Type IV pili (TFP) are very unique appendages on the bacterial surface. They are not only required for microbial adherence but also involved in bacterial movement, such as social gliding motility in Myxococcus xanthus and twitching motility in Pseudomonas and Neisseria species (33). How bacterial pili are involved in cellular movement has long been a mystery. Recent studies have revealed that TFP are motility apparatuses that extend and retract TFP filaments at the cellular pole (20, 29, 31). Furthermore, in Myxococcus and Synechocystis spp., the TFP action was found to be controlled by a chemotaxis-like system (3, 31). These findings provide insights at the molecular level into the involvement of TFP in cellular motility. They also suggest possible roles of TFP in directed cell movement, biofilm formation, guided tissue invasion, and other pathogenesis-related events.
TFP
Pili are fibrous organelles that are expressed on the surface of gram-negative bacteria (for a review, see reference 16). Historically, different families have been assigned on the basis of the specificity of host-receptor recognition and the seroreactivity of antibodies against pilin proteins. More recently, pili have been grouped on the basis of the deduced amino acid sequence of pilin genes and their assembly mechanisms. Although they differ in morphology and structure, most pili are adhesins involved in mediating bacterial interactions with the environment or other cells. For example, the type I and type P pili are assembled by the chaperone-usher pathway, expressed on the surface of uropathogenic strains of Escherichia coli and mediate bacterial attachment to host cells through specific carbohydrate binding proteins (25). TFP are conserved in their major pilin amino acid sequences. They are assembled through the general secretion pathway and are expressed in a divergent collection of gram-negative bacteria, including Pseudomonas aeruginosa, Neisseria gonorrhoeae, Neisseria meningitidis, Moraxella bovis, Eikenella corrodens, Vibrio cholerae, the cyanobacterium Synechocystis sp., enteropathogenic E. coli (EPEC), and enterotoxigenic E. coli (16, 30). TFP are 6- to 7-nm thick, rod-like fibers of variable length and relative flexibility and are usually polarly located.
Biochemical and genetic studies of TFP, mainly in P. aeruginosa and N. gonorrhoeae, have identified important components of TFP biogenesis. These components are homologous to those of the general (type II) secretion pathway, including pilin, prepilin peptidase, soluble proteins with or without essential nucleotide-binding motifs, inner membrane proteins, and outer membrane secretins that form oligomeric doughnut-shaped structures (for reviews on TFP biogenesis, see references 1, 13, 30, and 32, and for a review on the general secretion pathway, see reference 24). TFP are assembled primarily from one subunit, the pilin protein. The pilins are highly conserved at the N terminus, which is highly hydrophobic. They are synthesized in a precursor form from which the short, basic, amino-terminal signal peptide is cleaved by a prepilin peptidase. The peptidases are bifunctional in that they also methylate the first amino acid (Phe or Met) of the mature pilin. Based on accumulated genetic and biochemical evidence, the current model of TFP biogenesis is as follows (13). Prior to polymerization into pilus fibers, the pilin molecules are anchored to the inner membrane via the conserved hydrophobic region in the N-terminal domain. The pilins are then polymerized, possibly driven by energy-prone self-assembly in the presence of biogenesis machinery proteins, such as PilB and PilC in P. aeruginosa and PilF in N. gonorrhoeae. The polymerized pilus fibers thrust through an outer membrane pore consisting of oligomeric PilQ, which is homologous with the secretins of the general secretion pathway (5, 36). Many other genes, such as the pilMNOPQ operon in P. aeruginosa, also participate in TFP biogenesis (1). PilT, which is a cytoplasmic protein with a nucleotide-binding domain, is not required for pilus biogenesis, although in some cases it appears antagonistic to other pil mutants (35, 36). Nevertheless, PilT has a very important role in TFP function (see below).
TFP-ASSOCIATED CELLULAR FUNCTIONS AND TFP-DEPENDENT CELLULAR MOTILITY
The cellular functions of TFP have been extensively studied in various bacteria. Many studies have demonstrated that TFP proteins function as adhesins and mediate bacterial interactions with the environment and/or host cells (19, 23, 30). Thus, TFP are considered virulence factors in several important human and animal pathogens including P. aeruginosa, N. gonorrhoeae, N. meningitidis, M. bovis, E. corrodens, V. cholerae, EPEC, and enterotoxigenic E. coli. In nonpathogenic bacteria such as M. xanthus, TFP also act as adhesins required for bacterial agglutination (38).
Besides serving as adhesins, TFP are also involved in the secretion and uptake of some macromolecules. For example, TFP of P. aeruginosa are essential components for toxin secretion (18). In N. gonorrhoeae, TFP are involved in transforming DNA into bacterial cells (34). These cellular functions may be related to the fact that the membrane portion of TFP may function as a general (type II) secretion pathway.
The most unique (and interesting) cellular function of TFP is their involvement in the bacterial movements on solid surfaces (33). It is now well documented that many bacteria can move on solid surfaces without the aid of flagella (7, 12, 22). Social gliding motility in M. xanthus and twitching motility in P. aeruginosa and N. gonorrhoeae are the best examples of these flagellum-independent movements. Interestingly, genetic and behavioral studies show that both the social gliding motility of Myxococcus and twitching motility of Pseudomonas and Neisseria are absolutely dependent on TFP (14, 15, 37). Some recent studies have shown that other bacteria (e.g., EPEC and Synechocystis sp.) engage in TFP-dependent cellular motility on solid surfaces (2, 4).
TFP ARE RETRACTABLE MOTILITY APPARATUSES FOR SURFACE MOVEMENT
How TFP are involved in cellular motility has long been a mystery. In 1970, Bradley found that pilus-specific phage particles which initially attach to pili later appear at the cell surface (6). Based on this observation, he proposed that TFP may generate movement through pilus retraction. Recent studies in three model bacteria, M. xanthus, N. gonorrhoeae, and P. aeruginosa, have provided more-direct evidence that TFP indeed retract to generate gliding or twitching motility (20, 29, 31). Sun et al. (31) developed a tethering assay for M. xanthus cells in a highly viscous medium (1% methylcellulose). Using this assay, they observed that some M. xanthus cells could be tethered perpendicular to a glass or polystyrene surface in a TFP-dependent manner. These tethered cells displayed a jiggling motion as the cell bodies were drawn closer to the tethering surface. Further analyses of the behavior of these tethered cells led to a theory that M. xanthus cells extrude their pili, which attach to a solid surface and retract, bringing the tethered cell bodies closer to the surface. The study also showed that pilT mutant cells, which are piliated but defective in gliding and twitching motility, could be tethered to a solid surface (38) but fail to retract closer to the tethering surface, suggesting that PilT is required for generating the retraction force.
Merz et al. (20) used laser tweezers to show that N. gonorrhoeae pili retract. In one of their experiments, individual N. gonorrhoeae cells were immobilized on latex beads via antibodies specific for the bacterium. Smaller beads were used to tether the TFP fibers through a monoclonal antibody specific for a pilus surface-exposed epitope. The smaller beads, held in optical tweezers, were repeatedly pulled toward the immobilized cells. pilT mutant cells, in contrast, were unable to generate such retractile forces.
Skerker and Berg (29) recently labeled the TFP of P. aeruginosa with an amino-specific Cy3 fluorescent dye and visualized them on a quartz slide using a total internal reflection microscope. They were able to directly observe the extension and retraction of TFP of P. aeruginosa. Frequently, the distal tip of a pilus was adsorbed to the substratum and the pilus was pulled taut. Occasionally, the cell body detached from the surface and was pulled forward by means of pilus retraction.
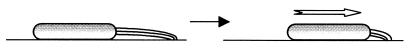
Through these studies, it is clear that TFP are motility apparatuses that extend pilus filaments, attach at their distal tips, and retract (Fig. 1). These findings greatly expand our knowledge about this mode of bacterial motility. Whereas flagellum-dependent motility works well in aquatic environments, TFP-dependent motility works well on solid surfaces. It would be interesting to discover how bacteria have evolved two very different motility mechanisms that are adapted for very different environments.
FIG. 1.
Model of TFP-dependent motility. Based on recent studies (20, 29, 31), bacteria such as M. xanthus and P. aeruginosa extend TFP filaments at one pole of the cell, attach them at the distal tips, and retract to generate motility.
TFP-DEPENDENT MOTILITY IN M. XANTHUS AND SYNECHOCYSTIS SP. IS GUIDED BY A CHEMOTAXIS-LIKE SYSTEM
Previous studies have clearly shown that TFP-dependent motility is not a random movement. For example, the cyanobacterium Synechocystis sp. PCC6803 directs its TFP-dependent motility to perform phototaxis (2, 3). Pseudomonas aeruginosa requires its TFP-dependent motility to build up a sophisticated biofilm structure (21). The best example for directed TFP-dependent motility is the involvement of social gliding motility in the fruiting body formation of M. xanthus.
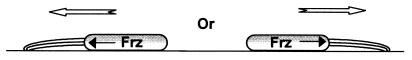
In response to starvation, hundreds of thousands of M. xanthus cells aggregate to form a multicellular structure called the “fruiting body,” which creates a unique environment that enables the starving cells to support each other. In this cellular process, starved cells not only perform TFP-dependent social gliding motility but also respond to environmental signals and cell density to modulate their reversal frequency and direct themselves to move toward aggregation centers (17, 27, 28, 37). Detailed genetic and behavioral analyses have identified a chemotaxis-like system (called the frz system in M. xanthus) that is involved in controlling TFP-dependent motility (26, 31). The frz system contains homologs of the genes encoding the methyl-accepting chemotaxis protein (MCP), cheA, cheY, cheW, cheB and cheR, and regulates cellular reversal (28). Further studies have revealed some insights at the molecular level into this control process (31). It appears that TFP in M. xanthus are usually located at one pole of the cell (17) but can switch from one pole to another (31). The reversal of gliding direction is associated with switching active TFP from one pole of the cell to another and is controlled by the frz system (31) (Fig. 2). Given the fact that the frz system contains MCP, which can sense various environmental signals, it is reasonable to assume that chemotaxis-guided TFP motility in M. xanthus can be achieved in the following way. (i) The frz system recognizes various chemical signals in the environment and relays the signals to the sites of pilus extrusion. (ii) The extruded pili coordinate cell movements in the direction of the chemical gradient as a consequence of pilus retraction. (iii) Switching the sites of pilus extrusion from one pole of the cell to the other results in cellular reversals in M. xanthus motility. (iv) The pilus switching frequency between the two poles of a cell is controlled by the frz chemosensory system.
FIG. 2.
Model of chemotaxis-guided TFP-dependent motility. Based on the study by Sun et al. (31), Frz chemotaxis homologs direct cell movements by controlling the sites of pilus extension.
Recent studies showed that chemotaxis-like genes are also required for regulating TFP-dependent motility to perform phototaxis in Synechocystis sp. PCC6803 (3, 8). Therefore, it is likely that many other TFP-dependent motility systems may also be controlled by similar chemotaxis-like systems (10). The chemotaxis system has been well characterized for its role in directing flagellum-dependent motility. In the flagellum-chemotaxis system, it is known that the chemotaxis signal is relayed through a motor switching complex containing FliM-FliN-FliG (11). These homologs have not been found in gliding bacteria as yet. It would be interesting to elucidate the components that relay the chemotaxis signal to the TFP motility system and to find out how similar chemotaxis systems have developed to interact with two totally different motility systems. It is also worthwhile to point out that the genome sequence of N. gonorrhoeae suggests that such che homologs may not be present. It is unclear whether the TFP-dependent motility in N. gonorrhoeae is being controlled by a chemotaxis-like system.
TFP-DEPENDENT MOTILITY AND ITS ROLE IN PATHOGENESIS
The recent advances in understanding TFP at the molecular level and their physiological functions will help us to further understand the role of TFP in bacterial pathogenesis. It has recently been shown that TFP are an essential component for bacterial biofilm formation (such as in P. aeruginosa) (21). Without TFP, bacterial cells are still able to attach to solid surfaces but fail to build up multicell layers of the biofilm structure (21). Now, considering that TFP are motility apparatuses that can pull cells forward or upward and that the action of TFP can be directed by a chemotaxis-like system, it becomes relatively easy to understand how TFP may be involved in forming sophisticated biofilm structures. Furthermore, chemotaxis-guided, TFP-dependent motility may enable the bacteria to seek and locate appropriate target cells or find a weak area within tissue for effective penetration and invasion. This review has focused on the motility of TFP. However, TFP retraction and its involvement in pathogenesis may be beyond the twitching-gliding motility. For example, TFP is required for DNA uptake, phage infection, and adherence pattern formation and has some interaction with the type III secretion system (6, 9, 19, 25). These aspects, together with TFP-dependent motility, go beyond the traditional sense of pilus, inspire new ideas, and open new fields for future exciting studies of pili.
Acknowledgments
We thank S. Hunt Gerardo for careful editing of the manuscript.
This work was supported by NIH grant GM54666 to W. Shi.
Editor: D. A. Portnoy
REFERENCES
- 1.Alm, R. A., and J. S. Mattick. 1997. Genes involved in the biogenesis and function of type-4 fimbriae in Pseudomonas aeruginosa. Gene 192: 89–98. [DOI] [PubMed] [Google Scholar]
- 2.Bhaya, D., N. R. Bianco, D. Bryant, and A. Grossman. 2000. Type IV pilus biogenesis and motility in the cyanobacterium Synechocystis sp. PCC6803. Mol. Microbiol. 37: 941–951. [DOI] [PubMed] [Google Scholar]
- 3.Bhaya, D., A. Takahashi, and A. R. Grossman. 2001. Light regulation of type IV pilus-dependent motility by chemosensor-like elements in Synechocystis PCC6803. Proc. Natl. Acad. Sci. USA 98: 7540–7545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bieber, D., S. W. Ramer, C. Y. Wu, W. J. Murray, T. Tobe, R. Fernandez, and G. K. Schoolnik. 1998. Type IV pili, transient bacterial aggregates, and virulence of enteropathogenic Escherichia coli. Science 280: 2114–2118. [DOI] [PubMed] [Google Scholar]
- 5.Bitter, W., M. Koster, M. Latijnhouwers, H. de Cock, and J. Tommassen. 1998. Formation of oligomeric rings by XcpQ and PilQ, which are involved in protein transport across the outer membrane of Pseudomonas aeruginosa. Mol. Microbiol. 27: 209–219. [DOI] [PubMed] [Google Scholar]
- 6.Bradley, D. E. 1972. Shortening of Pseudomonas aeruginosa pili after RNA-phage adsorption. J. Gen. Microbiol. 72: 303–319. [DOI] [PubMed] [Google Scholar]
- 7.Burchard, R. P. 1981. Gliding motility of prokaryotes: ultrastructure, physiology, and genetics. Annu. Rev. Microbiol. 35: 497–529. [DOI] [PubMed] [Google Scholar]
- 8.Chung, Y. H., M. S. Cho, Y. J. Moon, J. S. Choi, Y. C. Yoo, Y. I. Park, K. M. Lee, K. W. Kang, and Y. M. Park. 2001. ctr1, a gene involved in a signal transduction pathway of the gliding motility in the cyanobacterium Synechocystis sp. PCC 6803. FEBS Lett. 492: 33–38. [DOI] [PubMed] [Google Scholar]
- 9.Comolli, J. C., A. R. Hauser, L. Waite, C. B. Whitchurch, J. S. Mattick, and J. N. Engle. 1999. Pseudomomas aeruginosa gene products PilT and PilU are required for cytotoxicity in vitro and virulence in a mouse model of acute pneumonia. Infect. Immun. 67: 3625–3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Darzins, A. 1994. Characterization of a Pseudomonas aeruginosa gene cluster involved in pilus biosynthesis and twitching motility: sequence similarity to the chemotaxis proteins of enterics and the gliding bacterium Myxococcus xanthus. Mol. Microbiol. 11: 137–153. [DOI] [PubMed] [Google Scholar]
- 11.DeRosier, D. J. 1998. The turn of the screw: the bacterial flagellar motor. Cell 93: 17–20. [DOI] [PubMed] [Google Scholar]
- 12.Ehlers, K. M., A. D. Samuel, H. C. Berg, and R. Montgomery. 1996. Do cyanobacteria swim using traveling surface waves? Proc. Natl. Acad. Sci. USA 93: 8340–8343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fernandez, L. A., and J. Berenguer. 2000. Secretion and assembly of regular surface structures in gram-negative bacteria. FEMS Microbiol. Rev. 24: 21–44. [DOI] [PubMed] [Google Scholar]
- 14.Henrichsen, J. 1975. The occurrence of twitching motility among gram-negative bacteria. Acta Pathol. Microbiol. Scand. Sect. B 83: 171–178. [DOI] [PubMed] [Google Scholar]
- 15.Henrichsen, J. 1983. Twitching motility. Annu. Rev. Microbiol. 37: 81–93. [DOI] [PubMed] [Google Scholar]
- 16.Hultgren, S. J., C. H. Jones, and S. Normark. 1996. Bacterial adhesins and their assembly, p. 2730–2756. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Echerichia coli and Salmonella, 2nd ed., vol. 2 ASM Press, Washington, D.C.
- 17.Kaiser, D. 1979. Social gliding is correlated with the presence of pili in Myxococcus xanthus. Proc. Natl. Acad. Sci. USA 76: 5952–5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu, H. M., S. T. Motley, and S. Lory. 1997. Interactions of the components of the general secretion pathway: role of Pseudomonas aeruginosa type IV pilin subunits in complex formation and extracellular protein secretion. Mol. Microbiol. 25: 247–259. [DOI] [PubMed] [Google Scholar]
- 19.Merz, A. J., C. A. Enns, and M. So. 1999. Type IV pili of pathogenic neisseriae elicit cortical plaque formation in epithelial cells. Mol. Microbiol. 32: 1316–1332. [DOI] [PubMed] [Google Scholar]
- 20.Merz, A. J., M. So, and M. P. Sheetz. 2000. Pilus retraction powers bacterial twitching motility. Nature 407: 98–102. [DOI] [PubMed] [Google Scholar]
- 21.O’Toole, G. A., and R. Kolter. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30: 295–304. [DOI] [PubMed] [Google Scholar]
- 22.Pate, J. L. 1985. Gliding motility in Cytophaga. Microbiol. Sci. 2: 289–290, 293–295. [PubMed] [Google Scholar]
- 23.Pujol, C., E. Eugene, M. Marceau, and X. Nassif. 1999. The meningococcal PilT protein is required for induction of intimate attachment to epithelial cells following pilus-mediated adhesion. Proc. Natl. Acad. Sci. USA 96: 4017–4022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Russel, M. 1998. Macromolecular assembly and secretion across the bacterial cell envelope: type II protein secretion systems. J. Mol. Biol. 279: 485–499. [DOI] [PubMed] [Google Scholar]
- 25.Sauer, F. G., M. A. Mulvey, J. D. Schilling, J. J. Martinez, and S. J. Hultgren. 2000. Bacterial pili: molecular mechanisms of pathogenesis. Curr. Opin. Microbiol. 3: 65–72. [DOI] [PubMed] [Google Scholar]
- 26.Shi, W., T. Kohler, and D. R. Zusman. 1993. Chemotaxis plays a role in the social behaviour of Myxococcus xanthus. Mol. Microbiol. 9: 601–611. [DOI] [PubMed] [Google Scholar]
- 27.Shi, W., F. K. Ngok, and D. R. Zusman. 1996. Cell density regulates cellular reversal frequency in Myxococcus xanthus. Proc. Natl. Acad. Sci. USA 93: 4142–4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi, W., and D. R. Zusman. 1995. The frz signal transduction system controls multicellular behavior in Myxococcus xanthus, p. 419–430. In J. A. Hoch, and T. Silhavy (ed.), Two-component signal transduction. American Society for Microbiology, Washington, D.C.
- 29.Skerker, J. M., and H. C. Berg. 2001. Direct observation of extension and retraction of type IV pili. Prot. Natl. Acad. Sci. USA 98: 6901–6904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Strom, M. S., and S. Lory. 1993. Structure-function and biogenesis of the type IV pili. Annu. Rev. Microbiol. 47: 565–596. [DOI] [PubMed] [Google Scholar]
- 31.Sun, H., D. R. Zusman, and W. Shi. 2000. Type IV pilus of Myxococcus xanthus is a motility apparatus controlled by the frz chemosensory system. Curr. Biol. 10: 1143–1146. [DOI] [PubMed] [Google Scholar]
- 32.Tonjum, T., and M. Koomey. 1997. The pilus colonization factor of pathogenic neisserial species: organelle biogenesis and structure/function relationships–a review. Gene 192: 155–163. [DOI] [PubMed] [Google Scholar]
- 33.Wall, D., and D. Kaiser. 1999. Type IV pili and cell motility. Mol. Microbiol. 32: 1–10. [DOI] [PubMed] [Google Scholar]
- 34.Wolfgang, M., P. Lauer, H. S. Park, L. Brossay, J. Hebert, and M. Koomey. 1998. PilT mutations lead to simultaneous defects in competence for natural transformation and twitching motility in piliated Neisseria gonorrhoeae. Mol. Microbiol. 29: 321–330. [DOI] [PubMed] [Google Scholar]
- 35.Wolfgang, M., H. S. Park, S. F. Hayes, J. P. van Putten, and M. Koomey. 1998. Suppression of an absolute defect in type IV pilus biogenesis by loss-of-function mutations in pilT, a twitching motility gene in Neisseria gonorrhoeae. Proc. Natl. Acad. Sci. USA 95: 14973–14978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wolfgang, M., J. P. van Putten, S. F. Hayes, D. Dorward, and M. Koomey. 2000. Components and dynamics of fiber formation define a ubiquitous biogenesis pathway for bacterial pili. Embo J. 19: 6408–6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu, S. S., and D. Kaiser. 1995. Genetic and functional evidence that type IV pili are required for social gliding motility in Myxococcus xanthus. Mol. Microbiol. 18: 547–558. [DOI] [PubMed] [Google Scholar]
- 38.Wu, S. S., J. Wu, and D. Kaiser. 1997. The Myxococcus xanthus pilT locus is required for social gliding motility although pili are still produced. Mol. Microbiol. 23: 109–121. [DOI] [PubMed] [Google Scholar]