Abstract
Cell biological, structural, and genetic approaches have demonstrated the presence of arabinogalactan proteins (AGPs) in the moss Physcomitrella patens and provided evidence for their function in cell expansion and specifically in the extension of apical tip-growing cells. Inhibitor studies indicated that apical cell expansion in P. patens is blocked by synthetic AGP binding β-glucosyl Yariv reagent (βGlcYR). The anti-(1→5)-α-l-arabinan monoclonal antibody LM6 binds to some AGPs in P. patens, to all plasma membranes, and to the cell wall surface at the most apical region of growing protonemal filaments. Moreover, LM6 labeling of cell walls at the tips of apical cells of P. patens was abolished in the presence of βGlcYR, suggesting that the localized movement of AGPs from the plasma membrane to the cell wall is a component of the mechanism of tip growth. Biochemical and bioinformatic analyses were used to identify seven P. patens ESTs encoding putative AGP core proteins from homology with Arabidopsis thaliana, Brassica napus, and Oryza sativa sequences and from peptide fragments isolated from βGlcYR-precipitated AGPs. Gene knockout by homologous recombination of one of these genes, P. patens AGP1, encoding a classical AGP core protein, resulted in reduced cell lengths in protonemal filaments, indicating a role for AGP1 in apical cell expansion in P. patens.
INTRODUCTION
The moss Physcomitrella patens is an established experimental bryophyte system for the study of genetics and development, and its study provides insight into land plant phylogeny and evolution (Rensing et al., 2002; Nishiyama et al., 2003). P. patens possesses an active somatic homologous recombination system that is highly suited to reverse genetic approaches to probing gene function (Schaefer and Zryd, 1997; Hiwatashi et al., 2001), has been the subject of significant EST generation programs (Rensing et al., 2002; Nishiyama et al., 2003), and is a highly accessible system for cell biological analysis (Reski, 1998; Schipper et al., 2002). For these reasons, P. patens is an attractive organism to use to elucidate processes fundamental to plant cell development.
Bryophyte and angiosperm evolution diverged more than 400 million years ago, but recent studies have shown that in terms of cell wall composition, considerable similarities exist (Ligrone et al., 2002; Popper and Fry, 2003; Kremer et al., 2004; Matsunaga et al., 2004). P. patens is therefore a useful species for research on the structure and functions of primary cell walls and their components. Arabinogalactan proteins (AGPs), a group of biochemically complex proteoglycans associated with plasma membranes and cell walls and belonging to a Hyp-rich glycoprotein superfamily, are defined on the basis of their core peptide sequence motifs, the presence of abundant type II arabinogalactan glycans, and their ability to specifically interact with a class of synthetic phenyl glycosides known as β-glycosyl Yariv reagents (Nothnagel, 1997; Gaspar et al., 2001; Showalter, 2001; Johnson et al., 2003a). Moreover, AGPs were the first known family of abundant plant proteins identified to have a glycosylphosphatidylinositol (GPI) anchor attachment to plasma membranes (Youl et al., 1998; Oxley and Bacic, 1999; Sherrier et al., 1999; Svetek et al., 1999). Four subclasses of GPI-anchored AGP backbones are currently recognized: classical AGPs with a characteristic Pro-rich module for the entire predicted protein between an N-terminal secretion signal sequence and a hydrophobic C terminus that is processed for GPI anchor attachment, AGPs with Lys-rich domains, arabinogalactan peptides with short protein backbones, and fasciclin-like AGPs (FLAs), a class of chimeric AGPs that contain both AGP motifs and a fasciclin domain. A further class of AGPs exists, known as nonclassical or chimeric AGPs that do not have a GPI anchor and AGP motifs that are restricted to a part of the predicted protein sequence (Schultz et al., 2000, 2002; Johnson et al., 2003a, 2003b). In addition to the complexity of protein components, most of an AGP molecule comprises complex branched glycans based on a (1→3)(1→6)-β-galactan framework decorated with a range of terminal sugars, including arabinose (Nothnagel, 1997; Gaspar et al., 2001; Tan et al., 2003, 2004). In some instances, short oligoarabinosides are also present (Goodrum et al., 2000). AGP glycan components are heterogeneous, and evidence from studies with antiglycan monoclonal antibodies indicates extensive regulation of AGP glycan structure in relation to cell development, although the functional significance of this is not yet known (Knox, 1997; Showalter, 2001).
Although AGPs are implicated in many aspects of cell development, including cell proliferation, cell expansion, organ extension, apoptosis, germination, and somatic embryogenesis, precise biochemical or cell biological functions have been difficult to elucidate (Majewski-Sawka and Nothnagel, 2000; Gaspar et al., 2001; Showalter, 2001). Approaches to dissect AGP function have included the addition of exogenous AGPs to developing systems, the use of β-glycosyl Yariv reagents (βGlcYRs) or anti-AGP antibodies as in vivo inhibitors of AGP action, and, recently, the genetic manipulation of AGP core proteins. Several lines of evidence implicate AGPs in cell expansion and organ elongation. The application of βGlcYR blocks the elongation of suspension-cultured carrot (Daucus carota) and tobacco (Nicotiana tabacum) cells (Willats and Knox, 1996; Vissenberg et al., 2001), indole-3-acetic acid–induced extension of cucumber (Cucumis sativus) hypocotyls (Darley et al., 2001), and cell elongation in Arabidopsis thaliana seedling roots (Willats and Knox, 1996; Ding and Zhu, 1997; McCartney et al., 2003). Treatment of lily (Lilium longiflorum) pollen tubes with βGlcYR uncoupled exocytosis of cell wall polymers from extension at the pollen tube apex, indicating a role for these proteoglycans in tip growth of pollen tubes (Roy et al., 1999; Mollet et al., 2002). Exogenous AGPs have been reported to promote development of somatic embryos (Showalter, 2001) and have been proposed to both promote and guide the tip growth of pollen tubes (Cheung et al., 1995; Wu et al., 2000). Arabidopsis mutants with reduced hypocotyl extension (Takahashi et al., 1995) and root growth (Ding and Zhu, 1997) have reduced levels or altered AGPs. The mur1 mutant has nonfucosylated AGPs, and this also results in reduced root growth (van Hengel and Roberts, 2002). A gibberellin-responsive gene expressed in cucumber hypocotyls encodes an AGP core protein that is involved in stem elongation, its overexpression promoting growth (Park et al., 2003). Overexpression of the gene encoding tomato (Lycopersicon esculentum) AGP1 resulted in changes to both growth and development (Sun et al., 2004). To date, mutations in genes encoding Arabidopsis AGP core proteins indicate extremely subtle effects on plant growth. Mutations causing the loss of Arabidopsis AGP17, FLA4 (SOS5), and AGP30 resulted in no altered phenotype under standard growth conditions, and distinct phenotypes only became apparent when grown in the presence of Agrobacterium tumefaciens, high salt, or abscisic acid, respectively (Nam et al., 1999; Shi et al., 2003; van Hengel and Roberts, 2003; Gaspar et al., 2004). The polar secretion of a hybrid AGP/lipid transfer protein has been identified as a component of cell–cell interactions during vascular development in Arabidopsis (Motose et al., 2004).
In P. patens, both the germination of haploid spores and the regeneration of protoplasts result in the development of networks of branched filaments known as protonemata. A protonema initially comprises chloronemal cells with characteristic perpendicular cross walls. Subsequently, caulonemal cells (with oblique cross walls) arise from chloronemal filaments. In turn, chloronemal cells arise from the branching of caulonemal cells. Both of these cell types grow by apical extension and serial division of the apical cell (Cove et al., 1997). Subapical cells do not extend, but they can divide to form side branch initials that develop either as caulonema, secondary chloronema, or into a bud that gives rise to a leafy shoot that is the gametophore (Cove et al., 1997). P. patens protonemata are grown readily on both solid and liquid media. Using cell biological, structural, and bioinformatic approaches, we have demonstrated the presence of AGPs in P. patens. Using both inhibitor and reverse genetic tools, we demonstrate a role for AGPs, specifically that of P. patens AGP1, which encodes a putative classical AGP core protein, in protonemal apical cell extension, the major mechanism of growth in the filamentous form of this organism.
RESULTS
Growth Characteristics of P. patens and Growth Inhibition by AGP Binding βGlcYR
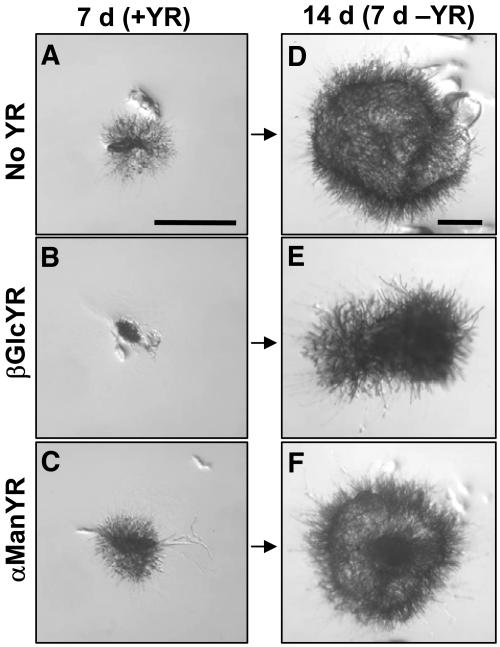
For ease of experimental manipulation, small inocula of P. patens cells can be subcultured vegetatively as an alternative to spore germination (Cove et al., 1997). This involves the removal of a cluster of ∼50 protonemal cells and their direct transfer to fresh media where they regenerate into new colonies within a few days. The resulting protonemata develop in a similar manner to those arising from spores, regardless of the initial inoculum size. Studies with AGP binding βGlcYR indicated that the growth of P. patens is sensitive to this reagent. The inclusion of 1 μM βGlcYR in solid growth medium resulted in a complete inhibition of colony growth over a 7-d period (Figure 1B). Growth was not disrupted by the presence of 1 μM α-mannosyl Yariv reagent (αManYR), a non-AGP binding analog of βGlcYR. The effect of βGlcYR was reversible as growth resumed upon transfer to fresh control media without βGlcYR for a further 7 d (Figure 1E).
Figure 1.
Reversible Inhibition of Growth of P. patens on Solid Media by βGlcYR.
P. patens cultures were inoculated on either BCD/AT medium alone (no YR control) (A) or medium with 1 μM βGlcYR (B) or 1 μM αManYR (C) and allowed to grow for 7 d. After 7 d of growth, all cultures were transferred to control media without YR for a further 7 d ([D] to [F]). Growth in the presence of the non-AGP binding YR, αManYR, was equivalent to growth on control medium. Colonies inhibited by βGlcYR resumed growth upon transfer of the culture to fresh medium (E). Bars = 1 mm.
βGlcYR Reversibly Blocks Apical Cell Extension over Short Time Periods
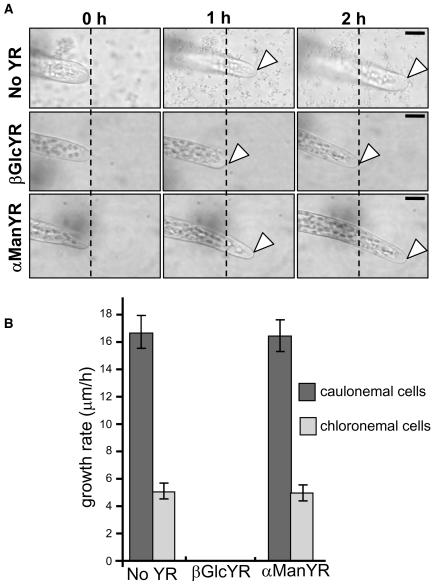
The small size of P. patens and its single-cell filamentous growth allowed observations on single extending apical cells in response to βGlcYR. Cultures, 3 d after initiation on agar, were transferred to perfusion chambers containing liquid media. Upon addition of 1 μM βGlcYR, an immediate cessation of apical cell extension (within 30 s, at the resolution of our time-lapse equipment) was observed (Figure 2A). In standard growth conditions with unilateral light of constant intensity, chloronemal cells exhibit an average extension rate of 5 μm/h, while caulonemal cells exhibit an average extension rate of ∼16 μm/h (Figure 2B). After addition of 1 μM βGlcYR, no cell extension was observed for either cell type. The equivalent application of 1 μM αManYR had no effect upon apical cell extension (Figure 2).
Figure 2.
Treatment with βGlcYR Inhibits Extension of Apical Cells of Protonemata.
(A) P. patens cultures inoculated in either liquid BCD media alone (no YR control) or with either 1 μM βGlcYR or 1 μM αManYR. Images of the apical caulonemal cell under observation were taken at 1-h intervals. The dotted black line through each micrograph indicates the most apical region of the cell at the start of observation. The arrowheads indicate the tip of apical cells after 1- and 2-h growth periods. In the presence of αManYR, growth is the same as in control media without YR. In the presence of βGlcYR, the apical cell did not extend. Bars = 10 μm.
(B) Histogram showing growth rates for caulonemal and chloronemal cells in P. patens cultures inoculated in either liquid BCD media alone (no YR control) or with 1 μM βGlcYR or 1 μM αManYR. Data shown are for six experiments per treatment, and the error bars indicate the standard error of the mean.
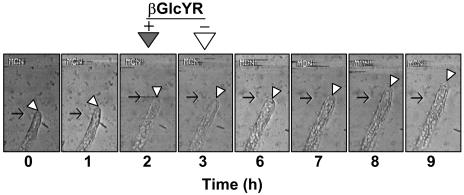
Analysis of cultures treated for 1 h with 1 μM βGlcYR, which blocked growth, followed by removal of βGlcYR by flushing the perfusion chamber with fresh control medium, indicated that a βGlcYR-inhibited apical cell was unable to extend for a period of ∼3 h after removal of the inhibitor, as shown in Figure 3. However, ∼4 h after removal of βGlcYR, the extension of the apical cell recommenced, and the apical cell reestablished a normal rate of cell extension (Figure 3).
Figure 3.
Time-Lapse Images Showing the Rapid Inhibition of Cell Extension by βGlcYR and the Recovery of Cell Extension Activity after Its Removal.
Cultures were inoculated into liquid BCD media in a perfusion chamber, and micrographs were taken of the same apical cells at 1-h intervals for up to 10 h. After 2 h, βGlcYR (1 μM) was supplied to the chamber and removed after a further 1 h (t =3). The apical cell was then observed for a further 6 h. The images shown are of a chloronemal apical cell, and the typical growth rate of such cells is 5 μm/h. Extension growth ceased immediately upon application of βGlcYR. The apical cell was unable to extend for up to 3 h following the removal of βGlcYR. After this time, growth recommenced, and a normal growth rate approaching 5 μm/h was achieved by 5 h after removal of βGlcYR. Black arrows indicate start points of the tip of the apical cell at t = 0 and also at point of application of βGlcYR (t = 2). White arrowheads indicate the position of the tip of the apical cell in each image.
Analysis of βGlcYR Binding AGPs in P. patens
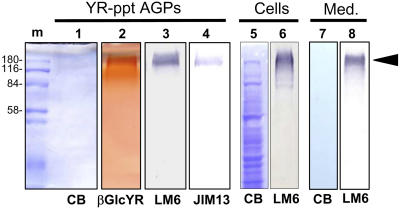
To identify the molecules that are the targets for βGlcYR action in inhibiting apical cell extension, βGlcYR was used to precipitate material from 7-d-old protonemata. Separation of the βGlcYR-precipitated material by SDS-PAGE is shown in Figure 4. No proteins were detectable by Coomassie blue staining at a gel loading of 10 μg carbohydrate. Staining of an equivalent gel directly with βGlcYR indicated material of high molecular weight in excess of 180 kD (lane 2, Figure 4). The immunoblotting of equivalent amounts of material with antiglycan monoclonal antibodies indicated that the most effective available antibody for βGlcYR-precipitated AGPs from P. patens was LM6, as shown in Figure 4 (lane 3). LM6 binds to (1→5)-α-l-arabinan (Willats et al., 1998), which in angiosperms is most commonly a structural feature of the pectic polysaccharide rhamnogalacturonan-I (Willats et al., 1998, 1999). The anti-AGP glycan monoclonal antibody JIM13 (Knox et al., 1991; Yates et al., 1996) also bound the βGlcYR-precipitated AGPs but less effectively than LM6 (lanes 3 and 4, Figure 4). LM6 immunoblots of a total extract of P. patens protonemata and of material secreted into the medium of a P. patens liquid culture indicated binding to material with the same properties as βGlcYR-precipitated AGPs (lanes 3 and 8, Figure 4).
Figure 4.
SDS-PAGE and Protein Gel Blots of βGlcYR-Precipitated Material, a Total Protein Extract of P. patens Grown on Solid Media, and Material Secreted into the Medium from P. patens in Liquid Culture.
βGlcYR-precipitated material, a total extract of protonemata (Cells) and a preparation of culture medium from a 14 d liquid culture of P. patens (Med.) were subjected to SDS-PAGE and protein gel blotting. βGlcYR-reactive material appeared as a smear on a denaturing gel with an apparent molecular mass in the region of 180 kD (lane 2), characteristic of AGPs. Coomassie blue (CB) staining of an equivalent gel did not detect any protein bands in YR-precipitated AGPs (lane 1) or culture medium (lane 7). Immunoblotting indicated that βGlcYR-precipitated AGPs were bound by anti-(1→5)-α-l-arabinan LM6 (lane 3) and anti-AGP-glycan JIM13 (lane 4). Loading per lane was 10 μg carbohydrate for YR-precipitated AGPs and medium preparation (Med.; lanes 7 and 8) and 10 μg protein for the total extract of P. patens cells (Cells; lanes 5 and 6). m, markers; YR-ppt, YR precipitate.
βGlcYR-precipitated AGPs were subjected to glycan monosaccharide linkage analysis. Results (Table 1) indicated that 1,5-linked Araf residues were a major feature of the βGlcYR-precipitated AGP glycans, confirming the observations made with the monoclonal antibody LM6. Other than the 1,5-linked Araf residues, the linkage composition, such as the presence of 3,6-linked-Galp residues, terminal Rhap and GlcAp residues, is characteristic of angiosperm type II AGP glycans (Table 1). Furthermore, the lack of 1,2-linked Rhap and 1,4-linked GalAp is further evidence that the βGlcYR-precipitated AGP fraction is essentially free of pectic polysaccharides.
Table 1.
Monosaccharide-Linkage Composition of βGlcYR-Precipitated AGPs from P. patens
| Monosaccharide | Deduced Linkage | Mol %a |
|---|---|---|
| Rhapb | Terminal | 18 |
| Araf | Terminal | 14 |
| 2- | 1 | |
| 3- | 1 | |
| 5- | 21 | |
| Manp | Terminal | tr |
| 4- | tr | |
| Glcp | Terminal | tr |
| 4- | 1 | |
| GlcApc | 4- | 11 |
| Galp | Terminal | 1 |
| 3- | 7 | |
| 6- | 2 | |
| 3,6- | 23 |
Note: low levels (<0.3 mol %) of Fucp (terminal), Xyl (terminal, 4-linked), Rha (2-, 4-, 2,4-linked), Glc (3-, 4,6-linked), Man (2,4-, 4,6-linked), and Gal (2,6-, 2,3-, 3,4-, 4,6-, 2,3,6-, 4,3,6-linked) were detected but not included in the table. tr, trace (<0.5 mol %).
Average of duplicate determinations.
Terminal Rha is deduced from 1,5-di-O-acetyl-6-deoxy-2,3,4-O-methyl hexitol, etc.
Deduced from the neutral glucosyl derivative derived from carboxyl reduction of the glucuronosyl reside.
Immunolocalization of P. patens AGPs in Planta
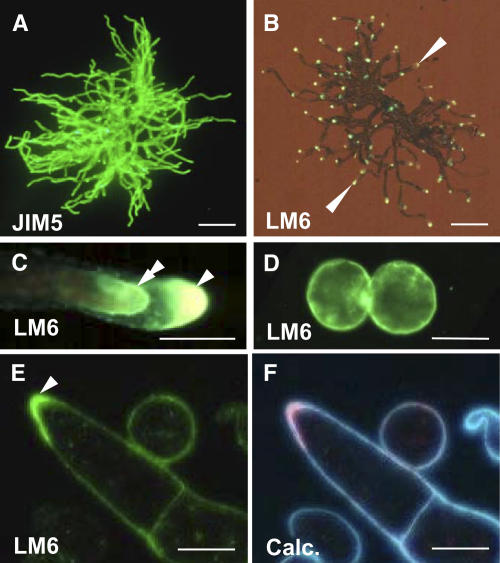
Immunolabeling of either whole mount or intact cell preparations is a useful technique to study the occurrence and distribution of cell wall polymers at the surface of organs or cells (Willats et al., 2001). The application of this technique to P. patens protonemata using an antihomogalacturonan monoclonal antibody JIM5 (Clausen et al., 2003) demonstrated that this cell wall epitope could be detected over the entire surface of a protonema (Figure 5A). In contrast with JIM5, immunolabeling of an intact protonema with anti-(1-5)-α-l-arabinan LM6, indicative of P. patens AGPs, shows that this epitope was restricted to the tip regions of apical cells where cell extension is taking place (Figure 5B). Incubation conditions that favor antibody binding (i.e., the presence of buffered saline) result in the partial plasmolysis of moss cells, and examination of the tips of apical cells revealed a dual location of the LM6 epitope at both the cell wall and on the plasma membrane (Figure 5C). The presence of the LM6 epitope on all P. patens plasma membranes was confirmed by the immunolabeling of protoplasts prepared from protonemata (Figure 5D). The LM6 epitope was detectable at the plasma membrane surface of all protoplasts immediately after protoplast isolation. As cell walls regenerated and covered the plasma membrane, the ability of LM6 to detect its epitope was generally lost until a growing tip formed (data not shown). We propose that it is only at the tip of growing apical cells that the cell wall is sufficiently permeable to allow the passage of the primary and secondary antibody molecules to access epitopes at the plasma membrane. Therefore, for most cells, the LM6 epitope cannot be detected at the plasma membrane using the whole mount procedure. The presence of the LM6 epitope at all plasma membranes was confirmed by the LM6 labeling of sections of resin-embedded material (Figure 5E). In some cases, the LM6 epitope was seen to be particularly abundant in the cell wall region at the tip of an apical cell (see Figure 5E), in addition to its presence at the plasma membrane.
Figure 5.
The Distribution of Pectic Polysaccharides and AGPs in P. patens.
(A) Indirect immunofluorescence labeling of an intact germinated P. patens spore (12 d) with antihomogalacturonan JIM5. The JIM5 epitope is abundant at the surface of all cell walls. Bar = 1 mm.
(B) Indirect immunofluorescence labeling of an intact germinated P. patens spore, equivalent to that shown in (A) with anti-(1→5)-α-arabinan LM6. In contrast with the occurrence of the JIM5 epitope, the LM6 epitope is most abundant at filament apices (arrowheads). Bar = 1 mm.
(C) Higher magnification of an apical cell from (B) displaying mild plasmolysis and showing a dual location of the LM6 epitope at the most apical region of both the plasma membrane (double arrowhead) and the cell wall (arrowhead). Bar = 20 μm.
(D) Indirect immunofluorescence labeling of P. patens protoplasts with LM6. LM6 bound to the plasma membrane surface of all protoplasts prepared from protonemata. Bar = 20 μm.
(E) Indirect immunofluorescence labeling of a section of resin-embedded protonemal cells with LM6. The LM6 epitope is present at all cell surfaces and is particularly abundant in the cell wall at the tip of an apical cell (arrowhead). Bar = 20 μm.
(F) The same section as shown in (E) stained with Calcofluor White (Calc.), which stains cellulose.
Application of βGlcYR Results in the Loss of LM6 Binding at the Surface of Apical Cells
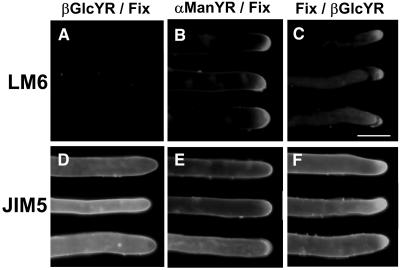
To investigate the events relating to the cessation of growth by βGlcYR, a series of experiments were performed in which cultures were incubated in the presence of 1 μM βGlcYR or 1 μM αManYR for 3 h. In one set of experiments, protonemata were fixed with 4% formaldehyde after treatment with Yariv reagent (YR). In a further set of experiments, protonemata were fixed with formaldehyde before treatment with YR for 3 h. Cells were then immunolabeled with the monoclonal antibodies JIM5 and LM6 using the whole mount immunolabeling protocol, and representative results are shown in Figure 6. The application of βGlcYR resulted in either the loss of the LM6 epitope or its accessibility at the surface of apical cells (Figure 6A), whereas the application of αManYR did not (Figure 6B). However, βGlcYR only resulted in the loss of the LM6 binding when applied to living cells. When protonemata were formaldehyde fixed prior to application of βGlcYR, there was no effect on LM6 binding (Figure 6C). The binding of JIM5 was unaffected by any of the treatments (Figures 6D to 6F). We conclude that the application of βGlcYR to living cells results in a disruption of the normal appearance of the LM6 binding AGPs in the cell wall at the tip.
Figure 6.
Immunofluorescence Analysis of the Effect of the Application of βGlcYR on the Occurrence of LM6 and JIM5 Epitopes in P. patens Protonemata.
P. patens cultures (14 d) were transferred to liquid media with either 1 μM βGlcYR ([A], [C], [D], and [F]) or 1 μM αManYR ([B] and [E]) for 3 h. One set of protonemata were fixed with 4% formaldehyde after transfer and incubation in presence of YR ([A], [B], [D], and [E]), and a second set were formaldehyde fixed prior to incubation with YR ([C] and [F]). After 3 h in the presence of YR, protonemata were immunolabeled with LM6 or JIM5. Incubation of living cells in the presence of βGlcYR resulted in the loss of the LM6 epitope from apical cells (A). The incubation of fixed, dead cells with βGlcYR had no effect on LM6 epitope occurrence (C). The JIM5 epitope was unaffected by growth in the presence of βGlcYR ([D] to [F]). Three representative filaments are shown for each treatment. Bar = 20 μm.
Biochemical Analyses of P. patens AGPs and Bioinformatic Identification of ESTs Encoding Putative P. patens AGP Core Peptides
To obtain information on the protein backbones of P. patens AGPs, two parallel approaches were adopted: an experimental approach with purified AGPs and a bioinformatic database approach. For the experimental approach, a sample of the βGlcYR-precipitated AGPs was deglycosylated with anhydrous hydrogen fluoride (HF), and the resulting peptides were separated by reverse-phase HPLC and N-terminal Edman sequenced. Four consensus peptide sequences were obtained. These peptide fragments showed characteristics of angiosperm AGPs, including the presence of Hyp and Ala. Using tBLASTn to search the National Center for Biotechnology Information database with the major amino acids and inserting a Pro residue in place of blank cycles during sequencing (see Table 2), matches to Arabidopsis AGP10 (At4g09030), AGP9 (At2g14890), and AGP18 (At4g37450) were found for sequences from P. patens peptides with retention times of 7.3, 9.1, and 15 min, respectively (Table 2). These correspond to two classical AGPs (Arabidopsis AGP9 and AGP10) and a Lys-rich Arabidopsis AGP (AtAGP18). Since the P. patens AGP preparation contained multiple peptides (as expected) and not all could be sequenced, we are as yet unable to assign which peptide(s) carries the LM6 epitope on the arabinogalactan chain. Although unlikely, it is formally possible that this epitope is also present on all peptides.
Table 2.
Sequence Alignment of Peptide Fragments Obtained from P. patens Protonemata βGlcYR-Precipitated Material with Arabidopsis AGP Core Protein Sequences and a Deduced Amino Acid Sequence in the PHYSCObase Contig Database
| N-Terminal Sequences | Alignmenta | Match (Accession No.) |
|---|---|---|
| XVPPPAGSPFQPVPS | 2 VPPPAGSP 8 | |
| * ****** | ||
| 36 VSPPAGSP 73 | AtAGP10 (At4g09030) | |
| 2 VPPPAGSPFQPVPS 15 | ||
| *********** * | ||
| 415 VPPPAGSPFQPPTS 456 | PHYSCObase Contig4134 | |
| SAGPVMAVVPPVTPPVTPSV | 1 SAGPVMAVVPPVT---PPVT 16 | |
| *********** | ||
| 19 SA-P-----PPVTTSPPPVT 32 | AtAGP9 (At2g14890) | |
| SPASSAGPPMGSPPSPAPA | 2 PASSAGPPMGSPPSPAP 18 | |
| ********* | ||
| 29 PTASASSPVESPKSPAP 45 | AtAGP18 (At4g37450) | |
| SPLTKAEVFYELKDLKGY | 5 KAEVF-YEL 12 | |
| ******* | ||
| 9 KAEIFGYEL 17 | At5g39930 (expressed protein) |
AGP backbone sequences were searched using the NCBI tBLASTn program and PHYSCObase contig database (http://moss.nibb.ac.jp) with the major amino acid sequences from the deglycosylated AGP as queries. Pro was used for the X amino acid, which is deduced from incomplete deglycosylation of Hyp leading to a blank (X) cycle during sequencing.
Asterisks indicate identical amino acids.
In a complementary bioinformatic strategy, a search of the P. patens EST databases using the entire angiosperm AGP core peptide sequences as queries was also performed. Six ESTs encoding putative AGP core peptides were identified, and the predicted sequences are shown in Figures 7 and 8. A P. patens sequence predicted to encode a classical AGP core protein, designated AGP1, was identified by homology to Arabidopsis AGP1 (At5g64310) and a Brassica napus AGP (T07975) (Figure 7). P. patens AGP1 has 29% amino acid identity with Arabidopsis AGP1 and 22% identity with B. napus AGP. These protein sequences are homologous throughout, except for a region of 16 amino acids adjacent to the predicted N-terminal signal sequence. Three related short P. patens arabinogalactan peptides, AGP3, AGP4, and AGP5, were identified by homology to Arabidopsis AGP12 (At3g13520) and AGP16 (At2g46330). Two fasciclin-like P. patens AGP sequences, FLA1 and FLA2, were identified with homology to Arabidopsis FLA1 (At5g55730), FLA2 (At4g12730), and a rice (Oryza sativa) FLA sequence (BAD01728). The sequences of P. patens AGP1 and AGP3-5 (PHYSCObase accession numbers Contig11049, Contig615, Contig6896, and Contig6913, respectively) were used to design primers to amplify the full sequence of these AGP genes from a cDNA library (a generous gift from Mitsuyasu Hasebe). The 5′ sequences for P. patens FLA1 and FLA2 were available in PHYSCObase (accession numbers Contig6808 and Contig9936). No 3′ sequences for either P. patens FLA1 or FLA2 were available in the EST databases, and so these were obtained by 3′ rapid amplification of cDNA ends. The amplified fragments were sequenced, and the products matched the predicted sequences from the initial bioinformatic search.
Figure 7.
Alignment of Classical AGP P. patens AGP1 Core Protein Sequences with Those of Arabidopsis AGP1 and B. napus AGP.
The putative protein sequence of classical AGP P. patens (Pp) AGP1 aligned with Arabidopsis (At) AGP1 (Arabidopsis Genome Initiative locus At5g64310) and B. napus (Bn) AGP (accession number T07975). Amino acids common to two sequences are shown in bold, and when common to all three sequences they are shown in bold underlined. The likely N-terminal signal sequence of P. patens AGP1 is underlined. The putative recognition site of P. patens AGP1 for the addition of a GPI anchor is shown with boxed outline, and the C-terminal hydrophobic region is shaded in gray.
Figure 8.
Putative Protein Sequences of P. patens AGPs.
The putative protein sequences of a classical AGP (AGP2) and three short arabinogalactan peptides AGP3-5, FLA1, and FLA2. AGP2 was identified from a tBLASTn search with peptide fragment 1 (Table 2) with sequence relating to the isolated peptide shown in bold. Sequences of AGP3-5 were identified by homology with Arabidopsis AGP12 (At3g13520) and AGP16 (At2g46330). FLA1 and FLA2 were identified by homology with Arabidopsis FLA1 (At5g55730) and FLA2 (At4g12730) and with rice FLA (BAD01728). Fasciclin domains are shaded in gray. Predicted N-terminal signal sequences and C-terminal hydrophobic regions predicted to be cleaved for GPI anchor attachment are underlined. aa, amino acids.
Shortly before the submission of this manuscript, a P. patens deduced amino acid sequence relating to peptide fragment 1 was identified in the PHYSCObase database (Table 2, Figure 8). This is a further classical AGP, designated P. patens AGP2 (PHYSCObase accession number Contig4134) (Figure 8). This list of putative P. patens AGP core peptides is clearly not complete, as it does not contain all the sequences of all of our biochemically identified P. patens AGP peptide fragments. The available Physcomitrella EST collections currently include >15,000 transcripts, but analyses have indicated that these libraries are not saturated (Nishiyama et al., 2003).
Knockout of P. patens AGP1 Reduces the Growth of P. patens
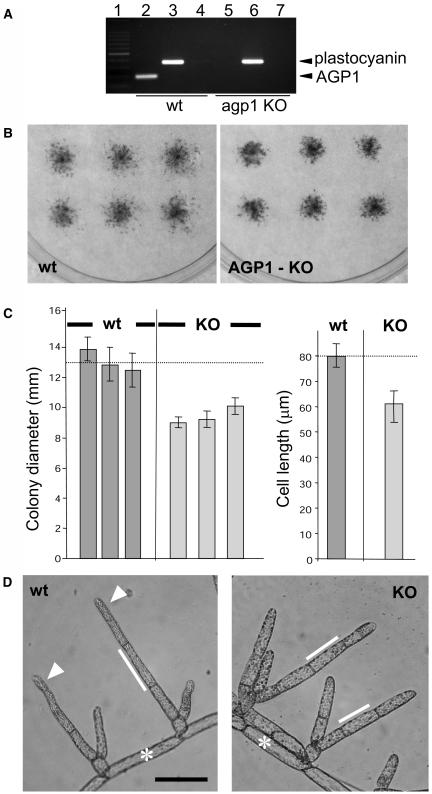
P. patens is suited to perform functional analysis by gene targeting; therefore, AGP1, the first full-length sequence we identified encoding a classical AGP backbone, was selected for manipulation. A knockout (KO) vector was designed to replace the endogenous AGP1 with a construct comprising a hygromycin phosphotransferase gene driven by the cauliflower mosaic virus 35S promoter, flanked by LoxP recombination sites (Sauer, 1998). This construct would replace the endogenous AGP1 by a dual recombination event 88 bp upstream from the ATG codon and 93 bp downstream from the stop codon of the gene. Plants were transformed by polyethylene glycol–mediated protoplast fusion with the AGP1:KO, and five stable transformant lines were isolated. PCR analysis indicated that two of these lines, which displayed similar phenotypes, were disrupted at the AGP1 locus. Analysis of the RNA extracted from the wild type and a KO line is shown in Figure 9A, indicating that no AGP1 transcript was detectable. Growth analysis of the KO lines was initially performed by studies of colony growth on solid media, and these indicated that the KO of AGP1 resulted in decreased growth of inocula (as measured by colony diameter) after 3 weeks on solid media (Figure 9B). The mean colony diameters were in the region of 75% of that of the wild type through several culture cycles. Examination of protonemata indicated that the basis of the reduction in colony size was reduced cell length (Figure 9C). Chloronemal cells, with less variation in cell length than caulonemal cells, were chosen for the cell length measurements. The transition of a chloronemal filament to caulonemal filament can take two or three cell divisions, and during this period, cells can show characteristics of both cell types and variable cell lengths, making caulonemal cells less suitable for analyses. Subapical cells of any position in the protonema were used for statistical analyses, with the exception of the first cell produced on side branch formation, which can often be very short. Wild-type subapical chloronemal cell lengths range between 70 and 100 μm. Subapical chloronemal cell lengths of the KO lines were consistently shorter than the wild type in the region of 50 to 70 μm. Representative protonemata from the wild type and a KO line are shown in Figure 9D. The apical cells of wild-type P. patens show a distal narrowing. This was absent in the KO lines.
Figure 9.
Expression of AGP1 and Phenotype of the Wild Type and a Transgenic KO Line of P. patens.
(A) RT-PCR analysis of wild-type and P. patens AGP1 KO cultures. Amplification of AGP1 in the wild type (lane 2) but not a KO line (lane 5). The control plastocyanin gene was amplified in both the wild type (lane 3) and the KO line (lane 6). The mock cDNA controls indicated that no contaminating genomic DNA was present in either sample (lanes 4 and 7). Lane 1 shows a DNA 100-bp ladder.
(B) Representative plates containing nine colonies for wild-type and KO lines after 3 weeks.
(C) Histogram of mean colony diameters for nine colonies from three wild-type plates and three plates of a KO line grown for 3 weeks on solid media. The horizontal dashed line indicates the mean colony diameter for the three wild-type plates. Similar colony diameter data were collected from three separate experiments, and the figure shows representative results from one experiment. Histogram of mean lengths of supapical chloronemal cells of the KO lines compared with the wild type. Error bars indicate standard deviation. Means were calculated from at least 35 subapical cells per line.
(D) Micrograph of wild-type P. patens protonemata showing the wild-type branching pattern of chloronemal filaments arising from caulonemal cells, distal narrowing of apical cells (arrowheads), and typical cell lengths. Micrograph of the P. patens AGP1 KO showing typical cell lengths and lack of narrowing of apical cells. Asterisks indicate caulonemal or intermediate caulonemal cells that give rise to chloronemal cells. White lines are aligned with chloronemal cells and indicate typical cell lengths. Bar = 100 μm.
DISCUSSION
P. patens AGPs
Previous studies have demonstrated the presence of AGPs in a range of leafy liverwort and moss bryophyte species, including P. patens, where they have been implicated in the transition from branching filaments to parenchymatous leafy shoots (Basile and Basile, 1987; Basile et al., 1999; Mignone and Basile, 2000). The presence of AGPs in P. patens has been shown here by the biological activity of βGlcYR and the isolation of βGlcYR-precipitable material with biochemical characteristics of AGPs. These observations, along with biochemical sequencing data and bioinformatic protein sequence data using Arabidopsis, Brassica, and Oryza sequences as queries, indicate the presence of at least 10 putative AGP core protein sequences in P. patens. Three of the four subclasses of GPI-anchored AGP core protein sequences (classical AGP, arabinogalactan peptide, and FLAs) have so far been identified in P. patens. It is of interest that after 400 million years of evolution, P. patens contains AGP core protein sequences that are homologous to those from angiosperms and suggests that AGPs play important roles in cellular processes that are common to bryophytes and angiosperms. The presence of three classes of AGP sequences in P. patens suggests that these classes of AGPs diverged before speciation events. The observation that not all the peptide sequences obtained by biochemical isolation of βGlcYR-precipitated AGPs relate to sequences identified by bioinformatic searches indicates that more AGP core protein genes remain to be identified in P. patens.
The behavior of βGlcYR-precipitated material on SDS-PAGE gels in terms of molecular mass and heterogeneity suggests properties similar to angiosperm AGPs. A significant and unusual feature of the βGlcYR-precipitated P. patens AGPs, confirmed by both biochemical analysis and immunochemistry, is the presence of (1→5)-α-l-arabinofuranosyl residues. This glycan structural feature is most often associated with the pectic polymer rhamnogalacturonan-I in angiosperms (Willats et al., 2001), although it has previously been identified in an Acacia AGP (Mollard and Joseleau, 1994).
AGPs and Plant Growth
We present evidence not only that AGPs are present in the moss P. patens but also that they function in apical cell extension that is the basis of protonemal growth. The growth inhibitor βGlcYR, which blocks cell extension, is likely to interact with a range of AGPs, suggesting that a range of AGPs may be involved in the cell expansion process. This is supported by the KO of one AGP core protein gene, P. patens AGP1, which resulted in an ∼25% reduction in both colony growth and cell length. βGlcYR has previously been used to inhibit the extension of tip-growing lily pollen tubes (Roy et al., 1999), and overexpression of a classical AGP core protein led to an increase in organ growth (Park et al., 2003). This article reports that the KO of a predicted classical AGP core protein gene can result in reduced growth of apically extending cells under normal growth conditions.
Cell Biological Function of AGPs in Cell Expansion
The presence of the LM6 epitope on P. patens AGPs at all plasma membrane surfaces and its localized occurrence at the cell wall surface at the tips of growing protonemata suggests that at these tip regions there is a localized loss and/or cleavage of AGPs from the plasma membrane and release into the cell wall that is a feature of apical cell extension. Furthermore, treatment with βGlcYR blocks apical extension and also the appearance and/or accessibility of the LM6 epitope in the cell wall at the tip of apical cells. This suggests that the tip-localized movement of AGPs into the cell wall may be required for apical cell extension. The putative P. patens AGP core protein genes identified to date all have predicted sites for a GPI anchor addition. Thus, a possible mechanism for the specific cleavage of AGPs from the plasma membrane and its release into the cell wall could be by the action of phospholipases (Oxley and Bacic, 1999; Gaspar et al., 2001). Components of the GPI anchor biosynthetic pathway have been shown to be required for pollen tube growth in Arabidopsis (Lalanne et al., 2004). We have observed that LM6-reactive AGPs specifically accumulate in the media of growing P. patens protonemata (Figure 4). The continuous tip-localized release of AGPs into the cell wall and then medium as a protonemal filament extends may be a requirement for growth. One possibility, supported by the suggestion that AGPs may act as plasticizers during cell wall extension (Lamport, 2001), is that movement of AGPs through the cell wall may modulate cell wall properties at the tip where considerable assembly and remodeling is taking place. Other possibilities include their involvement in the localized unloading of Golgi-derived vesicles at the tip or deposition of cell wall polymers. In the lily system, the application of βGlcYR has been shown to block cell extension but not secretion of cell wall polymers (Roy et al., 1999).
It is now becoming clear that βGlcYR binding glycomodules can be appended to protein motifs with diverse functions, indicating that experiments with βGlcYR need to be interpreted with care (Mashiguchi et al., 2004; Motose et al., 2004). The properties that the presence of AGP glycomodules impart to proteins and the extent of the occurrence of hybrid molecules containing AGP glycomodules in P. patens remain to be determined. The potential for combined cell biological and genetic approaches in P. patens should allow the elucidation of precise functions of AGP1 in the apical cell extension process. In summary, this work has established P. patens as an excellent system for the study of AGPs and their functions in the context of evolution.
METHODS
Plant Material and Culture Media
The wild-type strain of Physcomitrella patens (Hedw.) was isolated in Gransden Wood, Huntingdonshire, UK as a single spore (Ashton and Cove, 1977). Cultures were grown in liquid medium containing inorganic salts and a nitrogen source (either nitrate, known as BCD, or ammonium, known as BCD/AT) or on the same medium solidified with 1% (w/v) high gel strength agar (Sigma-Aldrich) under constant irradiation from fluorescent light of intensities ranging from 5 to 20 Wm−2 and at 22 to 25°C as described elsewhere (Knight et al., 2002). If cultures were to be removed from solid media for any reason, they were inoculated onto solid media overlaid with sterile cellophane disks (W.E. Canning) for ease of transfer.
Analysis of Moss Growth and Apical Cell Extension
For the analysis of apical cell extension in perfusion chambers, cultures were prepared by inoculating protonemal tissue fragments on solid media overlaid with cellophane. Following 3 d of growth, cultures were transferred to a glass slide overlaid with a Coverwell Perfusion Chamber and immersed in 300 μL of liquid BCD media. Cultures were treated with a 1-h pulse of 1 μM βGlcYR or αManYR in liquid media by pipetting through the access hole in the top of the perfusion chamber and displacing the chamber's existing contents. YRs were removed after 1 h by addition of fresh media through the access hole. All observations were made using an Olympus CKE microscope fitted with an RGB CCD video camera and monitor. Recordings were made using a Panasonic/National NV-8050 time-lapse video recorder. A video pointer (VP-380; FOR-A) provided a cursor that could be repositioned after YR addition/removal. Growth rates were determined by marking the position of the filament apex on the monitor screen at hourly intervals, followed by reference to a slide graticule.
Preparation of Plant Material for Light Microscopy and Immunolabeling
Material under investigation (except resin-embedded sectioned material) was fixed in 4% (w/v) paraformaldehyde and processed for whole mount immunolabeling as described elsewhere (McCartney et al., 2003). Rat monoclonal antibodies anti-(1→5)-α-l-arabinan LM6 (Willats et al., 1998) and antihomogalacturonan JIM5 (Clausen et al., 2003) were used as unpurified hybridoma supernatants. For resin embedding and sectioning, cultures were fixed in 2.5% (w/v) glutaraldehyde (in 0.1 M sodium phosphate buffer, pH 7.4, for 2 h at 4°C) and then processed for embedding in LR White resin and indirect immunofluorescence microscopy as described elsewhere (McCartney et al., 2003).
YR Preparation and Extraction and Analysis of AGPs
βGlcYR and αManYR were prepared as described elsewhere (Yariv et al., 1962). The method for the precipitation of AGPs with βGlcYR was that of Schultz et al. (2000). Protonemal tissue was grown for 7 d on cellophane overlaid on BCD/AT media, harvested, and then snap-frozen in liquid nitrogen. AGPs were quantified in terms of carbohydrate concentration by the phenol-sulphuric acid method (Dubois et al., 1956) with glucose as the standard. For total extracts, material (4 g fresh weight) was grown for 7 d on BCD/AT medium, snap-frozen in liquid nitrogen, and then lyophilized in a Modulyo 4K freeze drier (Edwards). Lyophilized tissue was homogenized in liquid nitrogen then added to 500 μL of extraction buffer (containing 50 mM Tris-HCl, 50 mM trans-1,2-cyclohexanediamine-N,N,N′,N′-tetraacetic acid, and 25 mM DTT, pH 7.8) in a 1.5-mL snap-top tube. The tube was vortexed briefly, then spun at maximum speed for 10 min in a microcentrifuge (MSE). The supernatant was applied to the gel as 10 μg of protein (as determined by Bio-Rad [Bradford] protein microassay). Material secreted into the culture medium was prepared from cultures transferred from cellophane overlay on solid medium to liquid BCD/AT medium in conical flasks on a shaking platform. Following 14 d of growth, the culture medium was separated from the protonemal tissue by filtration through Miracloth (Calbiochem), poured into a 12- to 14-kD cutoff dialysis membrane (Medicell International) sealed at both ends with plastic clips, and then dialyzed (3× 1 liter of 10 mM TrisHCl, pH 7.5, containing 5 mM β-mercaptoethanol). Dialyzed culture medium was snap-frozen in liquid nitrogen and lyophilized in a Modulyo 4K freeze drier (Edwards). Freeze-dried material was added to 1 mL of deionized water. Material was applied to the gel as a loading containing 10 μg of carbohydrate. SDS-PAGE analysis and immunoblotting were performed as described elsewhere (Willats and Knox, 1996).
Monosaccharide-Linkage Analysis of P. patens AGP Glycan
βGlcYR-precipitated AGP was carboxyl reduced according to the method reported by Sims and Bacic (1995) prior to methylation to detect uronic acids. Monosaccharide-linkage analysis was performed by methylation of the carboxyl-reduced AGP with methyl iodide in the presence of sodium hydroxide (Ciucanu and Kerek, 1984), followed by trifluoroacetic acid (TFA) hydrolysis (2.5 M at 100°C, 6 h), reduction with sodium borodeuteride (0.5 M in 1 M NaOH overnight at room temperature), and acetylation with acetic anhydride (100°C, 2.5 h). Partially methylated alditol acetates were recovered in CH2Cl2, separated on a BPX70 column (SGE International) using a Hewlett-Packard HP 6890 gas chromatograph, and detected by electron impact ionization (70 eV) mass spectrometry with a Hewlett-Packard 5973 mass selective detector (mass-to-charge ratio of 100 to 350). Uronic acids were quantified by the corresponding dideutero-bearing fragment ions of C-6.
Deglycosylation of AGPs by Anhydrous Hydrogen Fluoride
Chemical deglycosylation of βGlcYR-AGPs (6 mg in 100 μL of super dry methanol) was performed using anhydrous HF (1 mL) for 3 h at room temperature, according to the method of Mort and Lamport (1977) as previously described (Schultz et al., 2000). The HF was removed under vacuum; then the sample was desalted on a HW40 column (10 × 310 mm; ToyaPearl). The desalted deglycosylated AGP protein backbone fractions (void volume) were dried and further fractionated by reverse-phase HPLC.
HPLC
Reverse-phase HPLC was performed with a C8 RP 300 column (2.1 × 100 mm; Aquapore) attached to a Beckman Systems Gold HPLC equipped with a diode array detector. AGPs in the bound material were eluted and collected from the column and equilibrated in solvent A (0.1% TFA) with a linear gradient of solvent B (0.1% TFA in 80% acetonitrile), 0 to 100% solvent B in 30 min, at a flow rate of 0.5 mL/min. Chromatography was monitored by absorption at 214 and 280 nm. Individual peaks (retention times = 7.3, 9.1, 15, and 17 min, respectively) were manually collected and subjected to N-terminal Edman sequencing as described below.
Edman Degradation Protein Sequencing
N-terminal protein sequencing was performed by automated Edman degradation sequencing on a sequencer (Model LF 3400; Beckman Instruments) with online analysis on a Beckman System Gold HPLC.
Cloning of P. patens AGP1 cDNA and Sequence Analysis
To identify AGP protein backbone genes from moss EST databases, the BLAST search program (Altschul et al., 1990) was used to search against Arabidopsis thaliana AGP amino acid sequences (Schultz et al., 2000; Gaspar et al., 2001) as queries. Three cDNA clones were retrieved (GenBank accession numbers AW599813, BJ203418, and BJ610018). BJ203418 and BJ610018 were from PHYSCObase, which is enriched by full-length cDNAs (Nishiyama et al., 2003), and correspond to Contig11049. This sequence was named AGP1. The putative open reading frame (ORF) of P. patens AGP1 was amplified by PCR from a normalized full-length cDNA library prepared from protonemal tissue of P. patens (a generous gift from Mitsuyasu Hasebe) using the following primers: 5′-ATAGTTACAATGGCCGTGTC-3′ and 5′-GCAGATCAGTACAAGAGGTA-3′. The PCR product was cloned into pGEM-Teasy (Promega). The prediction of the N-terminal signal sequence, GPI anchor signal sequence, and the subcellular localization of P. patens AGP1 was performed using the PSORT program (http://psort.ims.u-tokyo.ac.jp/).
Cloning of P. patens AGP1 Genomic Sequence
The primers for the amplification of the ORF of P. patens AGP1 cDNA were used for the PCR amplification of the genomic sequence. The 5′ and 3′ sequence flanking the P. patens AGP1 ORF was obtained by the inverse PCR method (Ochman et al., 1988; Triglia et al., 1988). Two micrograms of Physcomitrella genomic DNA was digested with EcoRI or HindIII, purified with the QIAquick PCR purification kit (Qiagen), and eluted with 30 μL of the elution buffer. Four microliters each of the elutes was used for the self-ligation with T4 DNA ligase and the supplied buffer (Invitrogen) at 14°C for 12 h, then purified with the QIAquick PCR purification kit and eluted with 30 μL of the elution buffer. One microliter each of the elutes was used for the first PCR amplification by KlenTaq (BD Biosciences) and the supplied buffer with 25 μM each dNTP and 3.5 mM MgCl2 by 30 cycles of 96°C for 1 min, 55°C for 1 min, and 68°C for 2 min. The PCR products were purified as described above and subjected to a second PCR amplification using the same conditions as the first PCR. The primers used for the first and second PCR were 5′-GACACGGCCATTGTAACTAT-3′ and 5′-TGAAGCAATTTGTATGGTGG-3′, and 5′-CAATACTCAGCGTTCAGCAA-3′ and 5′-TCCTAACCCAATGGAGTCTG-3′, respectively. The amplified fragments after the second PCR were cloned into pGEM-Teasy and subjected to DNA sequencing using the M13 forward and reverse primers and BigDye Terminator mix (Applied Biosystems). The sequence analysis revealed the presence of a HindIII site and an EcoRI site 2.0 kb upstream from the ATG codon and 1.7 kb downstream from the stop codon, respectively. The 4.4-kb HindIII-EcoRI genomic fragment was amplified from a P. patens genomic DNA extract by PCR using primers 5′-AAGCTTATCTATAAAATATTATCCAATTAATTTAC-3′ and 5′-GAATTCTTTTTGGTCCAATTGTGCA-3′. The PCR program was 35 cycles of 96°C for 1 min, 55°C for 30 s, and 68°C for 10 min. The amplified fragment was cloned into pGEM-Teasy and subjected to sequencing analysis as described above.
DNA Constructs and Transformation of P. patens
The KO construct of P. patens AGP1 was made as follows. A SpeI site and a ClaI site were located in the AGP1 genomic sequence, 88 bp upstream from the ATG codon and 93 bp downstream from the stop codon, respectively. These sites were used to remove the AGP1 ORF sequence by restriction enzyme digestion. Then, a hygromycin-selectable marker cassette, flanked by LoxP recombination sites (Sauer, 1998), was inserted by blunt-end ligation. The transformation of P. patens protoplasts and the isolation and growth of transgenic lines were performed as described elsewhere (Bezanilla et al., 2003).
RT-PCR Analysis
Total RNA was extracted from wild-type and AGP1 KO P. patens tissue (100 mg each) using the RNeasy plant mini kit (Qiagen) according to the manufacturer's instructions. Total RNA (1 μg from each tissue sample) was treated with DNase I (Sigma-Aldrich) according to the manufacturer's instructions prior to cDNA synthesis using the ThermosScript RT-PCR system (Invitrogen Life Technologies). The first strand of cDNA was synthesized by a PCR reaction at 50°C for 1 h using 1 μg of RNA from each tissue sample with 50 pmol of the supplied oligo(dT)20 primer with 15 units of Thermoscript reverse transcriptase in a 20-μL reaction mixture. A control reaction was also performed where reverse transcriptase was omitted (mock cDNA control). Following cDNA synthesis, the RNA template was degraded with 2 units of Escherichia coli RNase H at 37°C for 20 min. Purified cDNAs were isolated using a QIAquick PCR purification column (Qiagen) according to the manufacturer's instructions.
PCR analysis of wild-type and AGP1 KO cDNAs was performed using primers for P. patens AGP1 (5′-GATTGTGCATCTGGCTTGCCTTGT-3′ and 5′-GGGTGCAACAATACTCAGCGTTCA-3′) and plastocyanin (5′-GCCTAATGAGAGCTTCTCC-3′ and 5′-GGTTTCAGGCATAGTTATCG-3′). PCR was performed as 30 cycles of 96°C for 1 min, 60°C for 30 s, and 70°C for 2 min.
Accession Numbers
Sequence data for P. patens AGP core proteins from this article can be found in the EMBL/GenBank data libraries under the following accession numbers: AGP1, BJ203418 (5′) and BJ961723 (3′); AGP2, BJ177746 (5′) and BJ601080 (3′); AGP3, BJ167100; AGP4, BJ172480; AGP5, BJ958679; FLA1, BJ198065 (5′) and BJ970317 (3′); FLA2, BJ198534 (5′) and BJ604841 (3′).
Acknowledgments
We thank the UK Biotechnology and Biological Sciences Research Council for a postgraduate studentship to support K.J.D.L. We (A.B., F.P., and S.-L.M.) acknowledge support from the Australian Research Council and the Cooperative Research Centre for Bioproducts. R.S.Q. acknowledges support from the National Science Foundation (IBN 0112461). We thank David Cove, Andrew Cuming, Yasuko Kamisugi, Iain Manfield, and Martin Kieffer for useful discussions.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: J. Paul Knox (j.p.knox@leeds.ac.uk).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.034413.
References
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Ashton, N.W., and Cove, D.J. (1977). The isolation and preliminary characterisation of auxotrophic and analogue-resistant mutants of the moss Physcomitrella patens. Mol. Gen. Genet. 154, 87–95. [Google Scholar]
- Basile, D.V., and Basile, M.R. (1987). The occurrence of cell wall-associated arabinogalactan proteins in the Hepaticae. Bryologist 90, 401–404. [Google Scholar]
- Basile, D.V., Basile, M.R., Salama, N., Peart, J., and Roberts, K. (1999). Monoclonal antibodies to arabinogalactan proteins (AGPs) released by Gymnocolea inflata when leaf and branch development is desuppressed. Bryologist 102, 304–308. [Google Scholar]
- Bezanilla, M., Pan, A., and Quatrano, R.S. (2003). RNA interference in the moss Physcomitrella patens. Plant Physiol. 133, 470–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen, M.H., Willats, W.G.T., and Knox, J.P. (2003). Synthetic methyl hexagalacturonate hapten inhibitors of anti-homogalacturonan monoclonal antibodies LM7, JIM5 and JIM7. Carbohydr. Res. 338, 1797–1800. [DOI] [PubMed] [Google Scholar]
- Cove, D.J., Knight, C.D., and Lamparter, T. (1997). Mosses as model systems. Trends Plant Sci. 2, 99–105. [Google Scholar]
- Cheung, A.Y., Wang, H., and Wu, H.-M. (1995). A floral transmitting tissue-specific glycoprotein attracts pollen tubes and stimulates their growth. Cell 82, 383–393. [DOI] [PubMed] [Google Scholar]
- Ciucanu, I., and Kerek, F. (1984). A simple and rapid method for the permethylation of carbohydrates. Carbohydr. Res. 131, 209–217. [Google Scholar]
- Darley, C.P., Forrester, A.M., and McQueen-Mason, S.J. (2001). The molecular basis of plant cell wall extension. Plant Mol. Biol. 47, 179–195. [PubMed] [Google Scholar]
- Ding, L., and Zhu, J.K. (1997). A role for arabinogalactan proteins in root epidermal cell expansion. Planta 203, 289–294. [DOI] [PubMed] [Google Scholar]
- Dubois, M., Gilles, K.A., Hamilton, J.K., Rebers, P.A., and Smith, F. (1956). Colorimetric method for determination of sugars and related substances. Anal. Chem. 28, 350–356. [Google Scholar]
- Gaspar, Y., Johnson, K.L., McKenna, J.A., Bacic, A., and Schultz, C.J. (2001). The complex structures of arabinogalactan proteins and the journey towards understanding function. Plant Mol. Biol. 47, 161–176. [PubMed] [Google Scholar]
- Gaspar, Y.M., Nam, J., Schultz, C.J., Lee, L.-Y., Gilson, P.R., Gelvin, S.B., and Bacic, A. (2004). Characterization of the Arabidopsis lysine-rich arabinogalctan protein AtAGP17 mutant (rat1) that results in a decreased efficiency of Agrobacterium transformation. Plant Physiol. 135, 2162–2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrum, L.J., Patel, A., Leykam, J.F., and Kieliszewski, M.J. (2000). Gum arabic glycoprotein contains glycomodules of both extensin and arabinogalactan glycoproteins. Phytochemistry 54, 99–106. [DOI] [PubMed] [Google Scholar]
- Hiwatashi, Y., Nishiyama, T., Fujita, T., and Hasebe, M. (2001). Establishment of gene-trap and enhancer-trap systems in the moss Physcomitrella patens. Plant J. 28, 105–116. [DOI] [PubMed] [Google Scholar]
- Johnson, K.L., Jones, B.J., Bacic, A., and Schultz, C.J. (2003. a). Non-enzymic cell wall glycoproteins. In The Plant Cell Wall, J.K.C. Rose, ed (Oxford: Blackwell Publishing), pp. 111–154.
- Johnson, K.L., Jones, B.J., Bacic, A., and Schultz, C.J. (2003. b). The fasciclin-like arabinogalactan proteins of Arabidopsis. A multigene family of putative cell adhesion molecules. Plant Physiol. 133, 1911–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight, C.D., Cove, D.J., Cuming, A.C., and Quatrano, R.S. (2002). Moss gene technology. In Molecular Plant Biology, Vol. 2, Practical Approach Series, P.M. Gilmartin and C. Bowler, eds (Oxford: Oxford University Press), pp. 285–301.
- Knox, J.P. (1997). The use of antibodies to study the architecture and developmental regulation of plant cell walls. Int. Rev. Cytol. 171, 79–120. [DOI] [PubMed] [Google Scholar]
- Knox, J.P., Linstead, P.J., Peart, J., Cooper, C., and Roberts, K. (1991). Developmentally-regulated epitopes of cell surface arabinogalactan proteins and their relation to root tissue pattern formation. Plant J. 1, 317–326. [DOI] [PubMed] [Google Scholar]
- Kremer, C., Pettolino, F., Bacic, A., and Drinnan, A. (2004). Distribution of cell wall components in Sphagnum hyaline cells and in liverwort and hornwort elaters. Planta 219, 1023–1035. [DOI] [PubMed] [Google Scholar]
- Lalanne, E., Honys, D., Johnson, A., Borner, G.H., Lilley, K.S., Dupree, P., Grossniklaus, U., and Twell, D. (2004). SETH1 and SETH2, two components of the glycosylphosphatidylinositol anchor biosynthetic pathway, are required for pollen germination and tube growth in Arabidopsis. Plant Cell 16, 229–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamport, D.T. (2001). Life behind cell walls: Paradigm lost, paradigm regained. Cell. Mol. Life Sci. 58, 1363–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligrone, R., Vaughn, K.C., Renzaglia, K.S., Knox, J.P., and Duckett, J.G. (2002). Diversity in the distribution of polysaccharide and glycoprotein epitopes in the cell walls of bryophytes: New evidence for the multiple evolution of water-conducting cells. New Phytol. 156, 491–508. [DOI] [PubMed] [Google Scholar]
- Mashiguchi, K., Yamaguchi, I., and Suzuki, Y. (2004). Isolation and identification of glycosylphosphatidylinositol-anchored arabinogalactan proteins and novel beta-glucosyl Yariv-reactive proteins from seeds of rice (Oryza sativa). Plant Cell Physiol. 45, 1817–1829. [DOI] [PubMed] [Google Scholar]
- Majewska-Sawka, A., and Nothnagel, E.A. (2000). The multiple roles of arabinogalactan proteins in plant development. Plant Physiol. 122, 3–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga, T., Ishii, T., Matsumoto, S., Higuchi, M., Darvill, A., Albersheim, P., and O'Neill, M.A. (2004). Occurrence of the primary cell wall polysaccharide rhamnogalacturonan II in pteridophytes, lycophytes, and bryophytes. Implications for the evolution of vascular plants. Plant Physiol. 134, 339–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCartney, L., Steele-King, C.G., Jordan, E., and Knox, J.P. (2003). Cell wall pectic (1→4)-β-D-galactan marks the acceleration of cell elongation in the Arabidopsis seedling root meristem. Plant J. 33, 447–454. [DOI] [PubMed] [Google Scholar]
- Mignone, M.M., and Basile, D.V. (2000). Evidence for the interrelated actions of auxin, ethylene and arabinogalactan proteins on the transition from non-apical to apical growth of Physcomitrella patens Hedw. (Funariaceae). In Cell and Developmental Biology of Arabinogalactan-Proteins, E.A. Nothnagel, A. Bacic, and A.E. Clarke, eds (New York: Kluwer Academic/Plenum Publishers), pp. 205–219.
- Mollard, A., and Joseleau, J.-P. (1994). Acacia senegal cells cultured in suspension secrete a hydroxyproline-deficient arabinogalactan protein. Plant Physiol. Biochem. 32, 703–709. [Google Scholar]
- Mollet, J.-C., Kim, S., Jauh, G.-Y., and Lord, E.M. (2002). Arabinogalactan proteins, pollen tube growth, and the reversible effects of Yariv phenylglycoside. Protoplasma 219, 89–98. [DOI] [PubMed] [Google Scholar]
- Mort, A., and Lamport, D.T.A. (1977). Anhydrous hydrogen fluoride deglycosylated glycoproteins. Anal. Biochem. 82, 289–309. [DOI] [PubMed] [Google Scholar]
- Motose, H., Sugiyama, M., and Fukuda, H. (2004). A proteoglycan mediates inductive interaction during plant vascular development. Nature 429, 873–878. [DOI] [PubMed] [Google Scholar]
- Nam, J., Mysore, K.S., Zheng, C., Knue, M.K., Matthysse, A.G., and Gelvin, S.B. (1999). Identification of T-DNA tagged Arabidopsis mutants that are resistant to transformation by Agrobacterium. Mol. Gen. Genet. 261, 429–438. [DOI] [PubMed] [Google Scholar]
- Nishiyama, T., Fujita, T., Shin, I.T., Seki, M., Nishide, H., Uchiyama, I., Kamiya, A., Carninci, P., Hayashizaki, Y., Shinozaki, K., Kohara, Y., and Hasebe, M. (2003). Comparative genomics of Physcomitrella patens gametophytic transcriptome and Arabidopsis thaliana: Implication for land plant evolution. Proc. Natl. Acad. Sci. USA 100, 8007–8012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothnagel, E.A. (1997). Proteoglycans and related components in plant cells. Int. Rev. Cytol. 174, 195–291. [DOI] [PubMed] [Google Scholar]
- Ochman, H., Gerber, A.S., and Hartl, D.L. (1988). Genetic applications of an inverse polymerase chain reaction. Genetics 120, 621–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oxley, D., and Bacic, A. (1999). Structure of the glycosyl-phosphatidylinositol membrane anchor of an arabinogalactan protein from Pyrus communis suspension-cultured cells. Proc. Natl. Acad. Sci. USA 6, 14246–14251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, M.H., Suzuki, Y., Chono, M., Knox, J.P., and Yamaguchi, I. (2003). CsAGP1, a gibberellin-responsive gene from cucumber hypocotyls (Cucumis sativus), encodes a classical arabinogalactan protein and is involved in stem elongation. Plant Physiol. 131, 1450–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popper, Z.A., and Fry, S.C. (2003). Primary cell wall composition of bryophytes and charophytes. Ann. Bot. (Lond.) 91, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing, S.A., Rombauts, S., Van de Peer, Y., and Reski, R. (2002). Moss transcriptome and beyond. Trends Plant Sci. 7, 535–538. [DOI] [PubMed] [Google Scholar]
- Reski, R. (1998). Development, genetics and molecular biology of mosses. Bot. Acta 111, 1–15. [Google Scholar]
- Roy, S.J., Holdaway-Clarke, T.L., Hackett, G.R., Kunkel, J.G., Lord, E.M., and Hepler, P.K. (1999). Uncoupling secretion and tip growth in lily pollen tubes: Evidence for the role of calcium in exocytosis. Plant J. 19, 379–386. [DOI] [PubMed] [Google Scholar]
- Sauer, B. (1998). Inducible gene targeting in mice using the Cre/lox system. Methods 14, 381–392. [DOI] [PubMed] [Google Scholar]
- Schaefer, D.G., and Zryd, J.P. (1997). Efficient gene targeting in the moss Physcomitrella patens. Plant J. 11, 1195–1206. [DOI] [PubMed] [Google Scholar]
- Schipper, O., Schaefer, D., Reski, R., and Fleming, A. (2002). Expansins in the bryophyte Physcomitrella patens. Plant Mol. Biol. 50, 789–802. [DOI] [PubMed] [Google Scholar]
- Schultz, C.J., Johnson, K.L., Currie, G., and Bacic, A. (2000). The classical arabinogalactan protein gene family of Arabidopsis. Plant Cell 12, 1751–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz, C.J., Rumsewicz, M.P., Johnson, K.L., Jones, B.J., Gaspar, Y.M., and Bacic, A. (2002). Using genomic resources to guide research directions. The arabinogalactan protein gene family as a test case. Plant Physiol. 129, 1448–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherrier, D.J., Prime, T.A., and Dupree, P. (1999). Glycosylphosphatidylinositol-anchored cell-surface proteins from Arabidopsis. Electrophoresis 20, 2027–2035. [DOI] [PubMed] [Google Scholar]
- Shi, H., Kim, Y., Guo, Y., Stevenson, B., and Zhu, J.-K. (2003). The Arabidopsis SOS5 locus encodes a putative cell surface protein and is required for normal cell expansion. Plant Cell 15, 19–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Showalter, A.M. (2001). Arabinogalactan-proteins: Structure, expression and function. Cell. Mol. Life Sci. 58, 1399–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims, I.M., and Bacic, A. (1995). Extracellular polysaccharides from suspension cultures of Nicotiana plumbaginifolia. Phytochemistry 38, 1397–1405. [Google Scholar]
- Sun, W., Kieliszewski, M.J., and Showalter, A.M. (2004). Overexpression of tomato LeAGP-1 arabinogalactan protein promotes lateral branching and hampers reproductive development. Plant J. 40, 870–881. [DOI] [PubMed] [Google Scholar]
- Svetek, J., Yadav, M.P., and Nothnagel, E.A. (1999). Presence of a glycosylphosphatidylinositol lipid anchor on rose arabinogalactan proteins. J. Biol. Chem. 274, 14724–14733. [DOI] [PubMed] [Google Scholar]
- Tan, L., Leykam, F., and Kieliszewski, M.J. (2003). Glycosylation motifs that direct arabinogalactan addition to arabinogalactan proteins. Plant Physiol. 132, 1362–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, L., Qiu, F., Lamport, D.T., and Kieliszewski, M.J. (2004). Structure of a hydroxyproline (Hyp)-arabinogalactan polysaccharide from repetitive Ala-Hyp expressed in transgenic Nicotiana tabacum. J. Biol. Chem. 279, 13156–13165. [DOI] [PubMed] [Google Scholar]
- Takahashi, T., Gasch, A., Nishizawa, N., and Chua, N.-H. (1995). The DIMINUTO gene of Arabidopsis is involved in regulating cell elongation. Genes Dev. 9, 97–107. [DOI] [PubMed] [Google Scholar]
- Triglia, T., Peterson, M.G., and Kemp, D.J. (1988). A procedure for in vitro amplification of DNA segments that lie outside the boundaries of known sequences. Nucleic Acids Res. 16, 8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hengel, A.J., and Roberts, K. (2002). Fucosylated arabinogalactan proteins are required for full root cell elongation in Arabidopsis. Plant J. 32, 105–113. [DOI] [PubMed] [Google Scholar]
- van Hengel, A.J., and Roberts, K. (2003). AtAGP30, an arabinogalactan protein in the cell walls of the primary root, plays a role in root regeneration and seed germination. Plant J. 36, 256–270. [DOI] [PubMed] [Google Scholar]
- Vissenberg, K., Feijo, J.A., Weisenseel, M.H., and Verbelen, J.P. (2001). Ion fluxes, auxin and the induction of elongation growth in Nicotiana tabacum cells. J. Exp. Bot. 52, 2161–2167. [DOI] [PubMed] [Google Scholar]
- Willats, W.G.T., and Knox, J.P. (1996). A role for arabinogalactan proteins in plant cell expansion: Evidence from studies on the interaction of β-glucosyl Yariv reagent with seedlings of Arabidopsis thaliana. Plant J. 9, 919–925. [DOI] [PubMed] [Google Scholar]
- Willats, W.G.T., Marcus, S.E., and Knox, J.P. (1998). Generation of a monoclonal antibody specific to (1→5)-α-L-arabinan. Carbohydr. Res. 308, 149–152. [DOI] [PubMed] [Google Scholar]
- Willats, W.G.T., McCartney, L., and Knox, J.P. (2001). In-situ analysis of pectic polysaccharides in seed mucilage and at the root surface of Arabidopsis thaliana. Planta 213, 37–44. [DOI] [PubMed] [Google Scholar]
- Willats, W.G.T., Steele-King, C.G., Marcus, S.E., and Knox, J.P. (1999). Side chains of pectic polysaccharides are regulated in relation to cell proliferation and cell differentiation. Plant J. 20, 619–628. [DOI] [PubMed] [Google Scholar]
- Wu, H., Wong, E., Ogdahl, J., and Cheung, A.Y. (2000). A pollen tube growth-promoting arabinogalactan protein from Nicotiana alata is similar to the tobacco TTS protein. Plant J. 22, 165–176. [DOI] [PubMed] [Google Scholar]
- Yariv, J., Rapport, M.M., and Graf, L. (1962). The interaction of glycosides and saccharides with antibody to the corresponding phenylazo glycosides. Biochem. J. 85, 383–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates, E.A., Valdor, J.-F., Haslam, S.M., Morris, H.R., Dell, A., Mackie, W., and Knox, J.P. (1996). Characterization of carbohydrate structural features recognized by anti-arabinogalactan protein monoclonal antibodies. Glycobiology 6, 131–139. [DOI] [PubMed] [Google Scholar]
- Youl, J.J., Bacic, A., and Oxley, D. (1998). Arabinogalactan proteins from Nicotiana alata and Pyrus communis contain glycosylphosphatidylinositol membrane anchors. Proc. Natl. Acad. Sci. USA 95, 7921–7926. [DOI] [PMC free article] [PubMed] [Google Scholar]