Abstract
Chloroplasts of maize (Zea mays) leaves differentiate into specific bundle sheath (BS) and mesophyll (M) types to accommodate C4 photosynthesis. Consequences for other plastid functions are not well understood but are addressed here through a quantitative comparative proteome analysis of purified M and BS chloroplast stroma. Three independent techniques were used, including cleavable stable isotope coded affinity tags. Enzymes involved in lipid biosynthesis, nitrogen import, and tetrapyrrole and isoprenoid biosynthesis are preferentially located in the M chloroplasts. By contrast, enzymes involved in starch synthesis and sulfur import preferentially accumulate in BS chloroplasts. The different soluble antioxidative systems, in particular peroxiredoxins, accumulate at higher levels in M chloroplasts. We also observed differential accumulation of proteins involved in expression of plastid-encoded proteins (e.g., EF-Tu, EF-G, and mRNA binding proteins) and thylakoid formation (VIPP1), whereas others were equally distributed. Enzymes related to the C4 shuttle, the carboxylation and regeneration phase of the Calvin cycle, and several regulators (e.g., CP12) distributed as expected. However, enzymes involved in triose phosphate reduction and triose phosphate isomerase are primarily located in the M chloroplasts, indicating that the M-localized triose phosphate shuttle should be viewed as part of the BS-localized Calvin cycle, rather than a parallel pathway.
INTRODUCTION
Leaves in C4 plants such as maize (Zea mays) form a classical Kranz leaf anatomy during their development (Edwards and Walker, 1983; Nelson and Langdale, 1992). In this Kranz anatomy, each vein is surrounded by a ring of bundle sheath (BS) cells, followed by one or more concentric files of mesophyll (M) cells. BS cells have thick cell walls and contain centrifugally arranged chloroplasts with large starch granules and unstacked thylakoid membranes, whereas the M cells contain randomly arranged chloroplasts with stacked thylakoids and little or no starch (Edwards and Walker, 1983). Each cell type accumulates a distinct set of C4 photosynthetic enzymes (reviewed in Langdale, 1998; Sheen, 1999).
Numerous studies have focused on the analysis of BS and M cell differentiation and the relationship to formation of the Kranz anatomy (Edwards et al., 2004). Transcript accumulation studies in BS and M cells using in situ hybridization have been successful in determining spatial patterns and developmental gradients for several specific genes in maize (Furumoto et al., 2000) and in sorghum (Sorghum bicolor; Wyrich et al., 1998). Signals required for plastid-specific gene expression of C4 enzymes involve promoter sequences and other DNA regulatory elements (Sheen, 1999). Collectively, these and other studies show that M cells in maize leaves develop in a C3 pattern by default and in a C4 pattern through the influence of closely neighboring veins. Specialization of BS cells seems to be a combination of their procambial lineage and their vein adjacent position (Smith et al., 1996; Jankovsky et al., 2001). The golden 2 nuclear transcription factor seems to play an important role to promote BS chloroplast development and plastid-specific protein accumulation (Cribb et al., 2001). Several M/BS differentiation mutants were analyzed, such as bundle sheath defective1 (bsd1) (Langdale and Kidner, 1994; Hall et al., 1998) and bsd2 (Roth et al., 1996; Brutnell et al., 1999).
The fully differentiated BS and M chloroplasts each accumulate a distinct set of photosynthetic enzymes and proteins that enables them to cooperate in carbon fixation (Hatch and Osmond, 1976; Edwards and Walker, 1983). For instance, ribulose-1,5-bis-phosphate carboxylase/oxygenase (Rubisco) (RBCL and RBCS) accumulates exclusively in BS chloroplasts, whereas pyruvate phosphate dikinase (PPDK) and photosystem II accumulate in M chloroplasts. Differential protein accumulation of several other chloroplast proteins has been determined using chloroplast fractionation techniques, in combination with protein gel blot analysis, in situ immunolocalization, and green fluorescent protein fusions (reviewed in Edwards et al., 2001). In addition, several BS- and M-specific enzymatic activities were determined using nonaqueous fractionation techniques and pulse labeling techniques (Slack et al., 1969). Collectively, these biochemical studies showed that M cells carry out linear photosynthetic electron transport that produces the ATP and NADPH needed for autotrophic growth, to import pyruvate from the BS cells to generate phosphoenolpyruvate, and to convert the C4 acid oxaloacetate into the C4 acid malate. In complementary fashion, BS cells import and decarboxylate malate into pyruvate, using the released NADPH and CO2 to generate reduced carbohydrates in the Calvin cycle and convert the surplus of carbohydrates into starch. Additional NADPH is imported from the M cells via the triosphosphate shuttle. Differentiation of many other chloroplast functions (reviewed in Neuhaus and Emes, 2000), such as synthesis of fatty acids, nucleotides, hormones, and amino acids, is largely unknown in C4 plants.
The high mass accuracy, sensitivity, and high-throughput possibilities of mass spectrometry (MS), together with the availability of genomes, now allow the rapid identification of large sets of proteins (Aebersold and Mann, 2003; Ferguson and Smith, 2003; Lin et al., 2003). Proteomes of different organelles, membranes, and tissues of Arabidopsis thaliana and Medicago truncatula have been analyzed to various degrees, providing a good starting point for many functional studies (e.g., Watson et al., 2003; Friso et al., 2004; Heazlewood et al., 2004; Nuhse et al., 2004; Peltier et al., 2004a; Shimaoka et al., 2004). The large EST assembly of maize and recent efforts in sequencing of the maize genome (Gomez et al., 2002; Palmer et al., 2003) now facilitate meaningful studies on the maize proteome (Porubleva et al., 2001; Hochholdinger et al., 2004; Lonosky et al., 2004).
Although cataloguing proteomes is useful, in the context of BS and M chloroplast differentiation, it is most important to compare protein accumulation in a quantitative manner. Initially, strategies based on two-dimensional gel electrophoresis (2-DE) were the main tool for comparative protein analysis. Examples of comparative 2-DE–based analysis in plants are protein expression profiling during seed filling and germination in Arabidopsis, M. trunculata, soybean (Glycine max), and pea (Pisum sativum; Gallardo et al., 2001, 2003; Schiltz et al., 2004; Hajduch et al., 2005). Very recently, several non-gel-based techniques have been developed that are based on differential labeling of proteomes with stable isotopes, followed by quantification in the mass spectrometer (Goshe and Smith, 2003; Ong et al., 2003). These non-gel-based methods allow better quantification and in principle much higher throughput analysis. In particular, the cleavable isotope coded affinity tag technique (cICAT) has been successfully employed for comparative proteome analysis (Hansen et al., 2003; Li et al., 2003; Tao and Aebersold, 2003; Conrads et al., 2004). However, these novel stable isotope techniques have not yet been applied in the analysis of plant proteomes.
We present here a systematic overview and a quantitative comparison of the soluble stromal proteomes of BS and M chloroplasts. The availability of the EST assembly and partial genome sequence allowed identification of specific members of protein families in many cases. This is important since different homologs within a protein family might have different functions and expression patterns, as will be shown in this study. The soluble, stromal proteomes of purified BS and M chloroplasts from young maize leaves were compared by a combination of 2-DE and image analysis, followed by identification of proteins by MS. To improve dynamic resolution, accuracy, and significance, we also used two non-gel-based techniques: a differential labeling with cICAT and a comparative analysis of unlabeled BS and M stromal proteomes by liquid chromatography electrospray mass spectrometry (LC-ESI-MS). In total, ∼400 accessions in the EST unigene assembly were identified, providing an excellent overview of the stromal chloroplast proteomes in maize. Differential expression was determined for 125 chloroplast proteins, providing numerous novel insights in BS and M functional differentiation. These data are available via our Plastid Proteome Database (PPDB) at http://ppdb.tc.cornell.edu/ and linked to homologous plastid proteins in Arabidopsis and rice (Oryza sativa).
RESULTS
Purification of BS and M Chloroplasts from Leaf Tips of Young Leaves
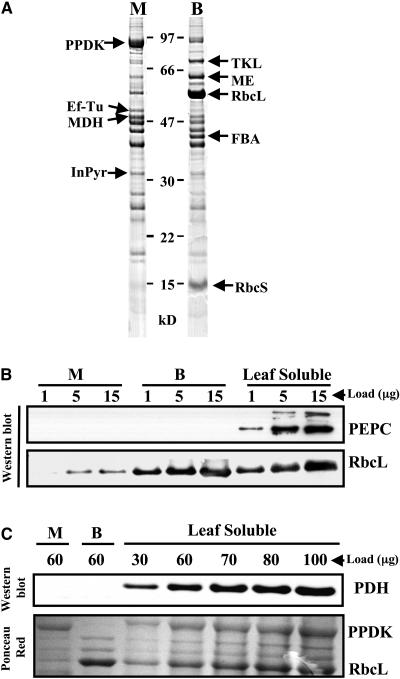
The objective of this study was to compare the stromal proteomes of M and BS chloroplasts that had completed their differentiation process. Therefore, WT-T43 maize plants were grown for 14 d in the growth chamber until the 4th leaf was emerging. The top 4-cm sections of the 2nd and 3rd leaves were harvested ∼2 h after the onset of the light period. We used a mechanical procedure for purification of M and BS chloroplasts, following suggestions from A. Barkan and R. Bassi and protocols adapted from Edwards and Black (1971) and Kannangara et al. (1977). This procedure is based on the much higher resistance of the BS cell wall to mechanical grinding as compared with the M cell wall. After BS and M chloroplast purification, both chloroplast types were broken by lysis and the membrane fractions removed by ultracentrifugation. The purity of M and BS chloroplast fractions was determined from stained SDS-PAGE gels using the abundant marker proteins Rubisco (RbcL at 54 kD) and PPDK (at 97 kD) for BS and M chloroplasts, respectively (Figure 1A). The one-dimensional gel electrophoresis (1-DE) analysis of BS and M purified stroma also clearly visualized additional BS/M-specific proteins, such as fructose biphosphate aldolase (FBA) and RBCS. This showed that our preparations were of high quality. For the analysis in this article, nine biologically independent preparations (pairs of BS and M) were used with cross-contamination levels between 5 and 15% but without any significant nonchloroplast contaminations. The cross-contamination of the BS chloroplasts by the M fraction was lower than we calculated, since it was shown that 10% of a light-independent PPDK form can accumulate in BS cells (Aoyagi and Nakamoto, 1985; Sheen and Bogorad, 1987a, 1987b). We deliberately did not use other types of BS/M preparations involving long enzymatic digestions of cell walls, likely resulting in unwanted induction of stress proteins.
Figure 1.
1-DE Gel Analysis of Chloroplast Stroma.
(A) 1-DE SDS-PAGE of stromal proteomes from isolated M and BS chloroplasts. The purity of each preparation is determined by the abundance of known marker proteins: PPDK for M and Rubisco (RbcL and RbcS) and NADP-malic enzyme (ME) for BS. Additional marker proteins are transketolase (TKL), translation elongation factor Tu (Ef-Tu), malate dehydrogenase (MDH), fructose biphosphate aldolase (FBA), and inorganic pyrophosphatase (InPyr).
(B) Protein gel blot analysis (Western) of BS and M chloroplast stroma to determine contamination levels with cytosolic phosphoenol pyruvate carboxylase (PEPC) and BS-localized Rubisco large subunit (RbcL). Gels were loaded with titrations (1, 5, and 15 μg of protein) of BS stroma, M stroma, and total soluble leaf proteins.
(C) Protein gel blot analysis (Western) of BS and M chloroplast stroma to determine contamination levels with mitochondrial pyruvate dehydrogenase (PDH). Gels were loaded with 60 μg of BS stroma and M stroma and a titration of total soluble leaf protein (30, 60, 70, 80, and 100 μg of protein). A corresponding stained (Ponceau Red) blot is shown as control for gel loading and to demonstrate the abundance of BS chloroplast RbcL and M chloroplast-localized PPDK.
To verify the contamination from non-chloroplast-localized proteins in the BS and M preparations, a protein gel blot analysis was performed using specific antisera against abundant proteins from the cytosol (phosphoenol pyruvate carboxylase [PEPC]; Figure 1B) and mitochondria (pyruvate dehydrogenase; Figure 1C). Gels were loaded with protein titrations of M stroma, BS stroma, and total soluble leaf proteome. As is apparent from the protein gel blot analyses, cross-contaminations of cytosolic and mitochondrial proteins were undetectable. The protein gel blot using antiserum against RBCL confirms that the M stroma contained ∼5% BS stroma (Figure 1B). A corresponding stained protein profile (Ponceau Red on membrane) of these protein samples is shown as control for protein loading and to demonstrate the abundance of RBCL and PPDK in the maize leaf proteome (Figure 1C).
Overview and Outline of the Comparative Proteome Analysis of BS and M Chloroplast Stroma
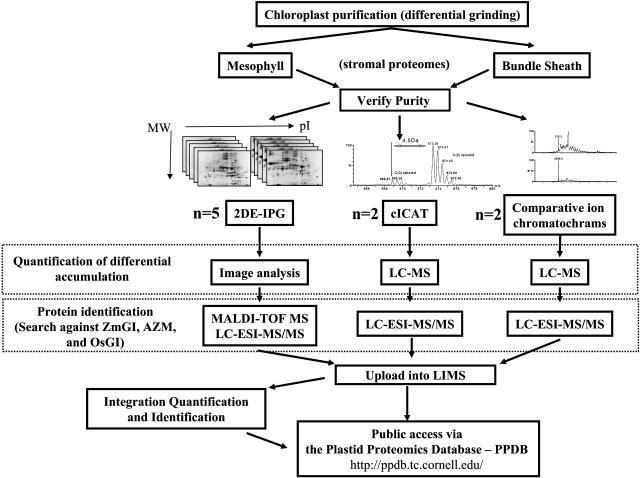
Quantitative comparison of BS and M chloroplast proteomes is challenging because (1) obtaining separated BS and M intact chloroplasts is not trivial, (2) the maize genome has not been fully sequenced, (3) the maize genome is replete with chromosomal duplications and repetitive DNA, and (4) the stromal chloroplast proteomes are dominated by a small set of proteins involved in primary carbon metabolism. Therefore, we used three complementary proteomics techniques to overcome the genome/proteome complexity, to maximize the accuracy of protein quantifications, and to maximize the dynamic resolution. In total, nine independent BS/M chloroplast preparations were analyzed. The three techniques involved comparative 2-DE gels (n = 5) and two non-gel-based comparative proteomics techniques using either cICAT (n = 2) or parallel LC-MS runs of unlabeled M and BS stromal proteomes (n = 2) (assigned in this study as parallel LC-MS), as outlined in Figure 2. Quantification of proteins was done by extensive image analysis in case of the 2-DE gel analysis and from integrated peak areas (extracted single ion chromatograms) in case of the non-gel-based methods.
Figure 2.
Schematic Overview of the Quantitative Comparative Analysis of Purified BS and M Chloroplast Stromal Proteomes.
BS and M chloroplasts were purified and their cross-contamination determined from 1-DE gel analysis. Subsequently, three complementary proteomics techniques were used to overcome the genome/proteome complexity, to maximize the accuracy of protein quantifications, and to maximize the dynamic resolution. In total nine independent BS/M chloroplast preparations were analyzed. The three techniques involved comparative 2-DE gels (n = 5) and two non-gel-based comparative proteomics techniques using either cICAT (n = 2) or parallel LC-MS runs of unlabeled M and BS stromal proteomes (n = 2). Quantification of proteins was done by extensive image analysis in case of the 2-DE gel analysis and from integrated peak areas (extracted single ion chromatograms) in case of the non-gel-based methods. All mass spectral data were searched against ZmGI, AZM, and OsGI. All verified protein identities are available via the PPDB. Since searching of the MS data against ZmGI gave the highest success rate, ZmGI was used for detailed protein quantification. Comparative proteomics information for all quantified proteins is available via PPDB as extractable tables as well as on the report page for each accession. This includes the peptide sequences with associated BS:M ratios in case of non-gel-based quantifications. Interactive 2-DE gels of M and BS stroma are available in the PPDB.
The MS or tandem MS (MS/MS) spectra were searched against the maize EST assembly from The Institute for Genomic Research (TIGR) (http://www.tigr.org; ZmGI, v1.4) and maize genome assemblies (AZM 4.0) as well as by homology-based search against the annotated rice genome (TIGR Rice Genome version 2).
Comparative BS and M Stromal Proteome Analysis by 2-DE
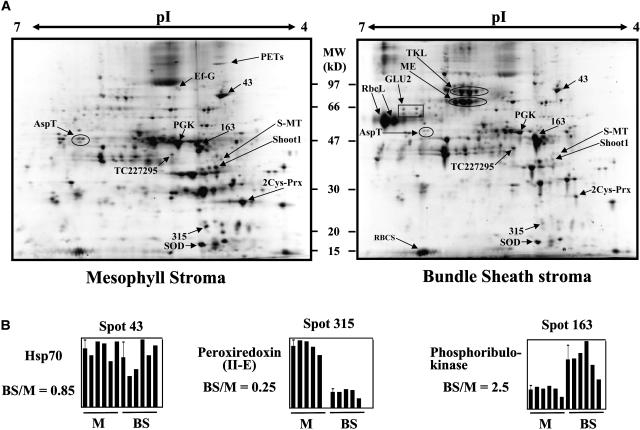
Representative 2-DE gels of BS and M stroma are shown in Figure 3A. Quantitative data are indicated for a few selected spots as an example of how quantification data were processed (Figure 3B). A complete listing for all quantified spots with bar diagrams for each spot can be found in Supplemental Table 2 online. Corresponding interactive gels with protein spot identities that were quantified are available through PPDB.
Figure 3.
2-DE Gels from BS and M Chloroplast Stroma and Examples of Quantifications.
(A) Purified BS and M stromal proteomes were first separated based on isoelectric point on immobilized pH gradient (IPG) strips, with a linear pH gradient from 4 to 7 (150 μg of protein/strip). The focused IPG strips were denatured, and proteins were separated based on mass by SDS-PAGE. The resulting 2-DE gels were stained with the fluorescent dye Sypro-Ruby, images were acquired with exposure times that minimized saturation (to ensure accurate quantification), and spots were detected, quantified, and normalized against the total spot volume. Proteins in the spots were identified by peptide mass fingerprinting using MALDI-TOF MS and/or by online nano-LC-ESI-MS/MS. The mass spectral data were searched against the maize EST assembly from TIGR (ZmGI v4.0) and maize genome assemblies from TIGR (AZM v1.0) as well as by homology-based search against the predicted rice proteome TIGR (OsGI v2.0). Spots identities are indicated for β-d-glucosidase (GLU2), Asp aminotransferase (AspT), unknown protein (TC227295), S-malonyltransferase (S-MT), phosphoglycerate kinase (PGK), unknown protein (Shoot1), 2-Cys peroxiredoxin (2Cys-Prx), superoxide dismutase (SOD), translation elongation factor G (Ef-G), polyprotein of Ef-Ts (PETs), transketolase (TKL), NADP-malic enzyme (ME), Rubisco large subunit (RbcL), and Rubisco small (RBCS) subunit.
(B) Examples of the quantifications of spots 43, 163, and 315. Five complete biological replicate experiments were performed with five pairwise comparisons of BS and M stromal chloroplast proteomes. The average BS:M ratio is indicated. Interactive gels with associated information can be found in PPDB, and all 2-DE gel data are available in Supplemental Table 2 online.
We identified 221 proteins in ZmGI, 184 in AZM, and 147 potential rice homologs from OsGI (see Supplemental Table 1 online). Pairwise BLAST alignment between the three data sets showed that, at a cutoff of E−50, most AZM and OsGI identifications were redundant to the ZmGI data set (data not shown). Therefore, we decided to use the ZmGI identifiers for relative quantifications and further analysis. The differential accumulation ratio in BS and M chloroplasts for each protein was determined for 152 spots (using the strict criteria explained in Methods). Twenty-eight spots of those 152 were only found in either BS or M proteome, and they were assigned a ratio of 10 (BS) or 0.1 (M). These 152 spots corresponded to 106 ZmGI accessions. Within those 106, it appeared that there was some redundancy or closely related gene products that could not easily be distinguished despite our experience with homolog identification in Arabidopsis (e.g., Friso et al., 2004; Peltier et al., 2004a) (see Supplemental Table 2 online).
Non-Gel-Based Quantification Using cICAT and Parallel Ion Chromatograms
The cICAT experiments identified 305 proteins (ZmGI) in BS and/or M chloroplasts (see Supplemental Table 1 online), of which 59 proteins could be quantified (see Supplemental Table 3 online). In total, 100 proteins (ZmGI) were identified with parallel LC-MS, with 69 proteins in BS stroma and 75 proteins in M stroma (see Supplemental Table 1 online). Forty-three of those proteins were identified in both plastid types and expression ratios could be quantified, while 19 proteins were only identified in BS and 31 only in M. All peptide sequences with BS:M ratios from both approaches can be found in Supplemental Table 3 online and are also available via PPDB, as extractable tables, as well as on the protein report page for each accession.
Analysis of the Collective Data Set: Identification, Quantification, and Function
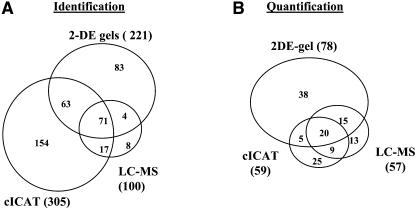
The data from the three methods described above were merged. Collectively, 400 ZmGI accessions were identified from searches against ZmGI, and a Venn Diagram for identified proteins by the three methods is shown in Figure 4A. Clearly, the methods were complementary in their identification success rates, with the cICAT experiments identifying most accessions (Table 1). In total, 327 proteins were identified by searching against the maize genome sequences (AZM), and 277 by homology-based searches against the predicted rice proteome (OsGI) (Table 1). A complete listing of all identified ZmGI, AZM, and OsGI accessions can be found in Supplemental Table 1 online.
Figure 4.
Identification and Quantification Success and Overlap of the Three Methods.
The data from the three methods (2-DE gels, cICAT, and parallel LC-MS) used in this study to analyze the BS and M stromal proteomes were compared in a Venn diagram.
(A) Collectively, 400 ZmGI accessions were identified from searches against ZmGI; aVenn diagram for identified proteins by the three methods is shown. Clearly, the methods were complementary in their identification success rates, with the cICAT experiments identifying most accessions and the parallel LC-MS experiments identifying 100 accessions.
(B) Collectively, we were able to assign differential BS:M chloroplast expression ratios for 125 ZmGI accessions. The expression ratios of 20 proteins were quantified by three methods and 29 proteins by two of the methods, while the remaining 76 proteins were quantified by only one method, in most cases 2-DE gels. Sixty-two percent of quantifications were obtained from 2-DE gel analysis, 47% from cICAT, and 46% by LC-unlabeled quantifications.
Table 1.
Overview of Identified Maize Accessions in Purified BS and M Chloroplast Stroma Identified by Mass Spectometry from the Three Complementary Methods
| 2-DE Gels
|
cICAT
|
LC-MS
|
||||||
|---|---|---|---|---|---|---|---|---|
| Database | BS | M | BS + M | BS + M | BS | M | BS + M | Total Nonredundanta |
| ZmGIb | 128 | 196 | 221 | 305 | 69 | 75 | 100 | 400 |
| AZMc | 108 | 161 | 184 | 238 | 58 | 63 | 76 | 327 |
| OsGId | 100 | 120 | 147 | 221 | 24 | 18 | 30 | 277 |
Total nonredundant identified accessions from the three different comparative proteomics methods.
Indentification by searching with the mass spectral data against the maize EST unigene collection (ZmGI v 4.0) assembled by TIGR.
Indentification by searching with the mass spectral data against the genome sequences (AZM v1.0) assembled by TIGR.
Indentification by searching with the mass spectral data against the predicted rice proteome (OsGI v 2.0) assembled by TIGR.
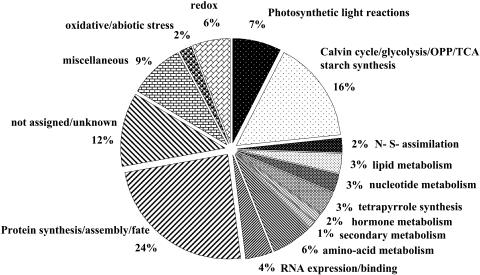
To determine the functions of the identified stromal proteomes, we used in-house BLAST alignments of the identified ZmGI accessions to the predicted rice and Arabidopsis proteomes. Proteins were then functionally classified using the nonredundant MapMan functional classification system (see http://gabi.rzpd.de/projects/MapMan/) developed for Arabidopsis (Thimm et al., 2004) as a basis. BLAST alignments of proteins identified in maize resulted in a set of Arabidopsis homologs, which we classified into 25 MapManBins covering a wide range of pathways and functions. This includes primary carbon metabolism and photosynthesis (Calvin cyle, oxidative phosphopentose pathway [OPPP], glycolysis, and starch metabolism) totaling 16%, amino acid metabolism (6%), lipid metabolism (3%), and nitrogen and sulfur assimilation (2%). Proteins involved in protein synthesis, folding, and degradation represented 24% of all identified proteins (Figure 5). BS- and M-localized enzymes of the C4 shuttle and the photosynthetic electron transport chain were also found. Several enzymes in tretrapyrrole synthesis and vitamin biosynthesis were identified. Of the identified proteins, 21% were classified in a miscellaneous category with a majority of unknown proteins. Thus, the analysis covered a wide range of plastid functions (Figure 5).
Figure 5.
Functional Summary of the 400 Identified Proteins.
To determine the functions of the identified stromal proteomes, we used in-house BLAST alignments of the identified ZmGI accessions to the predicted rice and Arabidopsis proteomes. Proteins were then functionally classified using the nonredundant MapMan functional classification system (see http://gabi.rzpd.de/projects/MapMan/) developed for Arabidopsis (Thimm et al., 2004) as a basis. BLAST alignments of proteins identified in maize resulted in a set of Arabidopsis homologs, which we classified into 25 MapMan Bins covering a wide range of pathways and functions.
Important for judging nonchloroplast contaminations was the fact that PEPC, an extremely abundant marker of maize cytoplasm, was identified only once (in a cICAT experiment) (see Supplemental Table 1 online). PEPC was not identified in the parallel LC-MS experiments nor on the 2-DE gels, showing that cytoplasmic contamination was extremely low. This is in agreement with the protein gel blot analysis (Figure 1B).
Collectively, we were able to assign differential BS:M chloroplast expression ratios for 125 ZmGI accessions (Figure 4B). The expression ratios of 20 proteins were quantified by three methods, and 29 proteins by two methods, while the remaining 76 proteins were quantified by only one method, in most cases 2-DE gels. The expression ratios for these 125 ZmGI accessions are summarized in Table 2 and plotted on a logarithmic scale, grouped by functional class (Figures 6 to 9). Rather than calculating one ratio for each quantified protein, we chose to report all of the values obtained from each method, providing an interval of calculated expression ratios. This gives a better sense of the data set and the biological significance than presenting a single average value for each protein. All detailed quantifications are also presented in Supplemental Table 4 online. Coherent expression patterns were observed for the large majority of identified proteins.
Table 2.
Summary of All Quantifications of Maize Chloroplast BS:M Protein Accumulation Ratios Determined by the Gel-Based and Non-Gel-Based Techniques
| No. for Figures 7 to 9a
|
Total Average BS:M Ratioe
|
BS:M Ratio (Average within a Technique)
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Figureb | Protein Namec | ZmGI Accessiond | 2-DE Gelf | 1-DE Gel cICATg | 2D-LC-cICATh | UN-LC1i | UN-LC2j | MapMan Bink | Arabidopsis Homologuel | cTPm | cTPn | ||
| 1 | 6 | Ferredoxin 2 (Fd2) | TC223586 | 3.5 | 4.10 | 2.90 | 1.1 PS.lightreaction | At1g60950.1 | C | C | |||
| 2 | 6 | Ferredoxin I (Fd1) | TC220059, TC238105 | 0.3 | 0.20 | 0.53 | 0.24 | 1.1 PS.lightreaction | At1g60950.1 | C | C | ||
| 3* | 6 | Ferredoxin reductase (FNR1) | TC219223TC219224TC219226 | 0.2 | 0.33 | 0.14 | 0.29 | 0.16 | 1.1 PS.lightreaction | At5g66190.1 | C | C | |
| 3* | 6 | Ferredoxin reductase (FNR1) | TC219223 | 0.1 | 0.10 | 0.13 | 1.1 PS.lightreaction | At5g66190.1 | C | C | |||
| 4 | 6 | Plastocyanin (PC2) | TC219293, TC219295 | 0.8 | 0.33 | 1.70 | 0.52 | 1.1 PS.lightreaction | At1g76100.1 | C | C | ||
| 5 | 6 | Oxygen evolving enhancer 2 (OEC23) | TC235206, TC235205 | 0.7 | 0.51 | 1.20 | 0.25 | 1.1.1 PS.lightreaction; photosystem II | At1g06680.1 | C | C | ||
| 6 | 6 | Oxygen evolving enhancer protein 1 (OEC33) | TC233304 | 0.1 | 0.04 | 0.26 | 1.1.1 PS.lightreaction; photosystem II | At3g50820.1 | C | C | |||
| 7 | 6 | Oxygen-evolving enhancer protein 3-1 (OEC16-1) | TC219937, TC238011 | 0.3 | 0.50 | 0.13 | 1.1.1 PS.lightreaction; photosystem II | At4g21280.1 | C | C | |||
| 8 | 6 | Photosystem II protein W-like protein (PsbW-like) | TC238911 | 0.7 | 0.71 | 1.1.1 PS.lightreaction; photosystem II | At4g28660.1 | C | C | ||||
| 9 | 6 | Thylakoid lumenal 29.8-kD protein (TL30) | TC226969 | 0.5 | 0.53 | 1.1.1 PS.lightreaction; photosystem II | At1g77090.1 | C | |||||
| 10 | 6 | ATP synthase β-chain (β-CF1) | TC224055 | 0.6 | 0.63 | 1.1.4 PS.lightreaction; ATP synthase | ATCG00480 | c-enc | c-enc | ||||
| 11 | 6 | Phosphoglycolate phosphatase (PGP) | TC229733 | 2.6 | 2.60 | 1.2 PS.photorepiration | At5g36790.1 | C | C | ||||
| 12 | 6 | Fructose-1,6-bisphosphatase (FBP) | TC224643, TC224642, TC247511 | 5.8 | 3.90 | 3.50 | 10.00 | 1.3 PS.calvin cyle | At3g54050.1 | C | C | ||
| 13 | 6 | Fructose-bisphosphate aldolase (FBA) | TC219359, TC219361 | 5.1 | 3.80 | 9.40 | 4.70 | 2.10 | 5.70 | 1.3 PS.calvin cyle | At4g38970.1 | C | C |
| 14 | 6 | Glyceraldehyde 3-phosphate dehydrogenase (GAP-A) | TC219897 | 2.2 | 6.20 | 1.50 | 1.10 | 0.67 | 1.40 | 1.3 PS.calvin cyle | At1g12900.1 | C | C |
| 15 | 6 | Glyceraldehyde-3-phosphate dehydrogenase (GAP-B) | TC234510 | 0.3 | 0.45 | 0.22 | 0.21 | 0.40 | 0.32 | 1.3 PS.calvin cyle | At1g42970.1 | C | C |
| 16* | 6 | Phosphoglycerate kinase (PGK) | TC219624, TC219625, TC219203 | 0.5 | 0.50 | 0.42 | 0.16 | 1.10 | 1.3 PS.calvin cyle | At1g56190.1 | C | C | |
| 16* | 6 | Phosphoglycerate kinase (PGK) | TC219624 | 1.1 | 1.10 | 1.10 | 1.3 PS.calvin cyle | At1g56190.1 | C | C | |||
| 17 | 6 | Phosphoribulokinase (PRK) | TC221089, TC225896 | 6.5 | 2.50 | 16.45 | 3.85 | 3.90 | 6.00 | 1.3 PS.calvin cyle | At1g32060.1 | C | C |
| 18 | 6 | Ribulose-5-phosphate-3-epimerase (RPE) | TC234954 | 8.8 | 10.00 | 6.30 | 10.00 | 1.3 PS.calvin cyle | At5g61410.2 | C | C | ||
| 19 | 6 | Rubisco activase (RCA) | TC223356, TC237072 | 4.4 | 1.85 | 10.10 | 2.50 | 3.30 | 1.3 PS.calvin cyle | At2g39730.1 | C | C | |
| 20 | 6 | Rubisco large subunit (RbcL) | TC233714 | 9.2 | 23.38 | 11.80 | 2.80 | 3.60 | 4.60 | 1.3 PS.calvin cyle | ATCG00490 | c-enc | c-enc |
| 21 | 6 | Rubisco small subunit (RBCS) | TC234038, TC234963 | 8.3 | 5.17 | 24.97 | 3.55 | 1.95 | 5.85 | 1.3 PS.calvin cyle | At1g67090.1 | C | C |
| 22 | 6 | Sedoheptulose-1,7-bisphosphatase (SBP) | TC239473 | 7.4 | 2.30 | 14.10 | 4.30 | 7.65 | 8.40 | 1.3 PS.calvin cyle | At3g55800.1 | C | C |
| 23 | 6 | Transketolase (TKL) | TC235000 | 5.0 | 4.40 | 7.20 | 3.20 | 3.90 | 6.10 | 1.3 PS.calvin cyle | At3g60750.1 | C | C |
| 24 | 6 | Aldose-1-epimerase (AE) | TC237704 | 7.1 | 10.00 | 5.40 | 5.90 | 3.5 minor carbohydrates, starch, OPP, glycolysis | At5g66530.1 | C | C | ||
| 25 | 6 | Ribose-5-phosphate isomerase (RPI) | TC221577 | 2.0 | 2.03 | 7.2 OPP.nonreductive PP | At3g04790.1 | C | C | ||||
| 26 | 6 | Triose phosphate isomerase (TIM) | TC233907, TC233905, TC233906, TC233912 | 0.2 | 0.19 | 0.14 | 0.16 | 0.25 | 0.19 | 4 glycolysis | At2g21170.1 | C | C |
| 27 | 6 | NADP-dependent malic enzyme (ME) | TC234846 | 7.0 | 8.15 | 14.20 | 3.80 | 3.40 | 5.50 | C4BS_8.2.10 TCA/org. transformation.other organic acid transformaitons.malic | At1g79750.1 | C | |
| 28 | 6 | Pyruvate orthophosphate dikinase (PPDK) | TC233444 | 0.2 | 0.37 | 0.15 | 0.19 | 0.09 | 0.29 | C4MS_6 gluconeogenese/glyoxylate cycle | At4g15530.1 | _ | |
| 29 | 6 | Malate dehydrogenase [NADP] (MDH) | TC220999 | 0.2 | 0.39 | 0.06 | 0.16 | 0.13 | 0.19 | C4MS_8.2 TCA/org. transformation.other organic acid transformaitons | At5g58330.1 | C | C |
| 30 | 7 | Acyl carrier protein I (ACP1) | TC223112, TC228772 | 1.3 | 1.30 | 11.1 lipid metabolism.FA synthesis and FA elongation | At4g25050.1 | C | C | ||||
| 31 | 7 | β-Hydroxyacyl-ACP dehydratase (β-ACPD) | TC240700 | 0.1 | 0.10 | 11.1 lipid metabolism.FA synthesis and FA elongation | At2g22230.1 | C | C | ||||
| 32 | 7 | S-malonyltransferase (S-MT) | TC223360 | 0.4 | 0.38 | 11.1 lipid metabolism.FA synthesis and FA elongation | At2g30200.1 | C | M | ||||
| 33 | 7 | Lipid transfer protein 7a2b (LTP) | AI966827 | 0.1 | 0.10 | 11.6 lipid metabolism.lipid transfer proteins etc | At5g59320.1 | S | |||||
| 34 | 7 | Ferredoxin-nitrite reductase (NiR) | TC222347, TC222348 | 0.3 | 0.67 | 0.17 | 0.17 | 0.40 | 0.25 | 12.1 N-metabolism.nitrate metabolism | At2g15620.1 | C | C |
| 35 | 7 | Ferredoxin-dependent Glu synthase (Fd-GOGAT) | TC236410 | 0.2 | 0.16 | 12.2 N-metabolism.ammonia metabolism | At5g04140.2 | C | C | ||||
| 36 | 7 | Gln synthetase (GS2) | TC220868 | 0.7 | 0.67 | 12.2 N-metabolism.ammonia metabolism | At5g35630.1 | C | C | ||||
| 37 | 7 | Asp transaminase (AspT) | TC219944 | 0.2 | 0.30 | 0.09 | 0.18 | 0.40 | 0.18 | 13.1.2.1 amino acid metabolism.central amino acid metabolism.aspartate.synthesis | At4g31990.2 | C | C |
| 38 | 7 | Lactoylglutathione lyase (LGHL) | TC234316 | 1.0 | 1.00 | 13.3.2.2 amino acid metabolism.aspartate family.threonine.degradation | At1g08110.1 | _ | |||||
| 39 | 7 | Ketol-acid reductoisomerase (KR) | TC219885 | 0.5 | 0.53 | 13.4.1.1 amino acid metabolism.alanine-valine-leucine group.valine.synthesis | At3g58610.1 | C | C | ||||
| 40 | 7 | 3-Isopropylmalate dehydrogenase (IMD) | TC220615, TC220616, TC234316 | 0.5 | 0.48 | 13.4.2.1 amino acid metabolism.alanine-valine-leucine group.leucine.synthesis | At1g80560.1 | C | C | ||||
| 41 | 7 | Cys synthase 1 (CS1) | TC235388, TC235390 | 0.6 | 0.63 | 13.5.3.1 amino acid metabolism.serine-glycine-cysteine group.cysteine.synthesis | At2g43750.1 | C | C | ||||
| 42 | 7 | ATP-sulfurylase 2 (ATP-S) | TC235924 | 2.0 | 1.95 | 14 S-assimilation | At1g19920.1 | C | C | ||||
| 43 | 7 | 1-Deoxy-d-xylulose-5-phosphate reductoisomerase (XRI) | TC238092 | 0.1 | 0.10 | 16.1.1 secondary metabolism.isoprenoids.nonmevalonate pathway (steroids) | At5g62790.1 | C | C | ||||
| 44 | 7 | TC227295 (unknown) | TC227295 | 6.5 | 6.45 | 17.2.3 hormone metabolism.auxin.induced-regulated-responsive-activated | At1g23740.1 | C | |||||
| 45 | 7 | Lipoxygenase (LOX1 and LOX2) | AW157962, TC234252, TC237970, TC237971 | 0.2 | 0.10 | 0.14 | 0.18 | 0.22 | 17.7.1 hormone metabolism.jasmonate.synthesis-degradation | At1g55020.1 | _ | ||
| 46 | 7 | δ-Aminolevulinic acid dehydratase (PS) | TC240729 | 0.8 | 0.77 | 19 tetrapyrrole synthesis | At1g69740.1 | C | C | ||||
| 47 | 7 | Glu-1-semialdehyde 2,1-aminomutase (GAM) | TC227768 | 0.4 | 0.40 | 20 tetrapyrrole synthesis | At5g63570.1 | C | C | ||||
| 48 | 7 | Magnesium-chelatase subunit I (ChlI) | TC223168 | 0.2 | 0.19 | 21 tetrapyrrole synthesis | At1g56190.1 | C | C | ||||
| 49 | 7 | ADP-glucose pyrophosphorylase (small) (APS) | TC232071 | 1.7 | 1.70 | 2.1.2 major CHO metabolism.synthesis.starch | At5g48300.1 | C | C | ||||
| 50 | 7 | ADP-glucose pyrophosphorylase (large) (APL) | TC222533, TC242174 | 7.3 | 4.55 | 10.00 | 2.1.2 major CHO metabolism.synthesis.starch | At5g19220.1 | C | C | |||
| 51 | 7 | Starch synthase (StS) | TC236897 | 3.3 | 3.30 | 2.1.2.02 major CHO metabolism.synthesis.starch.starch synthase | At5g24300.1 | C | C | ||||
| 52 | 7 | Unknown pollen signaling protein (TC235467) | TC235467 | 1.1 | 1.10 | 20.1 stress.biotic | At3g14460.1 | _ | C | ||||
| 53 | 7 | Hsp82 (HSP90) | TC221632 | 0.4 | 0.43 | 20.2.1 stress.abiotic.heat | At2g04030.1 | C | C | ||||
| 54 | 7 | Ferredoxin-thioredoxin reductase subunit A (FTR-A) | AZM4_47023 | 0.1 | 0.10 | 21.1 redox.thioredoxin | At5g08410.1 | C | C | ||||
| 55* | 7 | Thioredoxin-M2, -M4, -F (Trx) | TC228810, TC221334, TC220464 | 0.6 | 0.49 | 1.43 | 0.24 | 0.38 | 21.1 redox.thioredoxin | At5g16400.1, At3g15360.1 | C | C | |
| 55* | 7 | Thioredoxin M4-1 (Trx) | TC224103 | 2.4 | 2.40 | 21.1 redox.thioredoxin | At3g15360.1 | C | C | ||||
| 55* | 7 | Thioreodxin M4-2 (Trx) | TC238760 | 0.5 | 0.45 | 21.1 redox.thioredoxin | At3g15360.1 | C | C | ||||
| 56 | 7 | Glutathione-disulfide reductase (GR) | TC227052 | 1.2 | 1.22 | 21.2 redox.ascorbate and glutathione | At3g54660.1 | C | C | ||||
| 57 | 7 | Glutaredoxin (Grx) | TC225992 | 0.3 | 0.29 | 21.4 redox.glutaredoxins | At2g38270.1 | C | C | ||||
| 58 | 7 | 2-Cys peroxiredoxin-like (2CysB-Prx) | TC234346, TC234345 | 0.4 | 0.39 | 0.48 | 0.31 | 0.59 | 21.5 redox.periredoxins | At5g06290.1 | C | C | |
| 59 | 7 | Peroxiredoxine-like (PrxII-E) | TC223042 | 0.4 | 0.20 | 0.38 | 0.71 | 21.5 redox.periredoxins | At3g52960.1 | C | C | ||
| 60 | 7 | Superoxide dismutase [Cu-Zn] (SOD) | TC237182 | 0.6 | 0.77 | 0.67 | 0.42 | 21.6 redox.dismutases and catalases | At2g28190.1 | C | C | ||
| 61 | 8 | Adenylate kinase (ADK) | TC236532, TC236534 | 0.3 | 0.27 | 0.45 | 0.28 | 23.4 nucleotide metabolism.phosphotransfer and pyrophosphatases | At5g47840.1 | C | C | ||
| 62 | 8 | Inorganic pyrophosphatase (InPyr) | TC218860 | 0.5 | 0.71 | 0.36 | 0.56 | 23.4 nucleotide metabolism.phosphotransfer and pyrophosphatases | At5g09650.1 | C | C | ||
| 63 | 8 | Nucleoside diphosphate kinase II (NDKII) | TC238875 | 0.2 | 0.18 | 23.4 nucleotide metabolism.phosphotransfer and pyrophosphatases | At5g63310.1 | C | C | ||||
| 64 | 8 | Thylakoid lumenal 17.4-kD protein (TL17) | TC224315 | 0.2 | 0.18 | 26.15 misc. Pentapeptide repeat (PPR) unknown function | At1g26220.1 | C | |||||
| 65 | 8 | β-d-glucosidase (β-GLU2) | TC220472 | 11.9 | 8.78 | 17.00 | 10.00 | 26.3 misc.gluco-, galacto- and mannosidases | At2g44450.1 | S | |||
| 66 | 8 | Plastid lipid-associated protein (PAP3) | TC237165 | 0.1 | 0.10 | 26.31* misc. fibrillins | At2g35490.1 | C | C | ||||
| 67 | 8 | Glutathione S-transferase (GST10) | TC222544 | 0.5 | 0.47 | 0.45 | 26.9 misc.glutathione S-transferases | At3g03190.1 | _ | ||||
| 68 | 8 | Unknown (SET1 domain protein) (TC224607) | TC224607 | 6.7 | 6.70 | 27.3.69 RNA.regulation of transcription.SET-domain transcriptional regulator family | At1g73100.1 | _ | |||||
| 69 | 8 | mRNA binding protein (Csp41a-like) | CF646019, AZM4_135414 | 1.9 | 1.90 | 28 (Regulation) transcription and translation; unspecified | At3g63140.1 | C | C | ||||
| 70 | 8 | Nucleic acid binding protein (Cp31) | TC235457, TC235458 | 0.5 | 0.51 | 0.56 | 29 (Regulation) transcription and translation; unspecified | At4g24770.1 | C | C | |||
| 71 | 8 | Nucleic acid binding protein (Cp33) | TC236626 | 0.5 | 0.32 | 0.56 | 0.50 | 30 (Regulation) transcription and translation; unspecified | At4g24770.1 | C | C | ||
| 72 | 8 | Plastid-specific ribosomal protein 2 (PSRP2) | TC238798 | 0.3 | 0.29 | 29.2.1 protein.synthesis.chloroplast; plastid ribosomal protein | At3g52150.1 | C | C | ||||
| 73 | 8 | Ribosomal protein L1 (RP-L1) | TC219835 | 1.0 | 1.00 | 29.2.1 protein.synthesis.chloroplast; plastid ribosomal protein | At3g63490.1 | C | C | ||||
| 74 | 8 | Ribosomal protein L12.1 (RP-L12.1) | TC222916 | 0.2 | 0.10 | 0.27 | 0.28 | 29.2.1 protein.synthesis.chloroplast; plastid ribosomal protein | At3g27850.1 | C | C | ||
| 75 | 8 | Ribosomal protein S1 (RP-S1) | TC221841 | 0.1 | 0.10 | 29.2.1 protein.synthesis.chloroplast; plastid ribosomal protein | At5g30510.1 | C | C | ||||
| 76 | 8 | Ribosomal protein S5 (RP-S1) | TC239133 | 0.6 | 0.91 | 0.32 | 29.2.1 protein.synthesis.chloroplast; plastid ribosomal protein | At2g33800.1 | C | C | |||
| 77 | 8 | Ribosomal protein L10 (RP-L10) | TC224551, BM079225 | 0.1 | 0.10 | 29.2.1 protein.synthesis.chloroplast; plastid ribosomal protein | At5g13510.1 | C | C | ||||
| 78 | 8 | Elongation factor P (Ef-P) | TC237934 | 0.9 | 0.91 | 29.2.3 protein.synthesis.initiation | At3g08740.1 | C | C | ||||
| 79 | 8 | Elongation factor Tu (Ef-Tu) | TC220034, TC222181, TC226641, TC222182 | 0.3 | 0.42 | 0.24 | 0.30 | 0.16 | 0.42 | 29.2.4 protein.synthesis.release | At4g02930.1 | C | |
| 80 | 8 | Elongation factor G (Ef-G) | TC222353 | 0.2 | 0.15 | 29.2.4 protein.synthesis.elongation | At1g62750.1 | C | C | ||||
| 81 | 8 | Polyprotein of Ef-Ts (PETs) | TC226754 | 0.1 | 0.14 | 29.2.99 protein.synthesis.misc | At4g29060.1 | C | C | ||||
| 82 | 8 | Ribosome recycling factor (RRF) | TC237618 | 0.8 | 0.83 | 29.2.99 protein.synthesis.misc | At3g63190.1 | C | C | ||||
| 83 | 8 | Membrane-associated 30-kD protein (Vipp1) | TC220737 | 0.1 | 0.10 | 29.3 protein.targeting | At1g65260.1 | C | C | ||||
| 84 | 8 | ClpP protease [ClpP(1?)] | TC245457, TC225197 | 0.4 | 0.59 | 0.30 | 29.5 protein.degradation | ATCG00670 | c-enc | c-enc | |||
| 85 | 8 | Chaperonin 60 (β-Cpn60) | TC219523, TC219522 | 1.0 | 1.02 | 0.91 | 1.10 | 29.6 protein.(un)folding (includes isomerases and chaperones, if not heat induced) | At1g55490.2 | C | C | ||
| 86 | 8 | Chaperonin 20 (Cpn21) | TC233810, TC236117 | 0.9 | 0.92 | 29.6 protein.(un)folding (includes isomerases and chaperones, if not heat induced) | At5g20720.2, At5g20720.1 | C | C | ||||
| 87 | 8 | Chaperonin 60 (α-Cpn60) | TC220350, TC235184, TC236031 | 0.7 | 0.72 | 29.6 protein.(un)folding (includes isomerases and chaperones, if not heat induced) | At2g28000.1 | C | C | ||||
| 88* | 8 | Chaperonin 70 (HSP70) | TC220215, TC235785 | 0.7 | 0.83 | 0.59 | 0.16 | 29.6 protein.(un)folding (includes isomerases and chaperones, if not heat induced) | At5g49910.1, At4g24280.1 | C | C | ||
| 88* | 8 | Chaperonin 70 (HSP70) | TC235785 | 0.2 | 0.16 | 29.6 protein.(un)folding (includes isomerases and chaperones, if not heat induced) | At5g49910.1, At4g24280.1 | C | C | ||||
| 89 | 8 | GrpE (GrpE) | TC222867 | 0.2 | 0.19 | 29.6 protein.(un)folding (includes isomerases and chaperones, if not heat induced) | At5g17710.2 | C | C | ||||
| 90 | 8 | Peptidyl-prolyl cis-trans isomerase (TLP21) | TC228225 | 0.3 | 0.30 | 29.6 protein.(un)folding (includes isomerases and chaperones, if not heat induced) | At5g13120.1 | C | C | ||||
| 91 | 8 | Peptidyl-prolyl cis-trans isomerase (TLP40) | TC239826 | 0.4 | 0.36 | 29.6 protein.(un)folding (includes isomerases and chaperones, if not heat induced) | At3g01480.1 | C | C | ||||
| 92 | 8 | ClpC Hsp100 (ClpC) | TC235372 | 0.4 | 0.40 | 29.6 protein.folding | At5g50920.1 | C | C | ||||
| 93 | 8 | FK506 binding protein 1 (FK506) | TC247336 | 5.1 | 5.10 | 29.8 protein assembly and cofactor ligation | At5g45680.1 | C | |||||
| 94 | 9 | Vacuolar ATP synthase subunit C (VATC) | TC249232 | 0.3 | 0.34 | 34.1 transport.p- and v-ATPases | At1g12840.1 | M | |||||
| 95 | 9 | ABC transporter putative (ABC-T) | TC223951 | 0.1 | 0.10 | 34.16 transport.ABC transporters and multidrug resistance systems | At3g10670.1 | C | C | ||||
| 96 | 9 | CP12 protein precursor (CP12) | TC223290 | 4.2 | 4.20 | 35.1 not assigned. no ontology | At2g47400.1 | C | C | ||||
| 97 | 9 | SHOOT1 protein (Shoot1) | TC238048 | 0.3 | 0.42 | 0.17 | 35.2 not assigned. unknown | At1g55480.1 | C | C | |||
| 98 | 9 | Unknown (TC236586) | TC236586 | 0.3 | 0.31 | 35.2 not assigned. unknown | At2g24020.1 | C | C | ||||
| 99 | 9 | Aldo/keto reductase family protein (TC220484) | TC220484 | 0.3 | 0.30 | 35.2 not assigned. unknown | At2g27680.1 | C | C | ||||
| 100 | 9 | Unknown (TC220929) | TC220929 | 0.0 | 0.04 | 35.2 not assigned. unknown | At4g21210.1 | C | |||||
| 101 | 9 | Fruit protein PKIWI502 (PKIWI502) | TC222257 | 0.4 | 0.32 | 0.30 | 0.71 | 35.2 not assigned. unknown | At1g15140.1 | C | C | ||
| 102 | 9 | Inositol monophosphate family protein (TC238795) | TC238795 | 10.0 | 10.00 | 35.2 not assigned. unknown | At1g31190.1 | C | |||||
| 103 | 9 | Uknown (TC230439) | TC230439 | 0.4 | 0.38 | 35.2 not assigned. unknown | At4g34420.1 | _ | |||||
| 104 | 9 | Y230_ARATH (TC235613) | TC235613 | 0.4 | 0.37 | 0.36 | 35.2 not assigned. unknown | At2g37660.1 | C | C | |||
| 105 | 9 | Uknown (TC220990) | TC220990 | 0.6 | 0.59 | 35.2 not assigned. unknown | At4g15940.1 | M | M | ||||
| 106 | 9 | Unknown (CF032674) | CF032674 | 1.0 | 1.00 | 35.2 not assigned. unknown | None | ||||||
Protein number in Figures 6 to 9.
Corresponding figure.
Assigned protein named based on information from BLAST alignments. Protein for which a differential expression of paralogues (where it was possible to distinguish them) are marked with an asterisk. Protein name abbreviations are indicated in parentheses (see Supplemental Table 4 online).
ZmGI accession identified by MS.
Average BS:M protein accumulation ratio across all experiments.
Average BS:M ratio for ZmGI accession based on quantification from 2-DE gels.
Quantificaton from cICAT labeling combined with 1-DE gel protein separation.
Quantificaton from cICAT labeling combined with online 2D-LC–based separation.
First BS:M accumulation ratio based on parallel LC-MS–based quantifications of unlabeled peptides.
Second experiments with BS:M accumulation ratio based on parallel LC-MS–based quantifications of unlabeled peptides.
Functional assigment based on the MapMan Bin system developed by Thimm et al. (2004).
Best Arabidopsis homologues as judged by BLAST E-value.
Predicted subcellular localization of Arabidopsis homologues using TargetP. C, chloroplast; M, mitochondria; S, signal peptide.
Predicted subcellular localization of Arabidopsis homologues using Predotar. C, chloroplast; M, mitochondria.
ZmGI accessions were quantified by one or more of the following techniques: 2-DE gels, 1-DE SDS-PAGE followed by cICAT (1-DE gel cICAT), cICAT followed by 2D-LC fractionation (2D-LC-cICAT), and 2-DE IPG and parallel LC-MS–based quantifications of unlabeled peptides (UN-LC1 and UN-LC2). For the non-gel-based quantifications, each BS:M ratio was often based on more than one peptide pair. For 2-DE gel-based identification, at least three quantified protein spots on each BS and M contributed to the BS:M ratio. Within each technique, typically more than one observation per accession number was made. The differences in protein accumulation between the M and BS are represented as BS:M ratio. The average BS:M ratio across all experiments is listed. The closest Arabidopsis homologues are indicated followed by their annotation in PPDB and predicted subcellular localization. The asterisks indicate very similar accessions that could not unambiguously be individually quantified were grouped together. In some cases, one of the three quantification methods allowed individual quantification and are marked with an asterisk.
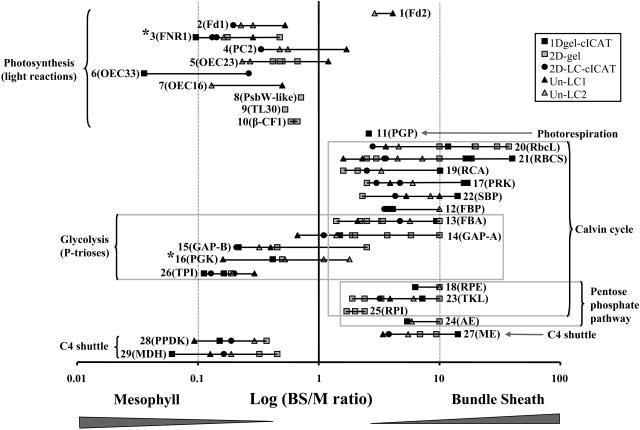
Figure 6.
Overview of BS:M Ratio Determinations by the Three Complementary Methods.
The 125 quantified maize proteins (ZmGI accessions) were divided in Figures 6 to 9 and are grouped by function; here, we show the first group. BS:M protein accumulation ratios obtained for each protein by three different methods (closed squares, 1-DE gel cICAT; circles, 2D-LC-cICAT; open squares, 2-DE gels; closed triangles, unlabeled LC1; open triangles, unlabeled LC2) were plotted on a logarithmic scale. The numbers represent the different quantified proteins. The actual accession numbers and associated information can be found in Table 2. A number of similar accessions that could not unambiguously be individually quantified were grouped together. In some cases, one of the three quantification methods allowed individual quantification and are marked with an asterisk. The protein name abbreviations are indicated as follows: 1, Ferredoxin 2 (Fd2); 2, Ferredoxin I (Fd1); 3, Ferredoxin reductase (FNR1); 4, plastocyanin (PC2); 5, Oxygen evolving enhancer 2 (OEC23); 6, Oxygen evolving enhancer protein 1 (OEC33); 7, Oxygen-evolving enhancer protein 3-1 (OEC16-1); 8, photosystem II protein W-like protein (PsbW-like); 9, thylakoid lumenal 29.8-kD protein (TL30); 10, ATP synthase β-chain (β-CF1); 11, phosphoglycolate phosphatase (PGP); 12, fructose-1,6-bisphosphatase (FBP); 13, fructose-bisphosphate aldolase (FBA); 14, glyceraldehyde 3-phosphate dehydrogenase (GAP-A); 15, glyceraldehyde-3-phosphate dehydrogenase (GAP-B); 16, phosphoglycerate kinase (PGK); 17, phosphoribulokinase (PRK); 18, ribulose-5-phosphate-3-epimerase (RPE); 19, Rubisco activase (RCA); 20, Rubisco large subunit (RbcL); 21, Rubisco small subunit (RBCS); 22, sedoheptulose-1,7-bisphosphatase (SBP); 23, transketolase (TKL); 24, aldose-1-epimerase (AE); 25, ribose-5-phosphate isomerase (RPI); 26, triose phosphate isomerase (TIM); 27, NADP-dependent malic enzyme (ME); 28, pyruvate orthophosphate dikinase (PPDK); 29, malate dehydrogenase [NADP] (MDH).
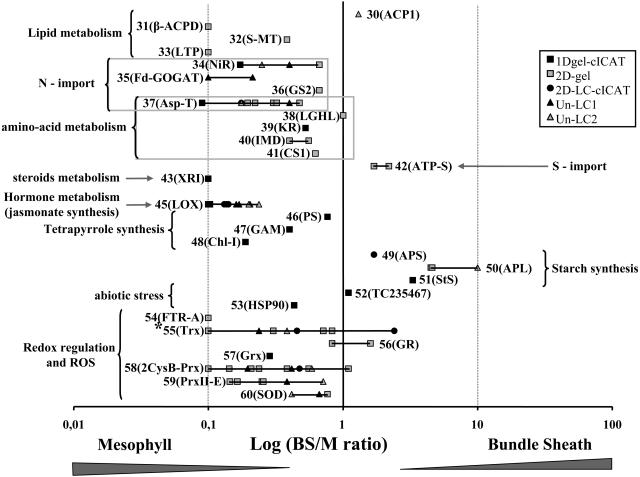
Figure 7.
Overview of BS:M Ratio Determinations by the Three Complementary Methods.
The 125 quantified maize proteins (ZmGI accessions) were divided in Figures 6 to 9 and are grouped by function; here, we show the second group. BS:M protein accumulation ratios obtained for each protein by three different methods (closed squares, 1-DE gel cICAT; circles, 2D-LC-cICAT; open squares, 2-DE gels; closed triangles, unlabeled LC1; open triangles, unlabeled LC2) were plotted on a logarithmic scale. The numbers represent the different quantified proteins. The actual accession numbers and associated information can be found in Table 2. A number of similar accessions that could not unambiguously be individually quantified were grouped together. In some cases, one of the three quantification methods allowed individual quantification and are marked with an asterisk. The protein name abbreviations are indicated as follows: 30, Acyl carrier protein 1 (ACP1); 31, β-hydroxyacyl-ACP dehydratase (β-ACPD); 32, S-malonyltransferase (S-MT); 33, lipid transfer protein 7a2b (LPT); 34, ferredoxin-nitrite reductase (NiR); 35, ferredoxin-dependent glutamate synthase (Fd-GOGAT); 36, Gln synthetase (GS2); 37, Asp transaminase (Asp-T); 38, lactoylglutathione lyase (LGHL); 39, ketol-acid reductoisomerase (KR); 40, 3-isopropylmalate dehydrogenase (IMD); 41, Cys synthase 1 (CS1); 42, ATP-sulfurylase 2 (ATP-S); 43, 1-deoxy-d-xylulose-5-phosphate reductoisomerase (XRI); 44, TC227295 (unknown); 45, Lipoxygenase (LOX1 and LOX2); 46, δ-aminolevulinic acid dehydratase (PS); 47, Glu-1-semialdehyde 2,1-aminomutase (GAM); 48, magnesium-chelatase subunit I (ChlI); 49, ADP-glucose pyrophosphorylase (small) (APS); 50, ADP-glucose pyrophosphorylase (large) (APL); 51, starch synthase (StS); 52, unknown pollen signaling protein (TC235467); 53, Hsp82 (HSP90); 54, ferredoxin-thioredoxin reductase subunit A (FTR-A); 55, thioredoxin-M2, -M4, and -F (Trx); 56, glutathione-disulfide reductase (GSR); 57, glutaredoxin (Grx); 58, 2-cys peroxiredoxin-like (2CysB-Prx); 59, peroxiredoxine-like (PrxII-E); 60, superoxide dismutase [Cu-Zn] (SOD).
Figure 8.
Overview of BS:M Ratio Determinations by the Three Complementary Methods.
The 125 quantified maize proteins (ZmGI accessions) were divided in Figures 6 to 9 and are grouped by function; here, we show the third group. BS:M protein accumulation ratios obtained for each protein by three different methods (closed squares, 1-DE gel cICAT; circles, 2D-LC-cICAT; open squares, 2-DE gels; closed triangles, unlabeled LC1; open triangles, unlabeled LC2) were plotted on a logarithmic scale. The numbers represent the different quantified proteins. The actual accession numbers and associated information can be found in Table 2. A number of similar accessions that could not unambiguously be individually quantified were grouped together. In some cases, one of the three quantification methods allowed individual quantification and are marked with an asterisk. The protein name abbreviations are indicated as follows: 61, adenylate kinase (ADK); 62, inorganic pyrophosphatase (InPyr); 63, nucleoside diphosphate kinase II (NDKII); 64, thylakoid lumenal 17.4-kD protein (TL17); 65, β-d-glucosidase (GLU2); 66, plastid-lipid associated protein (PAP3); 67, glutathione S-transferase (GST10); 68, unknown (SET1 domain protein) (TC224607); 69, mRNA binding protein (Csp41a-like); 70, nucleic acid binding protein (Cp31); 71, nucleic acid binding protein (Cp33); 72, plastid-specific ribosomal protein 2 (PSRP2); 73, ribosomal protein L1 (RP-L1); 74, ribosomal protein L12.1 (RP-L12.1); 75, ribosomal protein S1 (RP-S1); 76, ribosomal protein S5 (RP-S1); 77, ribosomal protein L10 (RP-L10); 78, elongation factor P (Ef-P); 79, elongation factor Tu (Ef-Tu); 80, elongation factor G (Ef-G); 81, polyprotein of Ef-Ts (PETs); 82, ribosome recycling factor (RRF); 83, membrane-associated 30-kD protein (Vipp1); 84, ClpP protease [ClpP(1?)]; 85, chaperonin 60 (β-Cpn60); 86, chaperonin 20 (Cpn21); 87, chaperonin 60 (a-Cpn60); 88, chaperonin 70 (HSP70); 89, GrpE (GrpE); 90, peptidyl-prolyl cis-trans isomerase (TLP21); 91, peptidyl-prolyl cis-trans isomerase (TLP40); 92, ClpC Hsp100 (ClpC); 93, FK506 binding protein 1 (FK506).
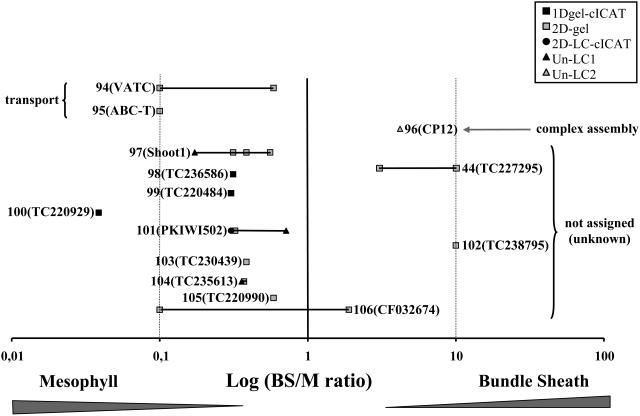
Figure 9.
Overview of BS:M Ratio Determinations by the Three Complementary Methods.
The 125 quantified maize proteins (ZmGI accessions) were divided in Figures 6 to 9 and are grouped by function; here, we show the fourth group. BS:M protein accumulation ratios obtained for each protein by three different methods (closed squares, 1-DE gel cICAT; circles, 2D-LC-cICAT; open squares, 2-DE gels; closed triangles, unlabeled LC1; open triangles, unlabeled LC2) were plotted on a logarithmic scale. The numbers represent the different quantified proteins. The actual accession numbers and associated information can be found in Table 2. A number of similar accessions that could not unambiguously be individually quantified were grouped together. In some cases, one of the three quantification methods allowed individual quantification and are marked with an asterisk. The protein name abbreviations are indicated as follows: 94, vacuolar ATP synthase subunit C (VATC); 95, ABC transporter putative (ABC-T); 96, CP12 protein precursor (CP12); 97, SHOOT1 protein (Shoot1); 98, unknown (TC236586); 99, aldo/keto reductase family protein (TC220484); 100, unknown (TC220929); 101, fruit protein PKIWI502 (PKIWI502); 102, inositol monophosphate family protein (TC238795); 103, unknown (TC230439); 104, Y230_ARATH (TC235613); 105, unknown (TC220990); 106, unknown (CF032674).
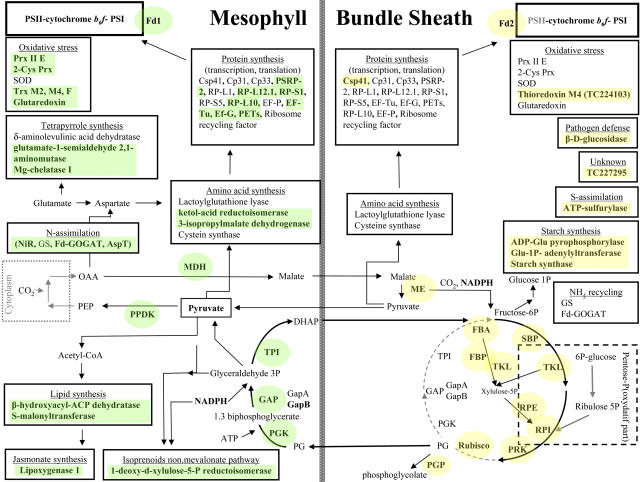
Our experimental data set was validated by comparison with accepted markers of M or BS chloroplasts (e.g., malate dehydrogenase [MDH] and PPDK in M; malic enzyme [ME] and several Calvin cycle enzymes in BS).
Compiled Expression Profiles and Functional Differentiation of BS and M Chloroplasts
In the remaining section of Results, we present the quantitative observations in terms of functions. We extensively compare the data with published literature, in an effort to pull together comparative BS/M maize (and sorgum) data resulting from diverse methodologies, such as transcript analysis, enzymatic measurements, and others. Importantly, new information of the BS/M distribution of less-studied metabolic pathways was determined, and strong quantitative differences in proteins involved in (regulation of) plastid gene expression and biogenesis are presented. The protein profiling data are summarized in Table 2 and corresponding Figures 6 to 9. We recommend using the protein report pages in PPDB to obtain a more interactive integration of the results.
C4 Carbon Shuttle, Carbon Fixation, OPPP, and Glycolysis
This category is composed of the most abundant stromal proteins identified on 2-DE gels, representing 34 and 65%, respectively, of the total spot volume in M and BS chloroplasts. Within the C4 carbon shuttle, we confirmed the known localization of MDH and PPDK in M chloroplasts and NADP-ME in BS. We quantified all 11 Calvin cycle enzymes, several of which overlap with the OPPP or glycolysis. Preferentially expressed in BS chloroplasts were the enzymes specific to the Calvin cycle (Rubisco [TC233714, TC234038, and TC234963], phosphoribulokinase [PRK; TC221089 and TC225896], and sedoheptulose-1,7-biphosphatase [TC239473]) and those shared between the Calvin cycle and the OPPP (ribulose 5-phosphate-3-epimerase [TC234954], ribulose-5-phosphate isomerase [TC221577], and transketolase [TC235000]). The BS preferential localization of Rubisco, ribulose-5-phosphate isomerase, PRK, FBA (TC219359 and TC219361), and ME (TC234846) do support the activity measurements of M and BS chloroplasts separated by nonaqueous fractionation (Slack et al., 1969). To our knowledge, the differential expression of ribulose 5-phosphate-3-epimerase and transketolase (involved in OPPP and Calvin cycle) has not been shown before. Rubisco activase is preferentially expressed in the BS chloroplasts, which was indirectly shown by in situ mRNA hybridization in maize (Ayala-Ochoa et al., 2004) and by differential cDNA screening in sorghum (Wyrich et al., 1998). Two homologs of triose phosphate isomerase (TC233907 and TC233912) involved in Calvin cycle and glycolysis were strongly expressed (between fivefold and ninefold) in the M chloroplasts. This settles the conflicting observations between transcript analysis in sorghum (Wyrich et al., 1998) and enzyme activity assays on total BS and M cellular extracts (Ku and Edwards, 1975).
The BS:M expression ratio of phosphoglycerate kinase (PGK) was different for two identified homologs (TC219624 and TC219625). The parallel LC quantifications were able to distinguish between the two isoforms. PGK-TC219625 was abundantly expressed in M; in each of the LC experiments, six peptides were quantified with twofold to sixfold higher accumulation in M chloroplasts. The expression of the other isoform (PGK-TC219624) was equally distributed (BS:M = 1.1) between M and BS. cICAT and 2-DE gel measurements did not distinguish between the two homologs since only shared peptides for the two PGK isoforms homologs were resolved. PGKs have a role in the reduction phase of the Calvin cycle as well as in glycolysis. The role of PGK-TC219625 is possibly to ensure high rates of triose phosphate reduction in the M chloroplast. We note that the (de)activation mechanisms for the two PGK isoforms are unknown but should be investigated; it is conceivable that the isoform more highly expressed in M chloroplasts has an activation mechanism specifically adapted to M localization. The activity of PGK was shown to be equally distributed between the M and BS chloroplasts prepared by nonaqueous chloroplast isolation (Slack et al., 1969) and by activity measurements on total cell extracts from M protoplasts and BS strands (Ku and Edwards, 1975). A subsequent article, however, suggested an increased M localization (Usuda and Edwards, 1980). These earlier conflicting reports are most likely due the activity and detection of the individual chloroplast PGK isoforms, with the background activity of the third cytoplasmic homolog.
The two non-gel-based methods consistently showed that GADPH-A (TC219897) was equally distributed between M and BS chloroplasts, while GADPH-B (TC234510) preferentially accumulated in the M chloroplast (2.5- to 4.8-fold up). The 2-DE analysis of the acidic GADPH-B also showed an increased accumulation in the M chloroplasts. Measurements of total GADPH activities showed equal distribution between M and BS chloroplasts (Slack et al., 1969; Ku and Edwards, 1975). In C3 plants, GAPDH-B can form a homotetramer in addition to formation of a heterotetramer with GAPDH-A1,2 (Baalmann et al., 1996). It is most likely that GAPDH oligomers in M maize chloroplasts differ in the GAPDH A/B protein ratio as compared with GAPDH complexes in BS chloroplasts. In C3 plants, GAPDH oligomers reversibly associate with PRK and a small CP12 linker protein to help activation of PRK (Baalmann et al., 1996; reviewed in Graciet et al., 2004). This likely regulates the carbon flow from the Calvin cycle to the oxidative pentose phosphate cycle (Tamoi et al., 2005). Interestingly, we quantified the maize CP12 (TC223290) homolog with a BS:M ratio of 4.2. The presence of CP12 in the BS fractions strongly suggests that formation of the GAPDH-CP12-PRK supercomplex takes place in the BS chloroplast (not in M chloroplast), in agreement with the role of the BS chloroplast in the Calvin cycle. The CP12 protein was not earlier detected in maize.
Starch Biosynthesis and Carbohydrate Metabolism
We found a preferential BS localization for several enzymes involved in starch synthesis in agreement with earlier observations (Spilatro and Preiss, 1987; Lunn and Furbank, 1997). The small subunit of ADP-glucose pyrophosphorylase (TC232071), the first committed step in starch synthesis, was 1.7-fold higher in BS. Isoforms of the large subunit ADP-glucose pyrophosphorylase glucose-1-phosphate adenyltransferase (TC222533 and TC242174) were also significantly higher in B chloroplasts, whereas aldose-1-epimerase (TC237704) and soluble starch synthase (TC236897) were 5- and 3.3-fold higher, respectively, in BS chloroplasts. It is important to point out, however, that we did observe starch accumulation in the M chloroplasts of older (20 d) maize leaves during chloroplast preparations (W. Majeran, unpublished data). This suggests that starch synthesis is mostly limited to BS chloroplasts when the leaf is a sink, but when leaf tissue becomes a source, M chloroplasts also have significant rates of starch synthesis.
Nitrogen Assimilation
Two nitrogen assimilation pathways can be distinguished in leaves. The primary N assimilation involves chloroplast nitrite reductase (NiR), Gln synthase 2 (GS2), ferredoxin-dependent Glu synthase (Fd-GOGAT), and Asp transaminase (AspT). Secondary N assimilation is performed by the enzyme couple Gln synthase 2 and Fd-GOGAT, which allows the reassimilation of ammonium produced as the last product of the photorespiration, in particular in C3 species (Weber and Flugge, 2002).
We were able to quantify the accumulation of four chloroplast enzymes (with one isoform each) implicated in N assimilation. Three enzymes were predominantly expressed in the M chloroplasts and are NiR (TC222347 and TC222348, 2.5- to 6-fold), Fd-GOGAT-1 (TC236410 or GLU1 in Arabidopsis, 5-fold or unique in M), and AspT (TC219944, 2- to 11-fold). GS (TC220868) synthase was identified only on 2-DE gels, and its BS:M ratio was close to one.
Several groups analyzed the localization of nitrogen assimilation in C4 plants and designated M cells as the compartment where primary N assimilation takes place (Rathnam and Edwards, 1975, 1976; Harel et al., 1977; Becker et al., 1993). However, conflicting conclusions were presented for the localization of GS and Fd-GOGAT involved in secondary N assimilation (Rathnam and Edwards, 1976; Harel et al., 1977; Becker et al., 1993, 2000). This might well have been the result of the lengthy preparations of M and BC cells using enzymatic digestion at 37C. A preferential localization of AspT activity in the M was previously described, but the authors associated it with the cytoplasmic fraction (Hatch and Mau, 1973). AspT catalyzes the fixation of ammonium on oxaloacetate to yield aspartate. It constitutes a metabolic link between N assimilation and amino acid synthesis. The increased accumulation of AspT in M chloroplasts is consistent with the fact that both the N assimilation (see above) and enzymes of amino acid synthesis pathways preferentially accumulate in M chloroplasts.
S-Assimilation
We identified one enzyme in the sulfur import pathway, the ATP-sulfurylase (TC235924, similar to Arabidopsis ATP SULFURYLASE2), which catalyzes the first step of sulfate import (sulfate fixation onto an adenyl-sulfate). Its accumulation was increased in the BS chloroplasts (twofold), which was consistent with previous localization of primary sulfur assimilation in the BS cells by activity measurements (Burgener et al., 1998). Cystein was proposed to be the transport factor for sulfur between M and BS cells (Burgener et al., 1998). A cystein synthase (TC235388 and TC235390) was slightly higher in the M chloroplast (1.6-fold), suggesting that secondary assimilation of H2S takes place in M chloroplasts.
Photorespiration
Chloroplast phosphoglycolate phosphatase (TC229733) accumulated preferentially in BS chloroplasts (BS:M = 2.6), in agreement with phosphoglycolate phosphatase activity measurements (Baldy and Cavalie, 1984). The significance and capacity of photorespiration in C4 photosynthesis in maize is, however, a matter of debate and is driven by the O2/CO2 partial pressures in BS cells. Clearly, it is lower in NADP-ME type C4 plants, such as maize, than in NAD-ME or phosphoenolpyruvate carboxykinase type C4 plants (Yoshimura et al., 2004).
Lipid Metabolism and Hormones
We quantified three proteins in lipid biosynthesis: acyl carrier protein 1 (ACP1; TC223112 and TC228772), β-hydroxyacyl-ACP dehydratase (TC240700), and S-malonyltransferase (TC223360). We also found a lipid transfer protein 7a2b (LTP-7a2b; AI966827). ACP1 showed an equal distribution between M and BS chloroplasts. The β-hydroxyacyl-ACP dehydratase was a low abundant spot in the 2-DE gel analysis, at the detection limit of the analysis. S-malonyltransferase was found to be preferentially expressed in the M chloroplasts (BS:M ratio 0.38), and the lipid transfer protein 7a2b showed a unique M localization.
We identified two types of enzymes, lipoxygenase 1 (LOX1) and LOX2 that are involved in the production of oxylipins, a diverse group of fatty acid derivatives that includes the hormone jasmonate and defensive metabolites (Feussner and Wasternack, 2002). LOX1 (AW157962) and LOX2 (TC234252, TC237970, and TC237971) all showed a high preference for M accumulation (fourfold up to ninefold). The high preferential accumulation of lipoxygenases in M chloroplasts has not been determined earlier and warrants further investigation.
Nucleotide Metabolism
Adenylate kinase (TC236532 and TC236534) and inorganic pyrophosphatase (InPyr; TC218860) both accumulated preferentially in the M chloroplast, with BS:M ratios of 0.3 and 0.5, respectively. Adenylate kinase removes and recycles AMP produced in the reversible PPDK reaction, while InPyr is also indirectly needed to activate PPDK through dephosphorylation. Thus, the relatively high M expression is likely to be dictated by the large flux through PPDK. InPyr also plays a role (by removal of pyrophosphate) to drive the forward reaction of plastid-localized ADP-glucose pyrophosphorylase toward starch biosynthesis. The observed expression ratios are consistent with their M localization based on activity assays in maize (Slack et al., 1969).
Tetrapyrrole Synthesis
We quantified three enzymes in the tetrapyrrole synthesis pathway implicated in three steps of the chlorophyll/heme biosynthesis pathway: (1) formation of the precursor 5-aminolevulinic acid (ALA), (2) synthesis of protoporphyrin, and (3) incorporation of Mg2+ into the porphyrin ring (for reviews, see Beale, 1999; Eckhardt et al., 2004). The formation of ALA is the limiting step for the rest of the pathway. We identified Glu-1-semialdehyde 2,1-aminomutase (TC227768), which catalyzes the rearrangement of Glu-1-semialdehyde into ALA. This stromal enzyme accumulated preferentially in M (BS:M 0.40). The subsequent enzyme, δ-aminolevulinic acid dehydratase (TC240729), which catalyzes the asymmetric condensation of two ALA to form porphobilinogen, also accumulated at a somewhat higher level in M chloroplasts. We identified the Mg-chelatase subunit I (TC223168), similar to Mg-chelatase I-2 from Arabidopsis (At5g45930.1), several enzymatic steps further downstream in the chlorophyll biosynthesis pathway. This enzyme showed a strong preferential M accumulation (BS:M 0.19), likely reflecting the higher demand of chlorophylls due to photosystem II (PSII) accumulation.
Defense
Hydroxamic acids (Hx) possess antifungal and insecticidal properties and are abundantly accumulating in rye (Secale cereale), wheat (Triticum aestivum), and maize cell walls as a glucoside-linked precursor (Hx-glc) (Niemeyer, 1988). Upon tissue damage, the β-d-glucosidases come in contact with Hx-glc and release the Hx that can be converted into a benzoxalolinone. IMBOA (2-O-β-d-glucopyranosyl-4-hydroxyl-7-methoxy-1,4-benzoxazin-3-one) is the main Hx-glc in maize and wheat.
Each of the three quantification methods used in this study showed that β-d-glucosidase (TC220472) accumulates at much higher levels in BS chloroplasts (BS:M ratio >6.7). This is in agreement with previous observations showing that β-d-glucosidases were found in maize plastids and proplastids of the BS cells (Nikus and Jonsson, 1999; Nikus et al., 2001). Although the major role of β-d-glucosidase was attributed to defense, it was also suggested to play a role in development and growth regulation, since transient expression of maize β-d-glucosidase in tobacco (Nicotiana tabacum) protoplasts was able to release cytokinin from an inactive cytokinin-O-glucoside (Brzobohaty et al., 1993).
Proteins Involved in Redox Regulation
Ferredoxins are not only essential in linking thylakoid-localized photosynthetic electron transport to NADPH production via Fd-NADP+ reductases (FNR), but also provide redox equivalents to the different thioredoxins (Trxf, -m, -x, and -y) via Fd-Trx reductase. These different Trx regulate several plastid-localized pathways by changing the redox state of thiol groups (S-S<→2SH) (Buchanan and Balmer, 2005).
We identified and quantified both Fd1 (TC220059 and TC238105) and Fd2 (TC223586). Fd1 accumulated predominantly in the M chloroplast (twofold to fivefold), while Fd2 accumulated predominantly in the BS chloroplast (threefold to fourfold). This is in agreement with transcript analysis of Fd1 and Fd2 (Matsumura et al., 1999; Furumoto et al., 2000). Preferential accumulation of Fd2 in BS chloroplasts was shown by immunoblots, while Fd1 was found to be equally distributed over both chloroplast types (Kimata and Hase, 1989). It seems that Fd1 and Fd2 have evolved to interact with different partners. They possess similar redox potentials but have different Km values for interaction with FNR, suggesting different cellular functions. Indeed, expression of maize Fd1 in Fd-deficient strains of cyanobacteria (Plectonema boryanum) led to a high light sensitive phenotype, while the strain expressing Fd2 showed a nitrogen deficiency phenotype (Kimata-Ariga et al., 2000).
We identified and quantified BS and M expression ratios for three FNR identifiers: TC219223, TC219224, and TC219226. All three sequences were similar to Arabidopsis FNR-2. All of them (FNR: TC219223, TC219224, and TC219226) showed a preferential localization in M chloroplasts (from twofold up to sevenfold). This is consistent with previous M localization of an FNR protein by rocket electrophoresis (Broglie et al., 1984).
The Trx play an important role in redox regulation (activation and deactivation) of many biosynthetic and other activities in plastids (Balmer et al., 2004). The distribution of the different Trx between the M and BS chloroplasts has not been reported earlier, which is somewhat surprising given the central role of the Trx in regulating chloroplast function. Based on their closest homologs in Arabidopsis, we identified and quantified Trx-m2, Trx-m4, and Trx-f2. For Trx-m4 (TC221334) and Trx-m2 (TC228810), only common peptides were identified showing their preferential accumulation in the M chloroplast (2.6 to 4). In the 2-DE spot 348 (only present in M stroma), Trx-f2 (TC220464) was identified; however, its quantification could not be exclusively determined as it comigrated with Trx-m4 (TC221334). The non-gel-based methods quantified unique peptides for two other Trx-m4 isoforms (TC238760 and TC224103), which showed preferential accumulation in M (BS:M ratio 0.45) and BS (BS:M ratio 2.4), respectively (Table 2; see Supplemental Table 4 online).
Proteins Involved in Detoxification of Reactive Oxygen Species
Chloroplasts have an elaborate enzymatic system to detoxify different reactive oxygen species (ROS), such as superoxide, hydrogen peroxide, and lipid peroxides. M chloroplasts have high rates of linear photosynthetic electron transport and high production rates of molecular oxygen through the water-splitting activity of PSII, whereas BS chloroplasts only carry out cyclic electron flow. It is thus quite likely that there are substantial differences in the quantity and quality of ROS in these two chloroplast types. However, differential expression levels of the BS and M chloroplast ROS defense systems are unknown. We quantified the BS/M chloroplast expression of Cu-Zn superoxide dismutase (SOD), glutathione disulfide reductase (GR), and several peroxiredoxins (Prx). GR (TC227052) and Cu-Zn SOD (TC237182) each accumulated in M and BS chloroplasts. We observed an equal distribution of GR over both plastid types. Both gel-based and non-gel-based analysis suggested that the BS:M expression ratio of CuZn SOD (TC237182) was between 0.4 and 0.7.
Several studies tried to measure SOD activity in BS and M maize cells (but not in purified chloroplasts), with inconsistent results between the different studies (Foster and Edwards, 1980; Doulis et al., 1997). Possibly this is due to the presence of different SOD homologs and different isolation procedures. Comparisons between the total maize leaf extracts and rapid M sap extractions showed that glutathione (Burgener et al., 1998), dehydroascorbate reductase, and GR were localized in the M, while ascorbate peroxidase was found in the total leaf extracts (Doulis et al., 1997). As GR mRNA was distributed in both compartments, a translational regulation mechanism of differential expression was proposed (Pastori et al., 2000).
Prx are Trx-dependent peroxidases that have a capacity to scavenge peroxides. They constitute an antioxidant system that operates in parallel to the ascorbate-glutathione system (reviewed in Dietz, 2003). In Arabidopsis, four types of Prx are targeted to plastids (Prx IIE, PrxQ, 2-Cys PrxA, and 2-Cys PrxB). We quantified the expression of maize EST products 2-CysPrx-TC234345 and 2-CysPrx-TC234346, which both showed high degrees of similarity to Arabidopsis 2-Cys PrxB (At5g06290.1), and TC223042, which was closest to Prx IIE (At3g52960.1). Both 2-Cys Prx (TC234345 and TC234346) and Prx IIE (TC223042) accumulated preferentially in M chloroplasts (1.8- up to 7-fold). The 2-Cys Prx were also identified on 2-DE gels in a small train of spots (spots 362, 459, and 125), migrating in the region between 47 and 53 kD at about twice the expected molecular mass (of 26 kD). These spots represented likely 2-Cys Prx dimers, despite the denaturing conditions of the 2-DE gel. The basic oligomeric state of Prx is a dimer that under reducing conditions can assemble into larger 2-Cys Prx oligomers (Konig et al., 2002). To our knowledge, the distribution of the different Prx homologs over M and BS chloroplasts was not determined in published literature. The preferential accumulation of Prx proteins in the M chloroplasts is consistent with the high linear electron flow and production of active oxygen species, such as H2O2 (Doulis et al., 1997) in M chloroplasts. A wide range of electron donors has been shown to interact with Prx. Some Prx can accept electrons from both Trx and glutaredoxins (Rouhier et al., 2001), whereas others can only use Trx as electron donor. It is interesting to note that the preferential expression of Prx (TC223042, TC234346, and TC234345) in M is correlated with the expression in M chloroplasts of two isoforms of Trx (TC238760 and TC221334) and one glutaredoxin (TC225992, 3.5-fold).
Chloroplast Chaperones, Isomerases, and Proteases
The two major housekeeping chloroplast chaperone families (Cpn60/20 and Hsp70/GrpE) were relatively abundant on the 2-DE gels. The Cpn60/20 complex is composed of three types of proteins, namely Cpn60α and Cpn60β (homologs to Escherichia coli GroEL) and Cpn20 (homolog to E. coli GroES). The identified Cpn60α homologs (TC220350, TC235184, and TC236031), Cpn60-β (TC219523 and TC219522), and Cpn20 (TC233810 and TC236117) all showed an equal distribution between M and BS chloroplasts, with BS:M ratios between 1 and 1.4. This clearly shows that the Cpn60/20 chaperone system is not exclusively functioning in Rubisco assembly (since Rubisco is absent in M chloroplasts).
We identified two highly homologous HSP70 homologs (TC220215 and TC235785). In the cICAT and 2-DE gel analysess, we were not able to distinguish the BS:M expression ratio for each of the HSP70 proteins individually. Collectively, their BS:M expression ratio ranged between 0.9 and 5.8, depending on the quantification method and protein spot. However, in the parallel LC-based quantifications, we were able to determine the BS:M expression ratios for each of the two homologs. This showed an equal distribution for HSP70-TC220215 and a strong (sixfold) preferential expression of HSP70-TC235785 in the M chloroplasts. It is noteworthy that one of the interacting partners of HSP70, the GrpE (TC222867) also showed a fivefold preferential M expression. This result is consistent with a previous sorghum cDNA library screening where HSP70 expression ranged from 1 up to 10 in M cells (Wyrich et al., 1998). This result suggests that besides the expression of chaperones in both plastid types as housekeeping proteins, HSP70-TC235785 seems to have a specific function in M chloroplasts.
Proteome analysis of chloroplasts in Arabidopsis showed accumulation of a relatively large number of protein peptidyl-prolyl cis-trans isomerases of different categories, many of which seem localized in the thylakoid lumen (Peltier et al., 2002; Schubert et al., 2002). Here, we identified and quantified BS:M expression ratios of three of these lumenal isomerases, apparently released into the chloroplast stroma. Tlp21-TC228225 and Tlp40-TC239826, homologs to the cyclophylins Tlp21 and Tlp40, both showed a BS:M expression ratio of 0.5 in the 2-DE gel experiments. FK506-TC247336, a homolog of Arabidopsis At5g45680 assigned FKBP13, was preferentially expressed in the BS chloroplasts (BS:M = 5). FKBP13 was found in yeast two-hybrid experiments as interacting with the cytochrome b6f complex Rieske subunit (Gupta et al., 2002).
Interestingly, we also quantified the expression ratio of ClpC-TC235372, a homolog of the HSP100 ClpC chaperone family. The BS:M expression ratio was 0.4. ClpC1 in Arabidopsis accumulates in the stroma likely delivering substrate to the Ser-type ClpP/R protease complex, while an envelope-associated form is involved in protein import. We also quantified two ClpP proteins (TC245457 and TC225197) with BS:M ratios ranging from 0.3 to 0.59. The alignment of TC245457 and TC225197 with the members of the ClpP/R family in Arabidopsis did not identify clear-cut homologs.
Plastid Protein Expression
Both BS and M chloroplasts contain the same plastid genome (with 104 genes), encoding for four rRNAs and 30 tRNAs and some 70 proteins involved in plastid gene expression (e.g., ribosomal proteins, tRNA synthetases, etc.) as well as photosynthesis (RBCL and thylakoid proteins) (Maier et al., 1995). Expression of these plastid genes likely involves hundreds of nuclear-encoded proteins, each imported into the chloroplast (reviewed in Barkan and Goldschmidt-Clermont, 2000).
We quantified BS/M expression for seven plastid ribosomal proteins. Ribosomal protein L1 (TC219835) was equally expressed in both compartments, while L12.1 (TC222916), S1 (TC221841), S5 (TC239133), and L10 (TC224551) predominantly or exclusively accumulated in M chloroplasts. This suggests that 70S ribosomes in M and BS chloroplasts are different in composition, in agreement with differential accumulation of chloroplast L2 in an earlier study (Zhao et al., 1999).
Homologs to Arabidopsis plastid-specific ribosomal proteins PSRP2 (TC238798) and polyprotein of Ef-Ts (PSRP7-EF-Ts fusion protein; TC226754) were preferentially expressed in the M chloroplasts, with BS:M ratios of 0.29 and 0.14, respectively. It was proposed that PSRP2 is part of a plastid translational regulatory module on the 30S ribosomal subunit structure for the possible mediation of nuclear factors on plastid translation (Yamaguchi et al., 2000). We identified TC226754 as a 125-kD protein on the 2-DE gels. It is likely a homolog of a polyprotein of Ef-Ts gene originally described in Chlamydomonas reinhardtii as a single-copy gene containing PSRP-7 and EF-Ts sequences (Beligni et al., 2004). Translation of this gene resulted in a 110-kD pro-protein, and the majority of this protein was likely posttranslationally processed into the 65-kD protein PSRP-7 and a 55-kD EF-Ts. Our MS data for this protein covered both the N- and the C-terminal parts containing the PSRP7 and EF-Ts sequences, respectively. We did not identify the 65- and 55-kD maturation products.
We found an equal BS/M distribution of a ribosome recycling factor (TC237618). In E. coli, it is implicated in the disassembly of the post-termination complex (Kaji et al., 1998), and a spinach (Spinacia oleracea) homolog was identified in chloroplasts (Rolland et al., 1999). An equal M and BS distribution was also observed for the elongation factor P (TC237934), which was shown to be an essential gene in E. coli (Aoki et al., 1997). Surprisingly, elongation factor G (TC222353) was expressed up to eightfold higher in the M than in BS chloroplasts. We identified one or two abundant elongation factors Tu, matching four ESTs (TC222181, TC226641, TC222182, and TC220034). Expression was twofold to sixfold higher in the M chloroplasts than in BS chloroplasts. Its high abundance as judged from the 2-DE gels suggested a role in addition to translation elongation. Indeed, elongation factor Tu has been characterized in maize as a heat-induced molecular chaperone, in addition to its role in translation (Bhadula et al., 2001; Rao et al., 2004).
In addition, searching the MS data against the maize genomic sequences (in AZM) identified a protein encoded by CF646019 (and AZM4_135414) similar to Arabidopsis CSP41, which was characterized as an mRNA binding protein in spinach chloroplasts with endoribonuclease activity (Yang et al., 1996) and is a homolog of C. reinhardtii RAP41 ribosome-associated protein (Yamaguchi et al., 2003). The expression of CSP41 (CF646019, AZM4_135414) shows a small preference for the BS chloroplasts (BS:M ratio = 1.8-fold).
We identified two abundant stromal ribonucleoproteins, CP31 (TC235457 and TC235458) and CP33 (TC236626). Their proposed role is to protect mRNAs that are not engaged in translation (Nakamura et al., 2001). Their accumulation levels were 1.8- to 3-fold higher in M chloroplasts.
Proteins with Unknown Function
We could not assign any obvious function (Table 2) to a dozen quantified proteins. Here, we just highlight a few of these proteins. We identified a tetratricopetide repeat (TPR) protein, TC238048, with an average BS:M accumulation ratio of 0.29. It is an homolog of the gene called Shoot1 of unknown function in Glycine max (Karakaya et al., 2002) and was quite abundant on the 2-DE gels (Figure 3). TPR proteins have been associated with numerous functions (Das et al., 1998). Characterized TPR proteins in the chloroplast play a role in stabilization of chloroplast mRNA (Vaistij et al., 2000; Felder et al., 2001) or in photosystem I assembly (Boudreau et al., 2000). TC236586 with a DUF149 motif with unknown function, TC220484 with an oxidoreductase or aldo/keto reductase domain, and TC222257 with an oxidoreductase NAD binding domain each accumulated with a BS:M ratio of 0.3. TC220929 with a DUF299 domain was predominantly expressed in M chloroplasts (BS:M ratio of 0.04) and is probably unique to M chloroplasts. The expression of TC238795 with similarity to inositol monophosphatase was five times higher in BS than in M chloroplasts. TC235613 showed a preferential (twofold or sevenfold) expression in the M chloroplast and was similar to a 3-β-hydroxy-δ5-steroid dehydrogenase, probably associated with steroid metabolism. Its Arabidopsis homolog (At2g37660) was identified numerous times in proteome analyses of thylakoids and stroma (see PPDB). On the BS side, we found in two spots (BS:M ratio = 2.9 and only found in BS) the unknown protein TC227295 containing the Qor motif (for NADPH quinone reductase and related Zn-dependent oxidoreductases) similar to At1g23740.
Peripheral and Lumenal Thylakoid Proteins Involved in Photosynthesis
The BS and M stromal analysis also identified several abundant members of the oxygen evolving complex of PSII (OEC16 [TC219937 and TC238011], OEC23 [TC235206 and TC235205], and OEC33 [TC233304]) and plastocyanin (TC219293 and TC219295) (see Supplemental Table 4 online). They are located on the lumenal side of the thylakoid membrane and were released into the stroma during chloroplast lysis. Their preferential accumulation in the M chloroplasts is consistent with the low but detectable accumulation levels of PSII in the BS chloroplasts (Meierhoff and Westhoff, 1993; Bassi et al., 1995). In this context, it is interesting to mention the identification of TC230967, the maize homolog of the Arabidopsis HCF136 protein in M chloroplasts. HCF136 is specifically involved in PSII assembly (Meurer et al., 1998), and its identification in M chloroplasts is consistent with its specialized role.
DISCUSSION
This article presents an extensive quantitative comparative analysis of cell-specific chloroplast proteome differentiation in maize, in which new quantitative protein profiling techniques were implemented and the data integrated with the published literature using various measures of enzyme activity as well as transcript accumulation. We provide an overview of the differential protein accumulation in the stroma of BS and M chloroplasts and identified some 400 proteins (ZmGI accessions) with few obvious nonchloroplast contaminants, covering a wide range of pathways and functions. After a brief comparison of the three comparative proteomics methods, we will focus this discussion on integration of the comparative data summarized in Figure 10.
Figure 10.
Overview of the Distribution of Different Chloroplast Functions over BS and M Chloroplasts.
The major metabolic pathways and associated proteins are listed, with some examples of other pathways. The proteins with differential accumulation are indicated in bold and are colored. General metabolic pathways are schematically indicated. Abbreviations are as follows: peroxiredoxines (Prx), superoxide dismutase (SOD), thioredoxins (Trx), magnesium chelatase (Mg-chelatase), nitrite reductase (NiR), glutamine synthase (GS), ferredoxin dependent glutamate synthase (Fd-GOGAT), aspartate transaminase (AspT), nucleic acid binding proteins (Cp33, Cp31), plastid specific ribosomal protein (PSRP), ribosomal protein (RP), polyprotein of Ef-Ts (PETs), elongation factors (Ef), glyceraldehyde dehydrogenase (GAP), fructose 1,6-biphosphatase (FBP), sedoheptulose-1,7-bisphosphatase (SBP), transketolase (TKL), ribose-5P-isomerase (RPI), phosphoglycerate kinase (PGK), triose phosphate isomerase (TPI), dihydroxyacetone (DHAP), phosphoglycerate (PG), ferredoxin (Fd), oxaloacetate (OAA), phosphoenolpyruvate (PEP) and glucose (Glu).
Evaluation of the Methodologies, Consistency of Quantification, and Variation
The three quantification methods used in this study were consistent in terms of BS/M expression patterns. Fifty percent of all quantified proteins were obtained by two or three independent quantification methods. The three methods were also complementary in that they each helped to quantify a number of proteins not observed by the other methods. Such complementarity is observed for identification of proteomes using different ionization methods (matrix-assisted laser-desorption ionization [MALDI] versus ESI) and fractionation techniques (e.g., gels, chromatography, etc.), but this phenomenon is less established for comparative proteome analysis. The reason for the complementarity observed in this study was severalfold. (1) The 2-DE gels only visualized proteins within the selected pI range and were only quantititative for those protein spots that contain only one (dominating) protein; we note that the selected 4 to 7 pI range does in fact cover the majority of the predicted plastid proteome (Sun et al., 2004). (2) The cICAT-based quantification requires the presence of one or more Cys residues, since the isotope tag is cross-linked to the peptides via a Cys cross-linker. This obviously excluded proteins that have no Cys residues and reduced the success rate for proteins with only one or two Cys residues. (3) The parallel LC-MS quantification on unlabeled proteomes seemed limited to the more abundant proteins but does not require any Cys residues. (4) Lower-abundance proteins with a near exclusive presence in either BS or M chloroplasts could not easily be quantified by the non-gel-based methods due to the relative complexity of the proteomes. By contrast, quantification of such proteins was possible and accurate on 2-DE gels in less complex regions of the gels. The MS analysis was sensitive enough to identify these low abundant proteins because of the strongly reduced complexity of proteins in gel spots.
In terms of BS:M expression ratios, we observed in a number of cases that the non-gel-based methods gave more pronounced ratios, indicating a stronger BS/M proteome differentiation than the 2-DE gel analysis. The reason is primarily that we used the highest quality BS and M preparation (with ∼5% cross-contamination) for the non-gel-based methods. This was done because these non-gel-based methods required less protein material (e.g., for parallel LC-MS, only 20 to 40 μg per sample), and the BS and M preparations of highest purity had the lowest yield. In a few cases (marked with an asterisk in Table 2 and Figures 6 to 9), we observed more variation in BS:M ratios, primarily because two isoforms or homologs could not be quantified separately in one or all of the quantification methods used. In case of 2-DE gel-based quantification, we also observed variation due to differential protein modifications.
We were pleased to find that our differential proteome expression data were generally consistent with published differential enzymatic activity data, transcript data, and immunoblots and in situ immunolocalization. This provided confidence in the significance and accuracy of our differential protein expression data.
General Conclusions for BS and M Chloroplast Differentiation
Five general conclusions can be extracted from the comparative proteomics data. First, many differences in metabolic differentiation can be explained as being driven by the production of NADPH in the M chloroplasts and a relative shortage of reducing equivalents in the BS chloroplasts. For instance, proteins involved in lipid biosynthesis and nitrogen import are primarily located in M chloroplasts, in addition to preferential M location of Calvin cycle enzymes involved in triose phosphate reduction. Second, many plastid functions are not exclusively located in one chloroplast type or the other, with some exceptions, such as β-d-glucosidase (TC220472) (BS:M >10) and LOX1 (AW157962, TC234252, TC237970, and TC237971). Third, differential accumulation occurred in a number of cases by differential expression of homologs within gene families; for instance, Fd1 (TC220059 and TC238105) accumulates mostly in M chloroplasts, while Fd2 (TC223586) accumulates mostly in BS. Fourth, our data set provides clear evidence for differential regulation of plastid gene expression, protein biogenesis, and protein fate. This is a relatively understudied area. The differential accumulation of VIPP1 (TC220737; high in M), for example, could help to accelerate our understanding of plastid protein biogenesis and the specific role of such proteins. Finally, we found several proteins of unknown function (e.g., KiwiFruit protein [TC222257] and Shoot1 [TC238048]), for which the relatively high abundance on the 2-DE gels suggested their participation in central functions, while their differential BS/M expression indicated their specialization. The role of these proteins in C4 metabolism awaits investigation.
In the remainder of this discussion, we integrate our observations in three parts, as follows: (1) differentiation of metabolic functions, (2) differentiation of redox regulation and antioxidative defense, and (3) differentiation of plastid gene expression and protein biogenesis/homeostasis. An overview of the observations and their connections are provided in Figure 10. All data are integrated in the PPDB and are linked to sequence and proteomics data from Arabidopsis.
Differentiation of Metabolic Chloroplast Functions
The C4 malate-pyruvate shuttle provides the net influx pathway for carbon from M to BS but only half of the required reducing equivalents for the Calvin cycle. The other half of the reducing equivalents needed for the Calvin cycle is provided through the triose phosphate shuttle. Our proteomics data are in agreement with earlier established differential localization and/or enzyme activities of these shuttles. Our study now provides specific ZmGI and AZM gene identifiers for these enzymes. Surprisingly, accumulation levels of triose phosphate isomerase (TC233907, TC233905, TC233906, and TC233912) are consistently (with all three proteomics methods) much higher in M chloroplasts than in BS chloroplasts. This suggests that the M-localized triose shuttle should be viewed as part of the BS-localized Calvin cycle rather than being viewed as an MS-localized shuttle operating in parallel to an exclusively BS-localized Calvin cycle. Our data also suggest that at least one GAPDH-B (TC234510) isoform (homolog) plays a more prominent role in M chloroplasts as part of the triose phosphate shuttle (or, less likely, in glycolysis).
As expected, the Calvin cycle–specific enzymes (Rubisco, PRK, and SBP) and CP12 [involved in assembly and (de)activation of PRK and GAPDH in C3 chloroplasts; Wedel et al., 1997] and Rubisco activase have high BS:M expression ratios. The differential accumulation of CP12 is an excellent tool to further define its role in regulation of carbon metabolism. Starch biosynthetic enzymes have higher accumulation levels in BS, which is a logical consequence of M localization of the Calvin cycle and is in agreement with earlier observations.
The possibility of an alternative NAD-ME C4 like pathway in maize (in addition to the established NADP+-ME C4 pathway) has been raised (Chapman and Hatch, 1981; Furumoto et al., 1999; Wingler et al., 1999), in which Asp could serve as the carbon carrier instead of malate, with AspT converting oxaloacetate into Asp (and several other differences). We did observe a preferential accumulation of AspT (TC219944) in M. It is conceivable that AspT participates in carbon transport, initiated not as postulated in the NAD-ME C4 pathway in the cytoplasm of the M cells (Hatch and Mau, 1973), but in chloroplasts. In this way, AspT could remove excess OAA from its main flow in situations where the MDH or the pool of malate was saturated.
Virtual lack of linear photosynthetic electron transport in BS thylakoids and high rates of linear electron transport in M thylakoids lead to an imbalance in supply and demand for NADPH (reducing equivalents) between the two chloroplast types. Many observed differences in metabolic differentiation outside of primary carbon metabolism can essentially be explained as being driven by this imbalance of redox equivalents. For instance, nitrogen import is a highly demanding pathway for energy (ATP) and reductive power (NADPH). Correspondingly, we observed NiR (TC222347 and TC222348), GS (TC220868), and Fd-GOGAT (TC236410) involved in the conversion of nitrite into Glu preferentially expressed in M chloroplasts. As a possible consequence of M-localized N-assimilation and availability of pyruvate as carbon backbone, we quantified several enzymes in Leu/Val synthesis (using pyruvate as carbon metabolite) and Asp/Thr (using oxaloacetate and Glu) with preference for M choroplasts (Figure 7). The preferred nitrogen assimilation into Glu in M cells also connects well with our observation that the enzyme Glu-1-semialaldehyde aminotransferase (TC227768) is preferentially located in the M chloroplasts. The high M accumulation ratio of Mg-chelatase subunit I (TC223168) suggests that overall chlorophyll synthesis rates are higher in the M chloroplasts, possibly to accommodate the high number of chlorophyll molecules in M-specific PSII.
The de novo fatty acid biosynthesis in plants is located in the chloroplast, whereas lipid biosynthesis is shared between different organelles, including chloroplasts (e.g., Ohlrogge and Browse, 1995; Ohlrogge and Jaworski, 1997). Reports about the differential contribution of BS and M cells and chloroplasts in fatty acid and lipid biosynthesis are scarce. Our finding that at least a part of the lipid biosynthesis pathway is located preferentially in M is consistent with the energy requirements of fatty acid biosynthesis. It is in M chloroplasts that reductive power and the carbon precursors like pyruvate and malate are present in abundance. The observation for preferential M accumulation of two lipoxygenases, LOX1 and LOX2 (AW157962, TC234252, TC237970, and TC237971), is intriguing and warrants further examination.
Differentiation of Redox Regulators and Antioxidative Defense Systems
Redox regulation plays a central role in many plastid functions, and chloroplasts undergo tremendous changes in redox potential during the day/night cycle and during variation in metabolic demand for NADPH and ATP. The production of ROS is tightly intertwined with this redox balance (Baier and Dietz, 2005). A multilayered anti-ROS detoxification system is in place in chloroplasts. These antioxidative systems and redox regulators and their connectivity have received lots of attention in Arabidopsis chloroplasts and other C3 species, as also evidenced by recent reviews on ROS (Noctor and Foyer, 1998; Mittler et al., 2004), Trx (Buchanan and Balmer, 2005; Gelhaye et al., 2005), and Prx (Dietz, 2003). The differentiation of maize BS and M chloroplasts in photosynthetic electron transport and carbon metabolism adds another level of complexity and dynamism.
We quantified two different Fds (Fd1 and Fd2), several FNRs, at least four Trx (Trx-f, Trx-m2, and two isoforms/homologs of Trx-m4), Fd-Trx reductase, components of the gluthatione defense system (gluthatione S-transferase and glutaredoxin), most known chloroplast Prx (Per II-E, 2-Cys PrxA, and 2-Cys PrxB), and Cu-Zn SOD. Expression levels for these different components were generally higher in M chloroplasts, with the exception of glutathione reductase and CuZn-SOD, each quite equally distributed over M and BS chloroplasts and a BS-specific isoform of Fd (Fd2) and Trx-m4. The novel observation of the preferential localization of two types of Prx in M raises interesting questions regarding the role and specificity of Prx.
Future challenges and questions in this area are severalfold. For instance, has the redox regulatory system adapted to the different redox states in BS and M chloroplasts? Do different isoenzymes/homologous proteins in metabolism with different BS/M expression levels have different redox activation levels? ROS seems to be emerging as an important signaling molecule (Apel and Hirt, 2004; Mittler et al., 2004). Does ROS contribute in BS/M development and/or gene regulation to maintain BS/M differentiation status? We believe that this comparative proteome analysis provides experimental starting points to address these questions.
Differentiation of Plastid Gene Expression and Protein Assembly and Fate
The comparison between protein machineries of BS and M chloroplast gene expression and chloroplast protein biogenesis/homeostasis provides a tool to understand many unresolved questions in this area. In particular it will be important to study (1) the influence of redox state on plastid gene expression since the two plastid types have such a different NADPH/NADP+ status and (2) the biogenesis of PSII and Rubisco, for example, since they each accumulate only or predominantly in one chloroplast type. The BS/M differentiation could narrow down the roles of proteins with unknown function, such as mRNA binding proteins and VIPP1, and also help to determine if regulators and chaperones, for example, are specific to particular complexes, such as HCF136 (Meurer et al., 1998).
The plastid genome encodes ∼50 proteins of the photosynthetic apparatus, of which 14 are PSII subunits, in addition to ribosomal subunits, four RNA polymerases (phosphoenolpyruvate), a Clp protease, and a few others (Maier et al., 1995). These 14 PSII subunits are highly abundant in M chloroplasts. PSII has also been shown (mostly in C3 species) to be particularly prone to photooxidative damage, requiring high protein synthesis rates for repair of the chloroplast-encoded reaction center protein D1 (Baena-Gonzalez and Aro, 2002). It is therefore likely that overall protein synthesis rates are much higher in M than in BS chloroplasts. Indeed, several ribosomal proteins were only identified in M chloroplasts and likely escaped identification in BS chloroplasts, indicative of generally higher accumulation levels of 70S ribosomes in M chloroplasts. We identified a number of proteins that preferentially accumulated in M chloroplasts, such as Ef-G (TC222353) and the PRSP7-Ef-Ts fusion protein (TC226754), which could fulfill M chloroplast-specific functions or are simply accommodating these higher translation rates.
In addition to plastid-encoded proteins, chloroplasts accumulate a few thousand nuclear-encoded proteins imported from the cytosol. It appears that the protein folding, assembly, and degradation machinery in the chloroplast stroma generally accommodates both chloroplast- and nuclear-encoded proteins. The accumulation pattern of the DnaK/DnaJ system or HSP70/GrpE was interesting in that one HSP70 homolog (TC220215) showed an equal distribution between BS and M chloroplasts, whereas the other HSP70 (TC235785) homolog and GrpE (TC222867) were clearly much higher in M chloroplasts. Interestingly, in the green alga C. reinhardtii, a HSP70/GrpE pair has been implicated in protection and repair of PSII, in particular under light stress conditions (Schroda et al., 1999). This would be the simplest explanation for their low BS:M ratio in maize chloroplasts.
The central focus of this article is to obtain a better understanding of the functional differentiation of BS and M chloroplasts. It is well established that BS and M cells and their respective chloroplasts each contain complementary sets of proteins facilitating C4 photosynthesis (for reviews, see Sheen, 1999; Miyao, 2003). However, it is less clear to what extent other chloroplast functions differentiate. This study brings a quantitative overview of differentiation not only of C4 functions but, most importantly, provides new insights in differentiation of other plastid functions. Moroever, the data set is robust, in particular since nine independent biological (pairwise) samples were analyzed. These data, including details of peptide sequences used for identification, are integrated into PPDB, along with several hundred additional maize chloroplast proteins that were identified but not quantified.
METHODS
Maize Genotype, Plant Growth, and Purification of BS and M Chloroplast Stroma
WT-T43 maize (Zea mays) plants were grown for 14 d in the growth chamber (16 h light/8 h dark, 400 μmol photons·m−2·s−1) until the 4th leaf was emerging. A new procedure for purification of M and BS chloroplasts was developed based on suggestions from A. Barkan and R. Bassi and on published protocols (Edwards and Black, 1971; Kannangara et al., 1977). The top 4-cm sections of the 2nd and 3rd leaves were harvested ∼2 h after the onset of the light period. Several hundreds of leaf tips (∼80 g of fresh tissue) were cut into 5- to 10-mm slices and gently ground by pestle and mortar in grinding buffer containing (350 mM sorbitol, 50 mM Hepes-KOH, pH 8, 2 mM EDTA, 5 mM ascorbic acid, and 5 mM l-cystein). The tearing effect applied by the pestle was sufficient to break M cells and release the M chloroplasts into the grinding buffer solution without excessively damaging the BS strands. The residual M cells attached to BS strands were removed by 10 min of blending (one-third of the maximal speed) in a modified Warring blender (WB1) with blunt blades. The purity of the BS strands was at this step assessed by light microscopy. After a wash with grinding buffer solution, the BS chloroplasts were released into the grinding buffer solution by short (2 s) high-speed pulses in a second modified Warring blender (WB2) in which the original blades were replaced by razor blades (as described in Kannangara et al., 1977). Subsequently, chloroplasts were further isolated, and the stromal proteome was released by osmotic shock, followed by removal of thylakoid and envelope membranes by ultracentrifugation, similarly as described by Peltier et al. (2004b). The cross-contamination of M and BS chloroplasts fractions was assessed from the presence of the M and BS markers (PPDK and Rubisco, respectively) visualized on Coomassie Brilliant Blue or silver-stained 1-DE SDS-PAGE gels. Chloroplast fractions showing <15% of cross-contamination were used, with the majority having a contamination of ∼10%. The yield of chloroplast preparations was inversely proportional to their purity. On average, preparations with <15% cross-contamination yielded ∼300 μg of protein for each M and BS fraction.
1-DE and 2-DE Gel Electrophoresis and Protein Gel Blotting
For 1-DE SDS-PAGE separation, proteins were equilibrated with SDS (0.2%), Na2CO3 (100 mM), DTT (100 mM), and sucrose (10%) and separated on 12% Tricine gels (Schägger and von Jagow, 1987). Gels were stained with fluorescent Sypro Ruby stain (Molecular Probes) or silver nitrate (Rabilloud et al., 1994). Protein gel blot analysis with polyclonal rabbit antisera and monoclonal mouse serum using chemoluminescence was essentially performed as described earlier (Friso et al., 2004).
Comparative BS and M Stromal Proteome Analysis by 2-DE
Purified M and BS stromal proteomes were first separated based on isoelectric point on IPG strips (150 μg of protein/strip), followed by a second dimension separation of SDS-PAGE. 2-DE-IPG was performed on 11-cm Immobiline Dry strips (Amersham Biosciences). Protein concentrations were determined by the Bradford method (Bradford, 1976). The IPG strips were rehydrated overnight with 150 μg of proteins solubilized in 9 M urea, 4% 3-([3-cholamidopropyl]dimethylammonio)-1-propane-sulfonate, 2 mM tributylphosphine, 2% pharmalyte, pH 4.0 to 7.0, 0.5% Triton X-100, and a few crystals of bromophenol blue (to evaluate the uniformity of the rehydration) in a reswelling tray at room temperature. Isoelectric focusing was conducted at 18°C in a Multiphor II (Amersham Biosciences) following the running conditions as described (Rouquie et al., 1997) for up to 72 kV/h. The focused strips were denaturated and reduced in an equilibration buffer (6 M urea, 30% glycerol, 50 mM Tris-HCl, pH 6.8, and 2% SDS) with addition of 5 mM tributylphosphine for 20 min and subsequently alkylated in equilibration buffer with 15 mM iodoacetamide for 20 min (Rabilloud et al., 1997; Herbert et al., 1998). The equilibrated IPG strips were loaded onto second dimension 10.5 to 14% Laemmli SDS gels (Criterium; Bio-Rad), and the protein separation was performed at constant current. Protein pIs on the resulting 2-DE-IPG gels were internally calibrated by mixing carbamylated standards (Pharmacia Biosciences) with the stromal extracts before 2-DE analysis. Denatured apparent molecular masses were determined from molecular mass markers loaded onto the second dimension gels.
Gel images were acquired with a Fluoro-S MultImager (Bio-Rad) using subsaturation exposure times. Five independent M and BS chloroplast preparations were resolved on five pairs of 2-DE gels. Image analysis was performed using Phoretix software (Nonlinear Dynamics). Approximately 400 spots per gel were detected, with spot volumes spanning five orders of magnitude. Spot selection was performed semiautomatically, with a large time investment in manual spot matching and verification. Ambiguous matches were resolved by protein identification from MS. After background removal and normalization of each spot volume to the total gel spot volume, virtual average M and BS gels were created, where each spot volume represented the average of volumes of matched spots in M or BS gels. The majority of spots (65% for M and 54% for BS gels) were represented at least three times either in M and/or in BS proteomes (45% for M and 39% for BS spots were represented at least four times). On average, we included gels only those matched spots that were present at least three times on M or BS gels. Corresponding interactive gels with protein spot identities that were quantified are available in the PPDB.
Sypro Ruby–stained spots were excised from the gel using a robot (ProPic; Genomic Solutions) or picked manually. The spots were automatically washed, digested with modified trypsin (Promega) (Shevchenko et al., 1996), and extracted using the ProGest robot (Genomic Solutions). The extracted peptides were dried and resuspended in 20 μL 5% formic acid. Protein identification was performed by peptide mass fingerprinting using MALDI time-of-flight (TOF) MS in reflectron mode (Voyager DE-STR; Perseptive Biosystems) and online LC-ESI-MS/MS (Q-TOF; Micromass). For MALDI-TOF analysis, 0.3 μL of each tryptic digest was crystallized in 0.3 μL of matrix solution (α-cyano-4-hydrocynnamic acid in 70% acetonitrile with 0.01% trifluoroacetic acid) on Teflon-coated MALDI-TOF target plates (96 wells; Perseptive Biosystems). MALDI-TOF MS spectra were internally calibrated using trypsin autodigestion peptides. Separation of peptide mixtures for nano-LC-ESI-MS/MS was achieved by collecting sample on a μ-Guard Pre-Column (inner diameter 300 μm, 1 mm long; LC Packings), followed by separation on a PepMap C18 Reverse Phase Nano Column (inner diameter 180 μm, 15 cm long; LC Packings). Peptides were eluted during a 45- to 60-min gradient using 95% water, 5% acetonitrile, and 0.1% formic acid as solvent A and 95% acetonitrile, 5% water, and 0.1% formic acid as solvent B at a flow rate of 0.75 μL/min.
The MS or MS/MS spectra were searched against the maize EST assembly from TIGR (http://www.tigr.org) (ZmGI, v1.4) and maize genome assemblies (AZM 4.0) as well as by homology-based searches against the annotated rice (Oryza sativa) genome (TIGR Rice Genome version 2), using an in-house installation of Mascot (www.matrixscience.com). Criteria for positive identification by MALDI-TOF MS peptide mass fingerprinting include five or more matching peptides with a narrow error distribution (clustering of errors) within 25 ppm and at least 15% sequence coverage. In exceptional cases, four matching peptides were considered as positive identification (e.g., proteins <20 kD and matching gel coordinates). Only those peptides were considered that had no missed cleavages (by trypsin) and no modifications, except for Met oxidation and carbamidomethylation (since gel samples were alkylated). For searches with MS/MS data, product ions must match within 0.8 D and precursor ions within 2 D. Criteria for positive identification by MS/MS were as follows: If only one or two matching sequence tags were found, clear partial Y-ion series and partial complementary B-ion series needed to be present (as determined by manual inspection) with MOWSE scores higher than 31 for one peptide identification and higher than 22 per peptide for two peptide identifications. If three or more sequence tags were found, manual inspection was not an absolute requirement, but the protein score must be well over 70, with only peptides with MOWSE scores over 21 contributing.
Comparative Proteomics by Stable Isotope Labeling Using cICAT
After purification of BS and M stroma, identical amounts of BS and M proteins (determined by the Bradford essay) were denatured and reduced, and all cysteins were labeled with the light (containing only 12C stable isotopes) or heavy (containing nine 13C stable isotope atoms) cICAT reagent, according to the manufacturer's instructions (Applied Biosystems). The differentially labeled BS and M stromal proteomes were mixed in a 1:1 ratio and proteins were analyzed in two different ways, as follows.
(1) Mixed and labeled BS/M stromal proteomes (200 μg for each proteome) were separated on a 1-DE Tricine-PAGE gel (12% acrylamide). After a short Coomassie Brilliant Blue (R-250) staining, gels were cut in 10-slices, and proteins were then in-gel digested by trypsin and resulting peptides extracted, using standard protocols. The biotin-tagged and cystein-containing peptides (useful for quantification) were then purified on avidin affinity columns, and the majority of the tag (containing the bulky biotin tag but not the isotope labels) was removed by acid hydrolysis, according to the manufacturer's instructions. Column materials were supplied as part of the cICAT labeling kit from Applied Biosystems. The flow-through containing the nonlabeled peptides was also collected since they were valuable for protein identification. cICAT-labeled and unlabeled peptides were identified by nano-LC-ESI-MS/MS, and cICAT-labeled peptides were quantified from nano-LC-ESI-MS runs.
(2) Mixed and labeled BS/M stromal proteomes were digested in solution by trypsin. Excess cICAT label was removed by strong offline cation exchange chromatography, followed by offline biotin-avidin affinity purification to collect the labeled peptides and removal of part of the tag by acid hydrolysis. The flow-through containing the nonlabeled peptides was also collected. After drying down and resuspension in 5% formic acid, cICAT-labeled peptides and peptides from the flow-through were analyzed separately by online two-dimensional nano-LC-ESI-MS/MS (both flow-through and cICAT-labeled peptides). The two-dimensional LC was a combination of strong cation exchange followed by reverse phase, as developed recently under the name MudPIT (Washburn et al., 2001; Wolters et al., 2001), and served to reduce complexity and maximize protein identification and quantification. Each cICAT-labeled sample was run twice under identical chromatographic conditions. Data-dependent acquisition (Masslynx 4.0) was employed to acquire MS spectra from the survey scan for quantification and tandem mass spectra for identification. In the first run, mass spectrometric parameters were setup favoring more time for the survey scans, and high-quality tandem MS spectra for peptides with higher signal intensity could also be obtained in this run. The second run was set up to obtain more tandem mass spectra, and peptides that had been analyzed by MS/MS in the first run were excluded in the second run.
All mass spectral data were searched against ZmGI, AZM, and OsGI for protein identification using in-house MASCOT search engine and additional filters using in-house written software (Q. Sun and K.J. van Wijk, unpublished data), followed by a manual verification steps for weaker identifications. For cICAT peptides, cICAT filter and variable Met oxidation were selected as Mascot search parameters. For 1-DE gel cICAT experiments, the non-Cys-containing peptides separated from both cICAT-labeled peptides were also purified by homemade reverse-phase columns packed with C18 beads, analyzed by LC-MS/MS, and searched against ZmGI to obtain protein identities. All verified protein identities using both unlabeled peptides and cICAT-labeled peptides are available via the PPDB. The three independent database searches showed that the EST assembly in ZmGI gave the best identification success rate, with the AZM and OsGI searches being virtually redundant (data not shown). Therefore, only ZmGI was used for detailed protein quantification.
The protein quantification was done by comparing the peak areas of corresponding peptides from BS or M. Masslynx 4.0 software (Waters) was employed to extract the peak areas of identified peptides of the corresponding single ion chromatograms. When needed, peak detection was manually adjusted to remove the influence of obvious satellite peak(s).
Comparative Proteomics by Parallel Ion Chromatograms from Online 1D LC-MS
As a third complementary approach, we used a non-gel-based quantitative approach that was independent of cystein labeling. For accurate quantification, it was critical that the BS and M samples were treated and analyzed in similar ways. This experiment was performed twice, first with 20 μg/g and then with 50 μg/g for each proteome, with independent biological samples (pairwise extractions of BS and M chloroplast proteomes). In the protein extraction and purification process, SDS was avoided and salt usage was kept low. The in-solution trypsin digest of the BS and M proteomes followed a procedure adapted from Chelius et al. (2003). Briefly, purified BS and M stromal proteomes were reduced with DTT and alkylated using iodoacetamide, followed by in-solution digestion by trypsin. Peptides were then dried down under vacuum, resuspended in 5% formic acid, and analyzed by nano-LC-ESI-MS for quantification and by nano-LC-ESI-MS/MS for peptide identification. Peptide identification and quantification procedures were similar to those in the cICAT experiment, except that (1) carbamidomethylation and partial Met oxidation were selected in the MASCOT search and (2) the single ion chromatograms of the corresponding peptides (from BS or M) were extracted from two different LC-MS runs (one for BS and one for M). The charge state, retention time, peak shape in single ion chromatograms, and tandem mass spectra of each peptide signal were strictly examined before quantification.
Functional Assignment of Identified Proteins
Many of the maize ZmGI and AZM accessions lack functional annotation. We functionally annotated all identified ZmGI and AZM accessions using a combination of BLAST alignments to the predicted rice proteome (OSGI) and the predicted Arabidopsis thaliana proteome (from The Arabidopsis Information Resource). In particular, we relied on the hierarchical, nonredundant classification system developed for MapMan (Thimm et al., 2004) (http://gabi.rzpd.de/projects/MapMan/), adjusted after manual verification and information from literature and incorporated into PPDB. The MapMan system has 35 main functional categories or Bins, with a larger number of subBins (subcategories) (see also http://ppdb.tc.cornell.edu/mapman.aspx). All best scoring directional pairwise BLAST alignment results between TAIR, OSGI, AZM, and ZMGI are available via PPDB. Where needed, functional assignments (Bins) were modified based on information from the published literature.
PPDB
The database engine for PPDB (http://ppdb.tc.cornell.edu/) is a Microsoft SQL server. The web interface for PPDB was developed on ASP.NET platform using C# language. The BLAST searches between maize (ZmGI and AZM), Arabidopsis (TAIR), and rice (OsGI) were performed and are available on the PPDB site. Comparative proteomics data can be obtained in two ways: (1) they can be downloaded as tables, and (2) they can be found on protein report pages for each quantified accession. In addition, representative 2-DE gels from BS and M chloroplast stroma with associated information can be interrogated.
Accession Numbers
Accession numbers for quantified genes discussed in this article are listed in Table 2.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table 1. Identification of Maize Proteins in Purified Bundle Sheath (BS) and Mesophyll (M) Chloroplast Stroma from Three Different Types of Experiments Involving 2-DE Gels, cICAT, and LC-MS Analysis of Unlabeled Peptides.
Supplemental Table 2. Summary of Identifications and Quantifications Obtained from the Analysis of 2-DE-IPG-PAGE for Each Protein Spot That Passed the Criteria for Reproducibility and That Was Used for Quantification.
Supplemental Table 3. Summary of the Non-Gel-Based Comparative Proteome Analysis of BS and M Chloroplast Stroma Using cICAT and Parallel LS-MS Runs.
Supplemental Table 4. Complete List of Quantifications for All Types of Comparative Proteomics Techniques Applied in This Study.
Supplementary Material
Acknowledgments
This project was supported by a grant from the National Science Foundation genome program (PGRP 0211935) and New York Science, Technology, and Academic Research to K.J.v.W. All database research was conducted using the resources of the Cornell Theory Center, which receives funding from Cornell University, New York state, federal agencies, foundations, and corporate partners. We thank J.J. Thelen for monoclonal antiserum against pyruvate dehydrogenase and M.E. Salvucci for polyclonal serum against PEPC. We thank members of the van Wijk laboratory, in particular G. Friso, L. Giacomelli, J.-B. Peltier, and J. Ytterberg for numerous discussions and advice. R. Bassi, A. Barkan, and T. Brutnell are acknowledged for suggestions in the initial stages of this study.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Klaas J. van Wijk (kv35@cornell.edu).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.035519.
References
- Aebersold, R., and Mann, M. (2003). Mass spectrometry-based proteomics. Nature 422, 198–207. [DOI] [PubMed] [Google Scholar]
- Aoki, H., Dekany, K., Adams, S.L., and Ganoza, M.C. (1997). The gene encoding the elongation factor P protein is essential for viability and is required for protein synthesis. J. Biol. Chem. 272, 32254–32259. [DOI] [PubMed] [Google Scholar]
- Aoyagi, K., and Nakamoto, H. (1985). Pyruvate, Pi dikinase in bundle sheath strands as well as in mesophyll cells in maize leaves. Plant Physiol. 78, 661–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apel, K., and Hirt, H. (2004). Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55, 373–399. [DOI] [PubMed] [Google Scholar]
- Ayala-Ochoa, A., Vargas-Suarez, M., Loza-Tavera, H., Leon, P., Jimenez-Garcia, L.F., and Sanchez-de-Jimenez, E. (2004). In maize, two distinct ribulose 1,5-bisphosphate carboxylase/oxygenase activase transcripts have different day/night patterns of expression. Biochimie 86, 439–449. [DOI] [PubMed] [Google Scholar]
- Baalmann, E., Scheibe, R., Cerff, R., and Martin, W. (1996). Functional studies of chloroplast glyceraldehyde-3-phosphate dehydrogenase subunits A and B expressed in Escherichia coli: Formation of highly active A4 and B4 homotetramers and evidence that aggregation of the B4 complex is mediated by the B subunit carboxy terminus. Plant Mol. Biol. 32, 505–513. [DOI] [PubMed] [Google Scholar]
- Baena-Gonzalez, E., and Aro, E.M. (2002). Biogenesis, assembly and turnover of photosystem II units. Philos. Trans. R. Soc. Lond. B Biol. Sci. 357, 1451–1459 (discussion 1459–1460). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baier, M., and Dietz, K.J. (2005). Chloroplasts as source and target of cellular redox regulation: A discussion on chloroplast redox signals in the context of plant physiology. J. Exp. Bot. 56, 1449–1462. [DOI] [PubMed] [Google Scholar]
- Baldy, P., and Cavalie, G. (1984). Compartimentation of photorespiratory enzymes in a C4 photosynthesis plant, Zea mays. Z. Pflanzenphysiol 114, 255–259. [Google Scholar]
- Balmer, Y., Koller, A., del Val, G., Schurmann, P., and Buchanan, B.B. (2004). Proteomics uncovers proteins interacting electrostatically with thioredoxin in chloroplasts. Photosynth. Res. 79, 275–280. [DOI] [PubMed] [Google Scholar]
- Barkan, A., and Goldschmidt-Clermont, M. (2000). Participation of nuclear genes in chloroplast gene expression. Biochimie 82, 559–572. [DOI] [PubMed] [Google Scholar]
- Bassi, R., Marquardt, J., and Lavergne, J. (1995). Biochemical and functional properties of photosystem II in agranal membranes from maize mesophyll and bundle sheath chloroplasts. Eur. J. Biochem. 233, 709–719. [DOI] [PubMed] [Google Scholar]
- Beale, S.J. (1999). Enzymes of chlorophyll biosynthesis. Photosynth. Res. 60, 43–73. [Google Scholar]
- Becker, T.W., Carrayol, E., and Hirel, B. (2000). Glutamine synthetase and glutamate dehydrogenase isoforms in maize leaves: Localization, relative proportion and their role in ammonium assimilation or nitrogen transport. Planta 211, 800–806. [DOI] [PubMed] [Google Scholar]
- Becker, T.W., Perro-Rechenmann, C., Suzuki, A., and Hirel, B. (1993). Subcellular and immunocytochemical localization of the enzymes involved in ammonia assimilation in mesophyll and bundle-sheath cells of maize leaves. Planta 191, 129–136. [Google Scholar]
- Beligni, M.V., Yamaguchi, K., and Mayfield, S.P. (2004). Chloroplast elongation factor ts pro-protein is an evolutionarily conserved fusion with the s1 domain-containing plastid-specific ribosomal protein-7. Plant Cell 16, 3357–3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhadula, S.K., Elthon, T.E., Habben, J.E., Helentjaris, T.G., Jiao, S., and Ristic, Z. (2001). Heat-stress induced synthesis of chloroplast protein synthesis elongation factor (EF-Tu) in a heat-tolerant maize line. Planta 212, 359–366. [DOI] [PubMed] [Google Scholar]
- Boudreau, E., Nickelsen, J., Lemaire, S.D., Ossenbuhl, F., and Rochaix, J.D. (2000). The Nac2 gene of Chlamydomonas encodes a chloroplast TPR-like protein involved in psbD mRNA stability. EMBO J. 19, 3366–3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford, M.M. (1976). A rapid a sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Broglie, R., Coruzzi, G., Keith, B., and Chua, N.H. (1984). Molecular biology of C4 photosynthesis in Zea mays: Differential localization of proteins and mRNAs in the two leaf cell types. Plant Mol. Biol. 3, 431–444. [DOI] [PubMed] [Google Scholar]
- Brutnell, T.P., Sawers, R.J., Mant, A., and Langdale, J.A. (1999). BUNDLE SHEATH DEFECTIVE2, a novel protein required for post-translational regulation of the rbcL gene of maize. Plant Cell 11, 849–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzobohaty, B., Moore, I., Kristoffersen, P., Bako, L., Campos, N., Schell, J., and Palme, K. (1993). Release of active cytokinin by a beta-glucosidase localized to the maize root meristem. Science 262, 1051–1054. [DOI] [PubMed] [Google Scholar]
- Buchanan, B.B., and Balmer, Y. (2005). Redox regulation: A broadening horizon. Annu. Rev. Plant Biol. 56, 187–220. [DOI] [PubMed] [Google Scholar]
- Burgener, M., Suter, M., Jones, S., and Brunold, C. (1998). Cyst(e)ine is the transport metabolite of assimilated sulfur from bundle-sheath to mesophyll cells in maize leaves. Plant Physiol. 116, 1315–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman, K.S.R., and Hatch, M.D. (1981). Aspartate decarboxylation in bundle sheath cells of Zea mays and its possible contribution to C4 photosynthesis. Aust. J. Plant Physiol. 8, 237–248. [Google Scholar]
- Chelius, D., Zhang, T., Wang, G., and Shen, R.F. (2003). Global protein identification and quantification technology using two-dimensional liquid chromatography nanospray mass spectrometry. Anal. Chem. 75, 6658–6665. [DOI] [PubMed] [Google Scholar]
- Conrads, K.A., Yu, L.R., Lucas, D.A., Zhou, M., Chan, K.C., Simpson, K.A., Schaefer, C.F., Issaq, H.J., Veenstra, T.D., Beck, G.R., Jr., and Conrads, T.P. (2004). Quantitative proteomic analysis of inorganic phosphate-induced murine MC3T3–E1 osteoblast cells. Electrophoresis 25, 1342–1352. [DOI] [PubMed] [Google Scholar]
- Cribb, L., Hall, L.N., and Langdale, J.A. (2001). Four mutant alleles elucidate the role of the g2 protein in the development of c(4) and c(3) photosynthesizing maize tissues. Genetics 159, 787–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das, A.K., Cohen, P., and Barford, D. (1998). The structure of the tetratricopeptide repeats of protein phosphatase 5: Implications for TPR-mediated protein-protein interactions. EMBO J. 17, 1192–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz, K.J. (2003). Plant peroxiredoxins. Annu. Rev. Plant Physiol. Plant Mol. Biol. 54, 93–107. [DOI] [PubMed] [Google Scholar]
- Doulis, A.G., Debian, N., Kingston-Smith, A.H., and Foyer, C.H. (1997). Differential localization of antioxidants in maize leaves. Plant Physiol. 114, 1031–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckhardt, U., Grimm, B., and Hortensteiner, S. (2004). Recent advances in chlorophyll biosynthesis and breakdown in higher plants. Plant Mol. Biol. 56, 1–14. [DOI] [PubMed] [Google Scholar]
- Edwards, G., and Walker, D.A. (1983). C3,C4: Mechanisms, and Cellular and Environmental Regulation, of Photosynthesis. (Berkeley, CA: University of California Press).
- Edwards, G.E., and Black, C.C.J. (1971). Isolation of mesophyll cells and bundle sheath cells from Digitaria sanguinalis (L.) scop. leaves and a scanning microscopy study of the internal leaf cell morphology. Plant Physiol. 47, 149–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards, G.E., Franceschi, V.R., Ku, M.S., Voznesenskaya, E.V., Pyankov, V.I., and Andreo, C.S. (2001). Compartmentation of photosynthesis in cells and tissues of C(4) plants. J. Exp. Bot. 52, 577–590. [PubMed] [Google Scholar]
- Edwards, G.E., Franceschi, V.R., and Voznesenskaya, E.V. (2004). Single-cell C(4) photosynthesis versus the dual-cell (Kranz) paradigm. Annu. Rev. Plant Biol. 55, 173–196. [DOI] [PubMed] [Google Scholar]
- Felder, S., Meierhoff, K., Sane, A.P., Meurer, J., Driemel, C., Plucken, H., Klaff, P., Stein, B., Bechtold, N., and Westhoff, P. (2001). The nucleus-encoded HCF107 gene of Arabidopsis provides a link between intercistronic RNA processing and the accumulation of translation-competent psbH transcripts in chloroplasts. Plant Cell 13, 2127–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson, P.L., and Smith, R.D. (2003). Proteome analysis by mass spectrometry. Annu. Rev. Biophys. Biomol. Struct. 32, 399–424. [DOI] [PubMed] [Google Scholar]
- Feussner, I., and Wasternack, C. (2002). The lipoxygenase pathway. Annu. Rev. Plant Biol. 53, 275–297. [DOI] [PubMed] [Google Scholar]
- Foster, J.G., and Edwards, G.E. (1980). Localization of superoxide dismutase in leaves of C3 and C4 plants. Plant Cell Physiol. 21, 895–906. [Google Scholar]
- Friso, G., Giacomelli, L., Ytterberg, A.J., Peltier, J.B., Rudella, A., Sun, Q., and Wijk, K.J. (2004). In-depth analysis of the thylakoid membrane proteome of Arabidopsis thaliana chloroplasts: New proteins, new functions, and a plastid proteome database. Plant Cell 16, 478–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furumoto, T., Hata, S., and Izui, K. (1999). cDNA cloning and characterization of maize phosphoenolpyruvate carboxykinase, a bundle sheath cell-specific enzyme. Plant Mol. Biol. 41, 301–311. [DOI] [PubMed] [Google Scholar]
- Furumoto, T., Hata, S., and Izui, K. (2000). Isolation and characterization of cDNAs for differentially accumulated transcripts between mesophyll cells and bundle sheath strands of maize leaves. Plant Cell Physiol. 41, 1200–1209. [DOI] [PubMed] [Google Scholar]
- Gallardo, K., Job, C., Groot, S.P., Puype, M., Demol, H., Vandekerckhove, J., and Job, D. (2001). Proteomic analysis of Arabidopsis seed germination and priming. Plant Physiol. 126, 835–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo, K., Le Signor, C., Vandekerckhove, J., Thompson, R.D., and Burstin, J. (2003). Proteomics of Medicago truncatula seed development establishes the time frame of diverse metabolic processes related to reserve accumulation. Plant Physiol. 133, 664–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelhaye, E., Rouhier, N., Navrot, N., and Jacquot, J.P. (2005). The plant thioredoxin system. Cell. Mol. Life Sci. 62, 24–35. [DOI] [PubMed] [Google Scholar]
- Gomez, S.M., Nishio, J.N., Faull, K.F., and Whitelegge, J.P. (2002). The chloroplast grana proteome defined by intact mass measurements from liquid chromatography mass spectrometry. Mol. Cell. Proteomics 1, 46–59. [DOI] [PubMed] [Google Scholar]
- Goshe, M.B., and Smith, R.D. (2003). Stable isotope-coded proteomic mass spectrometry. Curr. Opin. Biotechnol. 14, 101–109. [DOI] [PubMed] [Google Scholar]
- Graciet, E., Lebreton, S., and Gontero, B. (2004). Emergence of new regulatory mechanisms in the Benson-Calvin pathway via protein-protein interactions: A glyceraldehyde-3-phosphate dehydrogenase/CP12/phosphoribulokinase complex. J. Exp. Bot. 55, 1245–1254. [DOI] [PubMed] [Google Scholar]
- Gupta, R., Mould, R.M., He, Z., and Luan, S. (2002). A chloroplast FKBP interacts with and affects the accumulation of Rieske subunit of cytochrome bf complex. Proc. Natl. Acad. Sci. USA 99, 15806–15811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajduch, M., Ganapathy, A., Stein, J.W., and Thelen, J.J. (2005). A systematic proteomic study of seed filling in soybean. Establishment of high-resolution two-dimensional reference maps, expression profiles, and an interactive proteome database. Plant Physiol. 137, 1397–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, L.N., Rossini, L., Cribb, L., and Langdale, J.A. (1998). GOLDEN 2: A novel transcriptional regulator of cellular differentiation in the maize leaf. Plant Cell 10, 925–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen, K.C., Schmitt-Ulms, G., Chalkley, R.J., Hirsch, J., Baldwin, M.A., and Burlingame, A.L. (2003). Mass spectrometric analysis of protein mixtures at low levels using cleavable 13C-isotope-coded affinity tag and multidimensional chromatography. Mol. Cell. Proteomics 2, 299–314. [DOI] [PubMed] [Google Scholar]
- Harel, E., Lea, P.J., and Miflin, B.J. (1977). The localization of enzymes of nitrogen assimilation in maize leaves and their acivities during greening. Planta 134, 195–200. [DOI] [PubMed] [Google Scholar]
- Hatch, M.D., and Mau, S.L. (1973). Activity, location, and role of asparate aminotransferase and alanine aminotransferase isoenzymes in leaves with C4 pathway photosynthesis. Arch. Biochem. Biophys. 156, 195–206. [DOI] [PubMed] [Google Scholar]
- Hatch, M.D., and Osmond, C.B. (1976). Compartmentation and transport in C4 photosynthesis. In Encyclopedia of Plant Physiology, U. Heber and C. Stocking, eds (Berlin: Springer-Verlag), pp. 144–184.
- Heazlewood, J.L., Tonti-Filippini, J.S., Gout, A.M., Day, D.A., Whelan, J., and Millar, A.H. (2004). Experimental analysis of the Arabidopsis mitochondrial proteome highlights signaling and regulatory components, provides assessment of targeting prediction programs, and indicates plant-specific mitochondrial proteins. Plant Cell 16, 241–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert, B.R., Molloy, M.P., Gooley, A.A., Walsh, B.J., Bryson, W.G., and Williams, K.L. (1998). Improved protein solubility in two-dimensional electrophoresis using tributyl phosphine as reducing agent. Electrophoresis 19, 845–851. [DOI] [PubMed] [Google Scholar]
- Hochholdinger, F., Guo, L., and Schnable, P.S. (2004). Cytoplasmic regulation of the accumulation of nuclear-encoded proteins in the mitochondrial proteome of maize. Plant J. 37, 199–208. [DOI] [PubMed] [Google Scholar]
- Jankovsky, J.P., Smith, L.G., and Nelson, T. (2001). Specification of bundle sheath cell fates during maize leaf development: Roles of lineage and positional information evaluated through analysis of the tangled1 mutant. Development 128, 2747–2753. [DOI] [PubMed] [Google Scholar]
- Kaji, A., Teyssier, E., and Hirokawa, G. (1998). Disassembly of the post-termination complex and reduction of translational error by ribosome recycling factor (RRF)-A possible new target for antibacterial agents. Biochem. Biophys. Res. Commun. 250, 1–4. [DOI] [PubMed] [Google Scholar]
- Kannangara, C.G., Gough, S., Hansen, B., Rasmussen, J.T., and Simpson, D. (1977). A homogenizer with replacable razor blades for bulk isolation of active barley platids. Carlsberg Res. Commun. 42, 431–439. [Google Scholar]
- Karakaya, H.C., Tang, Y., Cregan, P.B., and Knap, H.T. (2002). Molecular mapping of the fasciation mutation in soybean, Glycine max (Leguminosae). Am. J. Bot. 89, 559–565. [DOI] [PubMed] [Google Scholar]
- Kimata, Y., and Hase, T. (1989). Localization of ferredoxin proteins in mesophyll and bundle sheath cells in maize leaf. Plant Physiol. 89, 1193–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimata-Ariga, Y., Matsumura, T., Kada, S., Fujimoto, H., Fujita, Y., Endo, T., Mano, J., Sato, F., and Hase, T. (2000). Differential electron flow around photosystem I by two C(4)-photosynthetic-cell-specific ferredoxins. EMBO J. 19, 5041–5050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig, J., Baier, M., Horling, F., Kahmann, U., Harris, G., Schurmann, P., and Dietz, K.J. (2002). The plant-specific function of 2-Cys peroxiredoxin-mediated detoxification of peroxides in the redox-hierarchy of photosynthetic electron flux. Proc. Natl. Acad. Sci. USA 99, 5738–5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku, S.B., and Edwards, G.E. (1975). Photosynthesis in mesophyll protoplasts and bundle sheath cells of various types of C4 plants. Z. Pflanzenphysiol. 77, 16–32. [Google Scholar]
- Langdale, J.A. (1998). Cellular differentiation in the leaf. Curr. Opin. Cell Biol. 10, 734–738. [DOI] [PubMed] [Google Scholar]
- Langdale, J.A., and Kidner, C.A. (1994). Bundle-sheath defective, a mutation that disrupts cellular differentiation in maize leaves. Development 120, 673–681. [Google Scholar]
- Li, J., Steen, H., and Gygi, S.P. (2003). Protein profiling with cleavable isotope-coded affinity tag (cICAT) reagents: The yeast salinity stress response. Mol. Cell. Proteomics 2, 1198–1204. [DOI] [PubMed] [Google Scholar]
- Lin, D., Tabb, D.L., and Yates, J.R. (2003). Large-scale protein identification using mass spectrometry. Biochim. Biophys. Acta 1646, 1–10. [DOI] [PubMed] [Google Scholar]
- Lonosky, P.M., Zhang, X., Honavar, V.G., Dobbs, D.L., Fu, A., and Rodermel, S.R. (2004). A proteomic analysis of maize chloroplast biogenesis. Plant Physiol. 134, 560–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunn, J.E., and Furbank, R.T. (1997). Localisation of sucrose-phosphate synthase and starch in leaves of C4 plants. Planta 202, 106–111. [DOI] [PubMed] [Google Scholar]
- Maier, R.M., Neckermann, K., Igloi, G.L., and Kossel, H. (1995). Complete sequence of the maize chloroplast genome: Gene content, hotspots of divergence and fine tuning of genetic information by transcript editing. J. Mol. Biol. 251, 614–628. [DOI] [PubMed] [Google Scholar]
- Matsumura, T., Kimata-Ariga, Y., Sakakibara, H., Sugiyama, T., Murata, H., Takao, T., Shimonishi, Y., and Hase, T. (1999). Complementary DNA cloning and characterization of ferredoxin localized in bundle-sheath cells of maize leaves. Plant Physiol. 119, 481–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meierhoff, K., and Westhoff, P. (1993). Differential biogenesis of photosytem II in mesophyll and bundle sheath cells of monocotyledonous NADP-malic enzyme-type C4 plants: The non-stoichiometric abundance of the subunits of photosystem II in the bundle sheath chloroplasts and the tranlational activity of the plastome-encoded genes. Planta 191, 23–33. [Google Scholar]
- Meurer, J., Plucken, H., Kowallik, K.V., and Westhoff, P. (1998). A nuclear-encoded protein of prokaryotic origin is essential for the stability of photosystem II in Arabidopsis thaliana. EMBO J. 17, 5286–5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler, R., Vanderauwera, S., Gollery, M., and Van Breusegem, F. (2004). Reactive oxygen gene network of plants. Trends Plant Sci. 9, 490–498. [DOI] [PubMed] [Google Scholar]
- Miyao, M. (2003). Molecular evolution and genetic engineering of C4 photosynthetic enzymes. J. Exp. Bot. 54, 179–189. [DOI] [PubMed] [Google Scholar]
- Nakamura, T., Ohta, M., Sugiura, M., and Sugita, M. (2001). Chloroplast ribonucleoproteins function as a stabilizing factor of ribosome-free mRNAs in the stroma. J. Biol. Chem. 276, 147–152. [DOI] [PubMed] [Google Scholar]
- Nelson, T., and Langdale, J.A. (1992). Developmental genetics of C4 photosynthesis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 43, 25–47. [Google Scholar]
- Neuhaus, H.E., and Emes, M.J. (2000). Nonphotosynthetic metabolism in plastids. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 111–140. [DOI] [PubMed] [Google Scholar]
- Niemeyer, H. (1988). Hydroxamic acids (4-hydroxy-1,4-benzoxazin-3-ones), defence chemicals in the gramineae. Phytochemistry 27, 3349–3358. [Google Scholar]
- Nikus, J., Daniel, G., and Jonsson, L.M. (2001). Subcellular localization of beta-glucosidase in rye, maize and wheat seedlings. Physiol. Plant 111, 466–472. [DOI] [PubMed] [Google Scholar]
- Nikus, J., and Jonsson, L.M. (1999). Tissue localization of β-glucosidase in rye, maize and wheat seedlings. Physiol. Plant 107, 373–378. [DOI] [PubMed] [Google Scholar]
- Noctor, G., and Foyer, C.H. (1998). Ascorbate and Glutathione: Keeping active oxygen under control. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 249–279. [DOI] [PubMed] [Google Scholar]
- Nuhse, T.S., Stensballe, A., Jensen, O.N., and Peck, S.C. (2004). Phosphoproteomics of the Arabidopsis plasma membrane and a new phosphorylation site database. Plant Cell 16, 2394–2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlrogge, J., and Browse, J. (1995). Lipid biosynthesis. Plant Cell 7, 957–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlrogge, J.B., and Jaworski, J.G. (1997). Regulation of fatty acid synthesis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 109–136. [DOI] [PubMed] [Google Scholar]
- Ong, S.E., Foster, L.J., and Mann, M. (2003). Mass spectrometric-based approaches in quantitative proteomics. Methods 29, 124–130. [DOI] [PubMed] [Google Scholar]
- Palmer, L.E., Rabinowicz, P.D., O'Shaughnessy, A.L., Balija, V.S., Nascimento, L.U., Dike, S., de la Bastide, M., Martienssen, R.A., and McCombie, W.R. (2003). Maize genome sequencing by methylation filtration. Science 302, 2115–2117. [DOI] [PubMed] [Google Scholar]
- Pastori, G.M., Mullineaux, P.M., and Foyer, C.H. (2000). Post-transcriptional regulation prevents accumulation of glutathione reductase protein and activity in the bundle sheath cells of maize. Plant Physiol. 122, 667–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier, J.B., Emanuelsson, O., Kalume, D.E., Ytterberg, J., Friso, G., Rudella, A., Liberles, D.A., Soderberg, L., Roepstorff, P., von Heijne, G., and van Wijk, K.J. (2002). Central functions of the lumenal and peripheral thylakoid proteome of Arabidopsis determined by experimentation and genome-wide prediction. Plant Cell 14, 211–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier, J.B., Ripoll, D.R., Friso, G., Rudella, A., Cai, Y., Ytterberg, J., Giacomelli, L., Pillardy, J., and Van Wijk, K.J. (2004. b). Clp protease complexes from photosynthetic and non-photosynthetic plastids and mitochondria of plants, their predicted three-dimensional structures, and functional implications. J. Biol. Chem. 279, 4768–4781. [DOI] [PubMed] [Google Scholar]
- Peltier, J.B., Ytterberg, A.J., Sun, Q., and van Wijk, K.J. (2004. a). New functions of the thylakoid membrane proteome of Arabidopsis thaliana revealed by a simple, fast, and versatile fractionation strategy. J. Biol. Chem. 279, 49367–49383. [DOI] [PubMed] [Google Scholar]
- Porubleva, L., Vander Velden, K., Kothari, S., Oliver, D.J., and Chitnis, P.R. (2001). The proteome of maize leaves: Use of gene sequences and expressed sequence tag data for identification of proteins with peptide mass fingerprints. Electrophoresis 22, 1724–1738. [DOI] [PubMed] [Google Scholar]
- Rabilloud, T., Adessi, C., Giraudel, A., and Lunardi, J. (1997). Improvement of the solubilization of proteins in two-dimensional electrophoresis with immobilized pH gradients. Electrophoresis 18, 307–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabilloud, T., Vuillard, L., Gilly, C., and Lawrence, J.J. (1994). Silver-staining of proteins in polyacrylamide gels: A general overview. Cell. Mol. Biol. (Noisy-le-grand) 40, 57–75. [PubMed] [Google Scholar]
- Rao, D., Momcilovic, I., Kobayashi, S., Callegari, E., and Ristic, Z. (2004). Chaperone activity of recombinant maize chloroplast protein synthesis elongation factor, EF-Tu. Eur. J. Biochem. 271, 3684–3692. [DOI] [PubMed] [Google Scholar]
- Rathnam, C.K., and Edwards, G.E. (1975). Intracellular localization of certain photosynthetic enzymes in bundle sheath cells of plants possessing the C4 pathway of photosynthesis. Arch. Biochem. Biophys. 171, 214–225. [DOI] [PubMed] [Google Scholar]
- Rathnam, C.K., and Edwards, G.E. (1976). Distribution of nitrate-assimilating enzymes between mesophyll protoplasts and bundle sheath cells in leaves of three groups of C4 plants. Plant Physiol. 57, 881–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland, N., Janosi, L., Block, M.A., Shuda, M., Teyssier, E., Miege, C., Cheniclet, C., Carde, J.P., Kaji, A., and Joyard, J. (1999). Plant ribosome recycling factor homologue is a chloroplastic protein and is bactericidal in Escherichia coli carrying temperature-sensitive ribosome recycling factor. Proc. Natl. Acad. Sci. USA 96, 5464–5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth, R., Hall, L.N., Brutnell, T.P., and Langdale, J.A. (1996). bundle sheath defective2, a mutation that disrupts the coordinated development of bundle sheath and mesophyll cells in the maize leaf. Plant Cell 8, 915–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhier, N., Gelhaye, E., Sautiere, P.E., Brun, A., Laurent, P., Tagu, D., Gerard, J., de Fay, E., Meyer, Y., and Jacquot, J.P. (2001). Isolation and characterization of a new peroxiredoxin from poplar sieve tubes that uses either glutaredoxin or thioredoxin as a proton donor. Plant Physiol. 127, 1299–1309. [PMC free article] [PubMed] [Google Scholar]
- Rouquie, D., Peltier, J.B., Marquis-Mansion, M., Tournaire, C., Doumas, P., and Rossignol, M. (1997). Construction of a directory of tobacco plasma membrane proteins by combined two-dimensional gel electrophoresis and protein sequencing. Electrophoresis 18, 654–660. [DOI] [PubMed] [Google Scholar]
- Schägger, H., and von Jagow, G. (1987). Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166, 368–379. [DOI] [PubMed] [Google Scholar]
- Schiltz, S., Gallardo, K., Huart, M., Negroni, L., Sommerer, N., and Burstin, J. (2004). Proteome reference maps of vegetative tissues in pea. An investigation of nitrogen mobilization from leaves during seed filling. Plant Physiol. 135, 2241–2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroda, M., Vallon, O., Wollman, F.A., and Beck, C.F. (1999). A chloroplast-targeted heat shock protein 70 (HSP70) contributes to the photoprotection and repair of photosystem II during and after photoinhibition. Plant Cell 11, 1165–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert, M., Petersson, U.A., Haas, B.J., Funk, C., Schroder, W.P., and Kieselbach, T. (2002). Proteome map of the chloroplast lumen of Arabidopsis thaliana. J. Biol. Chem. 277, 8354–8365. [DOI] [PubMed] [Google Scholar]
- Sheen, J. (1999). C-4 gene expression. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 187–217. [DOI] [PubMed] [Google Scholar]
- Sheen, J.Y., and Bogorad, L. (1987. a). Regulation of levels of nuclear transcripts for C-4 photosynthesis in bundle sheath and mesophyll cells of maize leaves. Plant Mol. Biol. 8, 227–238. [DOI] [PubMed] [Google Scholar]
- Sheen, J.Y., and Bogorad, L. (1987. b). Differential expression of C-4 pathway genes in mesophyll and bundle sheath cells of greening maize leaves. J. Biol. Chem. 262, 11726–11730. [PubMed] [Google Scholar]
- Shevchenko, A., Wilm, M., Vorm, O., and Mann, M. (1996). Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68, 850–858. [DOI] [PubMed] [Google Scholar]
- Shimaoka, T., Ohnishi, M., Sazuka, T., Mitsuhashi, N., Hara-Nishimura, I., Shimazaki, K., Maeshima, M., Yokota, A., Tomizawa, K., and Mimura, T. (2004). Isolation of intact vacuoles and proteomic analysis of tonoplast from suspension-cultured cells of Arabidopsis thaliana. Plant Cell Physiol. 45, 672–683. [DOI] [PubMed] [Google Scholar]
- Slack, C.R., Hatch, M.D., and Goodchild, D.J. (1969). Distribution of enzymes in mesophyll and parenchyma-sheath chloroplasts of maize leaves in relation to the C4-dicarboxylic acid pathway of photosynthesis. Biochem. J. 114, 489–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, L.G., Hake, S., and Sylvester, A.W. (1996). The tangled-1 mutation alters cell division orientations throughout maize leaf development without altering leaf shape. Development 122, 481–489. [DOI] [PubMed] [Google Scholar]
- Spilatro, S., and Preiss, J. (1987). Regulation of starch synthesis in the bundle sheath and mesophyll of Zea mays L.: Intercellular compartmentalisation of enzymes of starch metabolism and the properties of the ADPglucose pyrophosphorylases. Plant Physiol. 83, 621–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Q., Emanuelsson, O., and van Wijk, K.J. (2004). Analysis of curated and predicted plastid subproteomes of Arabidopsis. Subcellular compartmentalization leads to distinctive proteome properties. Plant Physiol. 135, 723–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamoi, M., Miyazaki, T., Fukamizo, T., and Shigeoka, S. (2005). The Calvin cycle in cyanobacteria is regulated by CP12 via the NAD(H)/NADP(H) ratio under light/dark conditions. Plant J. 42, 504–513. [DOI] [PubMed] [Google Scholar]
- Tao, W.A., and Aebersold, R. (2003). Advances in quantitative proteomics via stable isotope tagging and mass spectrometry. Curr. Opin. Biotechnol. 14, 110–118. [DOI] [PubMed] [Google Scholar]
- Thimm, O., Blasing, O., Gibon, Y., Nagel, A., Meyer, S., Kruger, P., Selbig, J., Muller, L.A., Rhee, S.Y., and Stitt, M. (2004). MAPMAN: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37, 914–939. [DOI] [PubMed] [Google Scholar]
- Usuda, H., and Edwards, G.E. (1980). Localization of glycerate kinase and some enzymes for sucrose synthesis in C3 and C4 plants. Plant Physiol. 65, 1017–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaistij, F.E., Boudreau, E., Lemaire, S.D., Goldschmidt-Clermont, M., and Rochaix, J.D. (2000). Characterization of Mbb1, a nucleus-encoded tetratricopeptide-like repeat protein required for expression of the chloroplast psbB/psbT/psbH gene cluster in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 97, 14813–14818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washburn, M.P., Wolters, D., and Yates III, J.R. (2001). Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19, 242–247. [DOI] [PubMed] [Google Scholar]
- Watson, B.S., Asirvatham, V.S., Wang, L., and Sumner, L.W. (2003). Mapping the proteome of barrel medic (Medicago truncatula). Plant Physiol. 131, 1104–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, A., and Flugge, U.I. (2002). Interaction of cytosolic and plastidic nitrogen metabolism in plants. J. Exp. Bot. 53, 865–874. [DOI] [PubMed] [Google Scholar]
- Wedel, N., Soll, J., and Paap, B.K. (1997). CP12 provides a new mode of light regulation of Calvin cycle activity in higher plants. Proc. Natl. Acad. Sci. USA 94, 10479–10484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingler, A., Walker, R.P., Chen, Z.H., and Leegood, R.C. (1999). Phosphoenolpyruvate carboxykinase is involved in the decarboxylation of aspartate in the bundle sheath of maize. Plant Physiol. 120, 539–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolters, D.A., Washburn, M.P., and Yates III, J.R. (2001). An automated multidimensional protein identification technology for shotgun proteomics. Anal. Chem. 73, 5683–5690. [DOI] [PubMed] [Google Scholar]
- Wyrich, R., Dressen, U., Brockmann, S., Streubel, M., Chang, C., Qiang, D., Paterson, A.H., and Westhoff, P. (1998). The molecular basis of C4 photosynthesis in sorghum: Isolation, characterization and RFLP mapping of mesophyll- and bundle-sheath-specific cDNAs obtained by differential screening. Plant Mol. Biol. 37, 319–335. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, K., Beligni, M.V., Prieto, S., Haynes, P.A., McDonald, W.H., Yates III, J.R., and Mayfield, S.P. (2003). Proteomic characterization of the Chlamydomonas reinhardtii chloroplast ribosome. Identification of proteins unique to th e70 S ribosome. J. Biol. Chem. 278, 33774–33785. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, K., von Knoblauch, K., and Subramanian, A.R. (2000). The plastid ribosomal proteins. Identification of all the proteins in the 30 S subunit of an organelle ribosome (chloroplast). J. Biol. Chem. 275, 28455–28465. [DOI] [PubMed] [Google Scholar]
- Yang, J., Schuster, G., and Stern, D.B. (1996). CSP41, a sequence-specific chloroplast mRNA binding protein, is an endoribonuclease. Plant Cell 8, 1409–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura, Y., Kubota, F., and Ueno, O. (2004). Structural and biochemical bases of photorespiration in C4 plants: Quantification of organelles and glycine decarboxylase. Planta 220, 307–317. [DOI] [PubMed] [Google Scholar]
- Zhao, Y.Y., Xu, T., Zucchi, P., and Bogorad, L. (1999). Subpopulations of chloroplast ribosomes change during photoregulated development of Zea mays leaves: Ribosomal proteins L2, L21, and L29. Proc. Natl. Acad. Sci. USA 96, 8997–9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.