidopsis by the floral dip method (Clough and Bent, 1998).
Isolation of RNA, Microarray Experiments, and Analysis
Total RNA was isolated from rosette leaves of 2-week-old Arabidopsis plants with Trizol, as described previously (Fukuda et al., 1991). For microarray analysis, Cy5- and Cy3-labeled cDNA probes were prepared and subjected to analysis with the Agilent Arabidopsis 2 Oligo Microarray kit (Agilent Technologies). All microarray experiments and analysis of data including calculation of P-values were performed according to the supplier's manual (available at http://www.chem.agilent.com/scripts/LiteraturePDF.asp?iWHID=37629) using the feature extraction and image analysis software (version A.6.1.1; Agilent Technologies). We eliminated spot data from the total sample of 22,500 Arabidopsis genes through a flag filter provided by the feature extraction software according to the following criteria: if the spot was signal saturated, if the signal was nonhomogenously distributed, if the signal was not significantly above background, or if the background-subtracted signal was less than background signal plus 2.6-fold of the standard deviation of background signal. Data of spots retained after filtering in both duplicated experiments (20,084) were subjected to subsequent analyses. The signal intensity of each pixel in a spot, except for outliers, was measured, and the mean and standard deviation of all the pixels in the spot were calculated. Normally, one spot contains >40 pixels. The value of the background signal of each spot was subtracted from the respective mean value of the spots and then dye normalized by the Lowess method. P-values for the difference of the log of the signal value in a spot between red and green was calculated by the following equation:
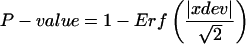 |
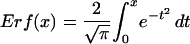 |
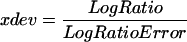 |
The feature extraction software calculated two kinds of P-values based on the Propagation of Pixel Level Error Model and the Universal Error Model (version A.6.1.1; Agilent Technologies), and the higher value was selected as the P-value. The former model calculates the propagation error of the standard deviation of all pixels in a spot. The latter model estimates the expected noise value of each spot using the actual noise values of microarrays that had been experimentally examined by Agilent. If the former model is used, then LogRatioError is calculated from the standard deviation of all the pixels in each spot for red and green channels and the value of background noise, and xdev is then calculated from the LogRatioError. When the Universal Error Model is used, xdev is calculated from the signal values of spots, multiplicative error factors, and additive error factors for red and green channels. LogRatioError is then calculated from the value of xdev.
To minimize a type-I family-wise error in multiple and simultaneous statistical tests, we adopted a strategy to suppress the number of false positives. For this, the Q-value to estimate false discovery rate was calculated from the P-value described above by QVALUE software (Storey and Tibshirani, 2003), with the default setting, and used as the criterion to assess significance. The description Q-value < 0.01 in this study means that Q-values of both duplicated experiments are under 0.01.
Comprehensive gene group analysis by Fisher's exact test was performed with the R program package (http://www.r-project.org/). The value cited for the extent of each difference in the comparison of 35S:NST1 with wild-type plants is the mean value from biological duplicate experiments. Those values for xylem versus nonvascular tissue were calculated from the data of Supplemental Table 1 in Zhao et al. (2005).
For analysis by RT-PCR, 5 μg of total RNA were treated with DNase I and subjected to first-strand cDNA synthesis. RT-PCR was performed with gene-specific primers (see Supplemental Table 2 online) for 25 to 30 cycles. Quantitative RT-PCR was performed by the SYBR green method using the ABI 7300 real-time PCR system (Applied Biosystems). Relative amounts of transcripts were calculated by an absolute quantification method, using the UBQ1 gene as an internal control. More than three replicates were included in each experiment.
Observations by Scanning Electron Microscopy
Flowers of 3- to 4-week-old plants were fixed in FAA solution (45% ethanol, 2.5% acetic acid, and 2.5% formalin) and dehydrated in ethanol. Fixed samples were subjected to critical point drying (WCP-2 Critical Point Dryer; Hitachi), coated with platinum palladium with an ion sputterer (Hitachi), and observed with a scanning electron microscope (JSM-6330F field emission scanning electron microscope; JEOL) at an accelerating voltage of 5 kV.
Light and Fluorescence Microscopy
For observations of lignin autofluorescence, we used a filter with the following specifications: glass, 365; dichroic mirror, 395; and long-pass, 400. To examine lignin deposition, we embedded inflorescence in Paraplast Plus (Sherwood) for preparation of 8-μm cross sections. After removal of paraffin, samples were stained with 2% (w/v) phloroglucinol in 95% ethanol for 2 min then washed in 10 N HCl for 1 min and mounted in 5 N HCl. To prepare 70- to 150-μm sections of inflorescence stems, the tissue was embedded in 3% agar and sectioned on a vibrating microtome (HM-650V; Microm).
Assays of GUS activity were performed with the T1 or T2 transgenic plants. Plant tissues were incubated in 100 mM sodium phosphate buffer, pH 7.0, that contained 0.1% Triton X-100, 1 mM 5-bromo-4-chloro-3-indolyl-β-d-glucuronide, and 0.5 mM potassium ferricyanide at 37°C for up to 12 h. Stained inflorescence stems were embedded in 3% agar and sectioned. For microscopy, stained tissues were bleached in several changes of 70% ethanol. All light and fluorescence microscopic observations except those of GFP fluorescence were performed with the Axioskop2 plus system (Carl Zeiss). The fluorescence from GFP was monitored with a confocal laser scanning microscope (Radiance2000; Bio-Rad).
Transient Effector-Reporter Analysis
Effector, reporter, and reference (Renilla LUC gene) plasmids were transiently introduced into rosette leaves of 2- to 4-week-old plants by particle bombardment, and relative luciferase activity was quantified and normalized as described previously (Fujimoto et al., 2000).
Accession Numbers and Data Deposition
NST1 and NST2 reported in this study correspond to the Arabidopsis Genome Initiative locus identifiers At2g46770 and At3g61910, respectively. Microarray data performed in this study can be found in the NCBI GEO data library under accession number GSE3363.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table 1. Data of Microarray Experiments.
Supplemental Table 2. Upregulated Genes Overlapping with Gene Groups Listed in Table 1.
Supplemental Table 3. Oligonucleotides Used in This Study.
Supplementary Material
Acknowledgments
We thank Sumie Ishiguro (Nagoya University, Japan) for providing seeds of the dad1 mutant, the ABRC for seeds of the NST1 and NST2 T-DNA–tagged plants, and Nobuko Kawanami and Yuko Sato (National Institute of Advanced Industrial Science and Technology) for cultivation of plants and skilled technical assistance.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Masaru Ohme-Takagi (m-takagi@aist.go.jp).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.036004.
References
- Aida, M., Ishida, T., Fukaki, H., Fujisawa, H., and Tasaka, M. (1997). Genes involved in organ separation in Arabidopsis: An analysis of the cup-shaped cotyledon mutant. Plant Cell 9, 841–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baima, S., Nobili, F., Sessa, G., Lucchetti, S., Ruberti, I., and Morelli, G. (1995). The expression of the athb-8 homeobox gene is restricted to provascular cells in Arabidopsis thaliana. Development 121, 4171–4182. [DOI] [PubMed] [Google Scholar]
- Baima, S., Possenti, M., Matteucci, A., Wisman, E., Altamura, M.M., Ruberti, I., and Morelli, G. (2001). The Arabidopsis ATHB-8 HD-zip protein acts as a differentiation-promoting transcription factor of the vascular meristems. Plant Physiol. 126, 643–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, D.M., Zeef, L.A.H., Ellis, J., Goodacre, R., and Turner, S.R. (2005). Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell 17, 2281–2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano-Delgado, A.I., Metzlaff, K., and Bevan, M.W. (2000). The eli1 mutation reveals a link between cell expansion and secondary cell wall formation in Arabidopsis thaliana. Development 127, 3395–3405. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Fujimoto, S.Y., Ohta, M., Usui, A., Shinshi, H., and Ohme-Takagi, M. (2000). Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell 12, 393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita, M., Fujita, Y., Maruyama, K., Seki, M., Hiratsu, K., Ohme-Takagi, M., Tran, L.S., Yamaguchi-Shinozaki, K., and Shinozaki, K. (2004). A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J. 39, 863–876. [DOI] [PubMed] [Google Scholar]
- Fukuda, H. (1997). Tracheary element differentiation. Plant Cell 9, 1147–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda, Y., Ohme, M., and Shinshi, H. (1991). Gene structure and expression of a tobacco endochitinase gene in suspension-cultured cells. Plant Mol. Biol. 16, 1–10. [DOI] [PubMed] [Google Scholar]
- Funk, V., Kositsup, B., Zhao, C., and Beers, E.P. (2002). The Arabidopsis xylem peptidase XCP1 is a tracheary element vacuolar protein that may be a papain ortholog. Plant Physiol. 128, 84–94. [PMC free article] [PubMed] [Google Scholar]
- Hardtke, C.S., and Berleth, T. (1998). The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J. 17, 1405–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsu, K., Matsui, K., Koyama, T., and Ohme-Takagi, M. (2003). Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 34, 733–739. [DOI] [PubMed] [Google Scholar]
- Hiratsu, K., Mitsuda, N., Matsui, K., and Ohme-Takagi, M. (2004). Identification of the minimal repression domain of SUPERMAN shows that the DLELRL hexapeptide is both necessary and sufficient for repression of transcription in Arabidopsis. Biochem. Biophys. Res. Commun. 321, 172–178. [DOI] [PubMed] [Google Scholar]
- Hiratsu, K., Ohta, M., Matsui, K., and Ohme-Takagi, M. (2002). The SUPERMAN protein is an active repressor whose carboxy-terminal repression domain is required for the development of normal flowers. FEBS Lett. 514, 351–354. [DOI] [PubMed] [Google Scholar]
- Ishiguro, S., Kawai-Oda, A., Ueda, J., Nishida, I., and Okada, K. (2001). The DEFECTIVE IN ANTHER DEHISCIENCE1 gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. Plant Cell 13, 2191–2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, J., and Fukuda, H. (2002). ZEN1 is a key enzyme in the degradation of nuclear DNA during programmed cell death of tracheary elements. Plant Cell 14, 3201–3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, L., Ennos, A.R., and Turner, S.R. (2001). Cloning and characterization of irregular xylem4 (irx4): A severely lignin-deficient mutant of Arabidopsis. Plant J. 26, 205–216. [DOI] [PubMed] [Google Scholar]
- Keijzer, C.J. (1987). The processes of anther dehiscence and pollen dispersal. the opening mechanism of longitudinally dehiscing anthers. New Phytol. 105, 487–498. [DOI] [PubMed] [Google Scholar]
- Ko, J.H., Han, K.H., Park, S., and Yang, J. (2004). Plant body weight-induced secondary growth in Arabidopsis and its transcription phenotype revealed by whole-transcriptome profiling. Plant Physiol. 135, 1069–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljegren, S.J., Ditta, G.S., Eshed, Y., Savidge, B., Bowman, J.L., and Yanofsky, M.F. (2000). SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature 404, 766–770. [DOI] [PubMed] [Google Scholar]
- Liljegren, S.J., Roeder, A.H., Kempin, S.A., Gremski, K., Ostergaard, L., Guimil, S., Reyes, D.K., and Yanofsky, M.F. (2004). Control of fruit patterning in Arabidopsis by INDEHISCENT. Cell 116, 843–853. [DOI] [PubMed] [Google Scholar]
- Manning, J.C. (1996). Diversity of endothecial patterns in the angiosperms. In The Anther: Form, Function and Phylogeny, W.G. Darcy, and R.C. Keating, eds (Cambridge, UK: Cambridge University Press), pp. 136–158.
- Matsui, K., Hiratsu, K., Koyama, T., Tanaka, H., and Ohme-Takagi, M. (2005). A chimeric AtMYB23 repressor induces hairy roots, elongation of leaves and stems, and inhibition of the deposition of mucilage on seed coats in Arabidopsis. Plant Cell Physiol. 46, 147–155. [DOI] [PubMed] [Google Scholar]
- Matsui, K., Tanaka, H., and Ohme-Takagi, M. (2004). Suppression of the biosynthesis of proanthocyanidin in Arabidopsis by a chimeric PAP1 repressor. Plant Biotechnol. J. 2, 487–494. [DOI] [PubMed] [Google Scholar]
- McConn, M., and Browse, J. (1996). The critical requirement for linolenic acid is pollen development, not photosynthesis, in an Arabidopsis mutant. Plant Cell 8, 403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda, Y., Mimura, T., and Hasezawa, S. (2005). Regulation of secondary cell wall development by cortical microtubules during tracheary element differentiation in Arabidopsis cell suspensions. Plant Physiol. 137, 1027–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta, M., Ohme-Takagi, M., and Shinshi, H. (2000). Three ethylene-responsive transcription factors in tobacco with distinct transactivation functions. Plant J. 22, 29–38. [DOI] [PubMed] [Google Scholar]
- Olsen, A.N., Ernst, H.A., Leggio, L.L., and Skriver, K. (2005). NAC transcription factors: Structurally distinct, functionally diverse. Trends Plant Sci. 10, 79–87. [DOI] [PubMed] [Google Scholar]
- Pearson, W.R., and Lipman, D.J. (1988). Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 85, 2444–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Amador, M.A., Abler, M.L., De Rocher, E.J., Thompson, D.M., van Hoof, A., LeBrasseur, N.D., Lers, A., and Green, P.J. (2000). Identification of BFN1, a bifunctional nuclease induced during leaf and stem senescence in Arabidopsis. Plant Physiol. 122, 169–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson, S., Wei, H., Milne, J., Page, G.P., and Somerville, C.R. (2005). Identification of genes required for cellulose synthesis by regression analysis of public microarray data sets. Proc. Natl. Acad. Sci. USA 102, 8633–8638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sablowski, R.W., and Meyerowitz, E.M. (1998). A homolog of NO APICAL MERISTEM is an immediate target of the floral homeotic genes APETALA3/PISTILLATA. Cell 92, 93–103. [DOI] [PubMed] [Google Scholar]
- Sanders, P.M., Lee, P.Y., Biesgen, C., Boone, J.D., Beals, T.P., Weiler, E.W., and Goldberg, R.B. (2000). The Arabidopsis DELAYED DEHISCENCE1 gene encodes an enzyme in the jasmonic acid synthesis pathway. Plant Cell 12, 1041–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa, S., Demura, T., Horiguchi, G., Kubo, M., and Fukuda, H. (2005). The ATE genes are responsible for repression of transdifferentiation into xylem cells in Arabidopsis. Plant Physiol. 137, 141–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid, M., Davison, T.S., Henz, S.R., Pape, U.J., Demar, M., Vingron, M., Scholkopf, B., Weigel, D., and Lohmann, J.U. (2005). A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37, 501–506. [DOI] [PubMed] [Google Scholar]
- Spence, J., Vercher, Y., Gates, P., and Harris, N. (1996). Pod shatter in Arabidopsis thaliana, Brassica napus and B. juncea. J. Microsc. 181, 195–203. [Google Scholar]
- Steiner-Lange, S., Unte, U.S., Eckstein, L., Yang, C., Wilson, Z.A., Schmelzer, E., Dekker, K., and Saedler, H. (2003). Disruption of Arabidopsis thaliana MYB26 results in male sterility due to non-dehiscent anthers. Plant J. 34, 519–528. [DOI] [PubMed] [Google Scholar]
- Storey, J.D., and Tibshirani, R. (2003). Statistical significance for genome-wide experiments. Proc. Natl. Acad. Sci. USA 100, 9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada, S., Hibara, K., Ishida, T., and Tasaka, M. (2001). The CUP-SHAPED COTYLEDON1 gene of Arabidopsis regulates shoot apical meristem formation. Development 128, 1127–1135. [DOI] [PubMed] [Google Scholar]
- Taylor, N.G., Howells, R.M., Huttly, A.K., Vickers, K., and Turner, S.R. (2003). Interactions among three distinct CesA proteins essential for cellulose synthesis. Proc. Natl. Acad. Sci. USA 100, 1450–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, N.G., Laurie, S., and Turner, S.R. (2000). Multiple cellulose synthase catalytic subunits are required for cellulose synthesis in Arabidopsis. Plant Cell 12, 2529–2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, N.G., Scheible, W.R., Cutler, S., Somerville, C.R., and Turner, S.R. (1999). The irregular xylem3 locus of Arabidopsis encodes a cellulose synthase required for secondary cell wall synthesis. Plant Cell 11, 769–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokimatsu, T., Sakurai, N., Suzuki, H., Ohta, H., Nishitani, K., Koyama, T., Umezawa, T., Misawa, N., Saito, K., and Shibata, D. (2005). KaPPA-View. A web-based analysis tool for integration of transcript and metabolite data on plant metabolic pathway maps. Plant Physiol. 138, 1289–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, S.R., and Hall, M. (2000). The gapped xylem mutant identifies a common regulatory step in secondary cell wall deposition. Plant J. 24, 477–488. [DOI] [PubMed] [Google Scholar]
- Turner, S.R., Taylor, N., and Jones, L. (2001). Mutations of the secondary cell wall. Plant Mol. Biol. 47, 209–219. [PubMed] [Google Scholar]
- Vroemen, C.W., Mordhorst, A.P., Albrecht, C., Kwaaitaal, M.A., and de Vries, S.C. (2003). The CUP-SHAPED COTYLEDON3 gene is required for boundary and shoot meristem formation in Arabidopsis. Plant Cell 15, 1563–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, D.X., Feys, B.F., James, S., Nieto-Rostro, M., and Turner, J.G. (1998). COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280, 1091–1094. [DOI] [PubMed] [Google Scholar]
- Xie, Q., Frugis, G., Colgan, D., and Chua, N.H. (2000). Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev. 14, 3024–3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, Z.H. (2002). Vascular tissue differentiation and pattern formation in plants. Annu. Rev. Plant Biol. 53, 183–202. [DOI] [PubMed] [Google Scholar]
- Zhao, C., Craig, J.C., Petzold, H.E., Dickerman, A.W., and Beers, E.P. (2005). The xylem and phloem transcriptomes from secondary tissues of the Arabidopsis root-hypocotyl. Plant Physiol. 138, 803–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, C., Johnson, B.J., Kositsup, B., and Beers, E.P. (2000). Exploiting secondary growth in Arabidopsis. Construction of xylem and bark cDNA libraries and cloning of three xylem endopeptidases. Plant Physiol. 123, 1185–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
NOTE ADDED IN PROOF
- While this manuscript was under review, Kubo et al. (2005) described that the NAC domain transcription factors VND6 (At5g62380) and VND7 (At1g71930) induced ectopic metaxylem- and protoxylem-like vessel elements, respectively, in Arabidopsis when overexpressed.
- Kubo, M., Udagawa, M., Nishikubo, N., Horiguchi, G., Yamaguchi, M., Ito, J., Mimura, T., Fukuda, H., and Demura, T. (2005). Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 19, 1855–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.